Abstract
Tryptase ϵ is a member of the chromosome 16p13.3 family of human serine proteases that is preferentially expressed by epithelial cells. Recombinant pro–tryptase ϵ was generated to understand how the exocytosed zymogen might be activated outside of the epithelial cell, as well as to address its possible role in normal and diseased states. Using expression/site-directed mutagenesis approaches, we now show that Lys20, Cys90, and Asp92 in the protease's substrate-binding cleft regulate its enzymatic activity. We also show that Arg-1 in the propeptide domain controls its ability to autoactivate. In vitro studies revealed that recombinant tryptase ϵ possesses a restricted substrate specificity. Once activated, tryptase ϵ cannot be inhibited effectively by the diverse array of protease inhibitors present in normal human plasma. Moreover, this epithelium protease is not highly susceptible to α1-antitrypsin or secretory leukocyte protease inhibitor, which are present in the lung. Recombinant tryptase ϵ could not cleave fibronectin, vitronectin, laminin, single-chain tissue-type plasminogen activator, plasminogen, or any prominent serum protein. Nevertheless, tryptase ϵ readily converted single-chain pro–urokinase-type plasminogen activator (pro-uPA/scuPA) into its mature, enzymatically active protease. Tryptase ϵ also was able to induce pro-uPA–expressing smooth muscle cells to increase their migration through a basement membrane–like extracellular matrix. The ability to activate uPA in the presence of varied protease inhibitors suggests that tryptase ϵ plays a prominent role in fibrinolysis and other uPA-dependent reactions in the lung.
Introduction
The gene that encodes tryptase ϵ (also known as protease serine S member 22 [PRSS22]; GenBank Locus ID 64063)1 resides on human chromosome 16p13.3 at the site that also includes the genes that encode the related serine proteases tryptase α, tryptase β1, tryptase β2, tryptase β3, tryptase δ, transmembrane tryptase (TMT)/tryptase γ/PRSS31, marapsin/pancreasin, EOS/PRSS33, and eosinophil serine protease-1/testisin/PRSS21.2-12 Its mouse ortholog resides on chromosome 17A3.3, along with the genes that encode 12 other tryptic proteases.13 There are 2 Xenopus proteases (designated as Xepsin and Xeps-1) that have been identified that are more similar to human tryptase ϵ than its other family members. Thus, a primordial tryptase ϵ–like gene probably was the first gene to develop at the locus.
Each functional member of this family of serine proteases contains a distinct set of amino acids in the 7 loops (designated loops A-D and 1-3) that form its substrate-binding cleft. Because of the unique features of their 3D structures,14-18 the substrate specificities of all members of the family that have been examined to date are more limited than that of pancreatic trypsin. For example, the amino acid sequences of human tryptase α and β2 are 93% identical, yet these 2 proteases are functionally distinct due primarily to an Asp/Gly difference in one of the loops that forms their substrate-binding clefts.18,19 Mast cell–deficient W/Wv mice are unable to combat bacteria infections effectively,20-23 and data from numerous in vitro and in vivo studies suggest that the mouse tryptases mouse mast-cell protease 6 (mMCP-6) and mMCP-7 work in concert with tumor necrosis factor α and probably with other factors in mast cell–mediated inflammatory reactions to control the efficient and selective extravasation of different types of granulocytes into bacteria-infected tissues.23-26 Recombinant mMCP-6 and human tryptase β1 also induce a prominent and selective extravasation of neutrophils into the lungs that enable W/Wv mice to combat life-threatening Klebsiella pneumoniae infections effectively.23 The tryptase locus is mutating at an unusually high rate in humans.27 These data imply that some of the evolutionary pressure to increase the number of serine protease genes on human chromosome 16p13.3 and mouse chromosome 17A3.3 is occurring because of their beneficial roles in immunity. The gene that encodes urokinase-type plasminogen activator (uPA) resides on human chromosome 10q24 rather than 16p13.3. Nevertheless, uPA also plays important roles in innate immunity. For example, this trypticlike protease is essential for combating life-threatening Cryptococcus neoformans infections in the lung.28 Clearance of Pseudomonas aeruginosa in the lung is also impaired in uPA-null mice and in CD87/uPA receptor (uPAR)–null mice.29
In contrast to most members of its family that are expressed in mast cells, epithelial cells are the only nontransformed cells that have been found so far to express tryptase ϵ mRNA and protein.1,13 The mouse and human trachea, esophagus, and skin contain high levels of tryptase ϵ mRNA, and this serine protease is constitutively exocytosed from cultured epithelial cells predominantly in its inactive zymogen form. However, nothing is known about the activation, catabolism, and function of this constitutively exocytosed serine protease in normal and disease states in any species. We now report that recombinant human tryptase ϵ can autoactivate and that the last residue in the protease's propeptide is required for this self-activation event. We show that the physical retention of the cleaved propeptide via a conserved Cys-9-Cys112 disulfide bond is required for optimal enzymatic activity. We also show that a conserved Lys and an unpaired Cys that reside in the respective A and C loops that help form the substrate-binding cleft control the spontaneous conversion of the human tryptase ϵ zymogen into an enzyme that is a potent and highly selective activator of pro-uPA (also known as single-chain uPA or scuPA). The finding that tryptase ϵ can activate pro-uPA efficiently in the presence of varied protease inhibitors has important biologic implications in terms of fibrinolysis, innate immunity, inflammation, angiogenesis, connective tissue remodeling, and adenocarcinomas.
Materials and methods
Generation of recombinant human tryptase ϵ in COS-7 and insect cells
A bioengineered derivative of human pro–tryptase ϵ was generated in mammalian cells; it contains at its C terminus a 45-mer peptide bearing the V5 and 6xHis epitopes. Transient transfections were performed in COS-7 cells (line CRL-1651; American Type Culture Collection, Manassas, VA) using SuperFect (Qiagen, Valencia, CA), according to the manufacturer's instructions. At 2 to 3 hours after transfection in the serum-enriched medium, the treated cells were washed extensively with phosphate-buffered saline. They were then cultured in serum-free, Opti-MEM I medium (Life Technologies, Rockville, MD) for another 24 to 48 hours. The resulting conditioned medium was centrifuged and concentrated approximately 5-fold using an Ultrafree-15 filtering device. Aliquots of the obtained material were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and the resulting protein blots were probed with anti-V5 antibody (Invitrogen, Carlsbad, CA) or anti–tryptase ϵ antibody.1 Other aliquots were evaluated for their enzymatic activities as noted below. A bioengineered form of recombinant human tryptase ϵ zymogen containing the 8-mer FLAG (D-Y-K-D-D-D-D-K) peptide at its C terminus rather than the V5 peptide also was generated in High 5 insect cells (Invitrogen) using a modification of the baculovirus/insect cell–expression system we previously developed for obtaining related recombinant mouse and human tryptases,19,23-25,30,31 except that an enterokinase-susceptibility site was not placed in the initially expressed tryptase ϵ zymogen.
Site-directed mutagenesis of human tryptase ϵ
Residues -1, +90, and +92 in pro–tryptase ϵ were mutated in order to understand how this serine protease regulates its own enzymatic activity. The single amino acid mutants R-1A and C90A, and the double amino acid mutant C90A/D92E, were generated using the QuikChange XL site-directed mutagenesis kit (Stratagene, Cedar Creek, TX) and were expressed in insect cells. The primers used in these experiments were 5′-CCCCAGCAGCTGAACGCGGTTGTGGGCGAG-3′ and 5′-CTCGCCCACAACCGCGTTCAGCTGCTGGGG-3′ for the R-1A mutant; 5′-TGGAAGGAAGGTGCCGCCGCAGACATTGCCCTG-3′ and 5′-CAGGGCAATGTCTGCGGCGGCACCTTCCTTCCA-3′ for the C90A mutant; and 5′-TGGAAGGAAGGTGCCGCCGCAGAAATTGCCCTG-3′ and 5′-CAGGGCAATTTCTGCGGCGGCACCTTCCTTCCA-3′ for the C90A/D92E mutant. Arg-1 is the last amino acid in the protease's propeptide and Asp92 is predicted to be a member of the protease's catalytic triad of amino acids. Thus, the R-1A and D92E mutants were created to prevent autoactivation. Cys90 is predicted to be an unpaired residue that controls access of proteins and peptides into the protease's substrate-binding cleft.1 An unpaired Cys near the catalytic Asp controls the activation of varied serine carboxypeptidases.32,33 Thus, the C90A and C90A/D92E mutants were generated to investigate the potential regulatory role of the unpaired Cys in tryptase ϵ.
Wild-type pro–tryptase ϵ, its C90A mutant, and its C90A/D92E mutant were placed in 20 mM Tris (tris(hydroxymethyl)aminomethane)–HCl, pH 7.5, and incubated at 37°C for 6 to 48 hours or at 4°C for up to 8 days. Wild-type pro–tryptase ϵ also was incubated with previously activated tryptase ϵ (20:1 ratio) for up to 2 hours at 37°C. Aliquots of the treated samples were evaluated for their enzymatic activities as noted below. Samples also were subjected to SDS-PAGE to evaluate the endogenous proteolytic fragmentation of tryptase ϵ. The SDS-PAGE–separated fragments were blotted onto Immun-Blot polyvinylidenefluoride membranes (Biorad, Hercules, CA), which were then stained with Ponceau S. The N-terminal amino acid sequences of the identified fragments were determined, as well as the molecular weights of the proteolytic fragments.
As noted in “Results,” wild-type human tryptase ϵ and its C90A mutant can activate pro-uPA efficiently, but not plasminogen or pro-tPA (also known as single-chain tPA and sctPA). A comparison of the amino acid sequences around their processing sites revealed that the P5′ residue in the respective proteases is Glu, Leu, and Cys. The 3D model of human tryptase ϵ predicted that the conserved Glu in mouse, rat, and human pro-uPA interacts with Lys20 in the substrate-binding clefts of human and mouse tryptase ϵ. Thus, an expression/site-directed mutagenesis approach also was used to convert this basic amino acid to Ala. Because the resulting recombinant protease could not autoactivate as predicted, a 5-mer enterokinase site (ie, Asp-Asp-Asp-Asp-Lys) was then added in between the natural propeptide and the catalytic domain of human tryptase ϵ as done for the expression of other family members.23-25
Tryptase ϵ becomes a 2-chain serine protease when proteolytically activated due to a disulfide bond that links Cys-9 and Cys112.1,13 The fact that this disulfide bond is present in mouse and human tryptase ϵ, as well as in all other group-1 family members, suggests that the physical association of the cleaved propeptide with the mature domain is functionally important. To evaluate this possibility, a C-9A/C90A/C112A mutant of human tryptase ϵ also was created; its activity was then compared with the C90A mutant.
Substrate specificity of tryptase ϵ and susceptibility of the enzyme to various protease inhibitors
The enzymatic activity of affinity-purified, insect cell–derived tryptase ϵ was evaluated using the trypsin-susceptible substrates Bz-Phe-Val-Arg-pNA, Boc-Leu-Gly-Arg-pNA, Z-Arg-Arg-pNA, H-Gly-Arg-pNA, H-Glu-Gly-Arg-pNA, H-D-Val-Leu-Arg-pNA, Suc-Ala-Ala-Pro-Arg-pNA, and H-D-Leu-Thr-Arg-pNA (Bachem, King of Prussia, PA), and the chymotrypsin-susceptible substrates H-Leu-pNA and Suc-Ala-Ala-Pro-Phe-pNA (Bachem). In these assays, approximately 1 μg affinity-purified tryptase ϵ was placed in each well of a 96-well plate with 0.1 μmol substrate in 40 μL 20-mM Tris-HCl, pH 7.5. After a 10-minute incubation at room temperature, the optical density of the digest was determined at 405 nm using a UVmax kinetic microplate reader (Molecular Devices, Sunnyvale, CA). Assays were carried out in triplicate. The enzymatic activity of COS-7 cell–derived tryptase ϵ also was evaluated on the basis of its ability to cleave H-D-Leu-Thr-Arg-pNA. For these experiments, supernatants (∼ 40 μL) from tryptase ϵ–expressing COS-7 cells were incubated with 0.1 μmol substrate at room temperature for varied times.
The protease/protease inhibitor balance in the lung is a major factor in the development of debilitating lung disorders such as chronic obstructive pulmonary disease and asthma. Thus, the ability of tryptase ϵ to degrade the extracellular matrix proteins fibronectin, vitronectin, and laminin (Sigma, St Louis, MO) were evaluated to address the biologic significance of tryptase ϵ in the lung. These 3 large proteins were selected because they are highly susceptible to trypsin and other neutral proteases. In these in vitro assays, activated tryptase ϵ was incubated with each extracellular matrix protein (∼ 1:7 weight ratio or ∼ 1:1 molar ratio) in a 20-μL volume for 2 hours at 37°C. To evaluate whether a prominent protein is present in human blood that is highly susceptible to tryptase ϵ, such as mMCP-7,24 1 μg active protease was added to 60 μL of a 2% solution of normal human plasma. After a 2-hour incubation at 37°C, the resulting samples were subjected to SDS-PAGE.
The ability of mature uPA to activate pro–tryptase ϵ was evaluated. For these studies, the latter zymogen was incubated at 37°C for 2 hours with enzymatically active uPA (Sigma) at an approximately 20:1 ratio. The ability of enzymatically active tryptase ϵ to activate plasminogen, pro-uPA, and pro-tPA was then evaluated. For these studies, enzymatically active tryptase ϵ was incubated at 37°C for up to 60 minutes with an approximately 20-fold excess of plasminogen (Calbiochem, San Diego, CA), pro-uPA (American Diagnostica, Greenwich, CT), or pro-tPA (Calbiochem). Conversion of pro-uPA to enzymatically active uPA was assessed by SDS-PAGE, N-terminal amino acid sequence analysis, and by monitoring the ability of the processed protease to cleave the uPA-susceptible chromogenic substrate H-Glu-Gly-Arg-pNA. The latter substrate was chosen because it is not cleaved by tryptase ϵ, which is also present in the reaction mixture, albeit at much lower concentrations. Due to the limited availability of pro-uPA, an indirect but more sensitive spectrophotometric assay was used to estimate the Michaelis-Menten constant (Km), the overall catalytic rate of an enzyme (kcat), and kcat/Km values for the tryptase ϵ–mediated conversion of pro-uPA to uPA. A fixed amount of enzymatically active tryptase ϵ was placed in a series of vials containing 100 μL buffer (137 mM NaCl, 27 mM KCl, and 10 mM sodium phosphate, pH 7.4) and variable amounts of pro-uPA. Each reaction was allowed to proceed for 45 minutes at 37°C. The amount of generated uPA was then determined by monitoring its ability to cleave H-Glu-Gly-Arg-pNA in a time-dependent manner.
Several physiologic and nonphysiologic protease inhibitors also were evaluated for their ability to inhibit the enzymatic activity of recombinant tryptase ϵ. The tested inhibitors included antipain, bestatin, pepstatin, aprotinin (Roche Diagnostics, Indianapolis, IN), purified human α1-antitrypsin (A1AT; also known as α1-proteinase inhibitor and SERPINA1; Sigma), and recombinant secretory leukocyte protease inhibitor (SLPI; Cell Sciences, Norwood, MA). In these assays, approximately 5 μg activated tryptase ϵ or 1 μg trypsin was preincubated with an approximately 10-fold excess of the protease inhibitor for 30 minutes. The ability of the treated proteases to cleave H-D-Leu-Thr-Arg-pNA was then measured after a 30-minute incubation at room temperature. In some cases, activated tryptase ϵ was preincubated for 30 minutes at room temperature or 37°C with human plasma prepared from a healthy individual (2% final concentration) before the enzymatic activity of the treated protease was evaluated.
Bronchial smooth-muscle–cell cultures
Cultured human bronchial smooth muscle cells constitutively secrete pro-uPA and plasminogen activator inhibitor-1 (PAI-1/SERPINE1). These cells also express CD87, which is the uPA receptor (uPAR). Thus, the ability of tryptase ϵ to activate smooth muscle cells in an uPA/uPAR-dependent manner in the presence of the other factors these mesenchymal cells produce was evaluated. Human bronchial smooth muscle cells (Cambrex, Walkersville, MD) were placed in 25-cm2 culture flasks containing 5 mL SmGM-2 BulletKit culture medium (Cambrex), which contains 5% fetal calf serum (FCS) and various cytokines. Confluent cultures were washed twice with Hanks balanced salt solution, and then 5 mL serum-free SmGM culture medium (Cambrex) was added followed by approximately 3.3 μg/mL wild-type tryptase ϵ, its C90A mutant, or its C90A/D92E mutant (final concentration, ∼ 100 nM). The supernatants from the treated cells were then concentrated approximately 5-fold and subjected to SDS-PAGE under nonreducing conditions to minimize dissociation of uPA/uPAR/PAI-1 macromolecular complexes. The resulting protein blots were probed with monoclonal antibodies specific for uPA, PAI-1, and uPAR (American Diagnostica). Aliquots of the supernatants also were evaluated for the presence of enzymatically active uPA using the synthetic substrate H-Glu-Gly-Arg-pNA.
Matrigel cell-invasion assay
Biocoat invasion chambers containing Matrigel-coated membranes with 8-μm pores (Becton Dickinson Labware, Bedford, MA) were used to evaluate the ability of tryptase ϵ–activated uPA to induce smooth muscle cells to migrate through a basement membrane–like extracellular matrix. In this in vitro assay, 5 × 104 human bronchial smooth muscle cells in 0.5 mL serum-free minimum essential Eagle medium were placed in the top chamber of each well of a 24-well culture dish. This was followed by no tryptase, enzymatically active tryptase ϵ (final concentration, ∼ 100 nM), or its inactive C90A/D92E mutant. FCS contains numerous factors that are chemotactic for smooth muscle cells. Thus, 0.75 mL culture medium containing 10% FCS was added to the lower chamber of each well. In some experiments, a small amount of pro-uPA (final concentration, ∼ 100 nM) also was added to the top chamber to minimize the problem of the uPA inhibitors in the FCS and conditioned medium. After a 36-hour incubation at 37°C, the noninvading cells were removed from the top chamber and the number of cells that extravasated through the extracellular matrix was determined at the light microscopic level by staining the bottom side of the membrane with Masson trichrome stain. Each assay was performed in duplicate. Chambers lacking Matrigel-coated membranes served as a control to ensure that the smooth muscle cells used in the experiments were responsive to the chemotactic factors in FCS.
The Matrigel-coated membranes were removed from the invasion chambers in the chemotaxis assays and incubated with Masson trichrome stain. Each treated membrane was placed on a slide, covered with a coverslip, and mounted with poly mount. The number of cells that extravasated through the membrane's 8-μm pores was measured using a Leica DMLB2 microscope equipped with a 10 ×/0.30 objective lens (final magnification, × 100) and a Leica Wild MPS52 camera (Leica, Heidelberg, Germany). Rather than acquisition software, Kodak film (Eastman Kodak, Rochester, NY) was used to save the images.
Molecular modeling and docking
The computer program MODELLER34 was used to build a 3D model of human tryptase ϵ based on the crystal structure of bovine pancreatic trypsin complexed to pancreatic trypsin inhibitor (Protein Data Bank code 2PTC).35,36 One of the advantages of the trypsin/trypsin inhibitor structure was that it allowed us to use a computational docking approach to predict how tryptase ϵ selectively recognizes the activation sequence in pro-uPA. To confirm the binding specificity inferred from our studies, the contribution to the binding energy due to electrostatics and the effect of solvation was computed using the DelPhi program.37,38
Results
Cys90-dependent autoactivation of human tryptase ϵ
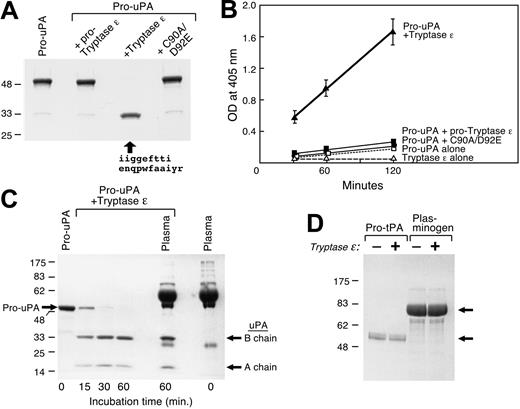
Whether expressed in COS-7 cells or insect cells, wild-type tryptase ϵ and its mutants were secreted into the conditioned medium in a time-dependent manner as inactive approximately 37-kDa zymogens (Figure 1). In contrast to all other studied members of the human chromosome 16p13.3 and mouse chromosome 17A3.3 families of serine proteases, wild-type tryptase ϵ zymogen was slowly converted into an approximately 31-kDa product that was enzymatically active. The final processed protease was recognized by anti–tryptase ϵ antibody but not by the C-terminal epitope tag. The sizes of the final products in the 2 expression systems predict that tryptase ϵ is cleaved slowly at its propeptide and then more quickly at a site that resides near its C terminus. Processed, enzymatically active tryptase ϵ was not obtained if residue -1 was mutated (Figures 1A-B). However, the C90A mutant was more susceptible to spontaneous activation, and the resulting material possessed greater enzymatic activity than wild-type tryptase ϵ incubated at either 37°C (Figure 1C) or 4°C (Figure 1D). The importance of Cys90 in the autoactivation process was seen more clearly when the zymogen was incubated at 4°C (Figure 1D). The inactive mutant C90A/D92E was barely processed to smaller forms, and no enzymatic activity was detected.
Generation of recombinant human tryptase ϵ in COS-7 and insect cells, and spontaneous activation of the recombinant protease. (A) Wild-type pro–tryptase ϵ and its activation-resistant R-1A mutant were expressed in COS-7 cells. After 24 and 48 hours, aliquots of the conditioned medium were subjected to SDS-PAGE and the resulting protein blots were probed with anti–tryptase ϵ antibody (top) or anti-V5 antibody (bottom). (B) Wild-type, COS-7 cell–derived tryptase ϵ (▵, ▴), and its R-1A mutant (▪, □) that had incubated for 24 (▵, □)or 48 (▴, ▪) hours also were evaluated for their enzymatic activity using the tryptase ϵ–susceptible synthetic substrate H-D-Leu-Thr-Arg-pNA. OD indicates optical density. (C-D) Recombinant pro–tryptase ϵ, its C90A mutant, and its C90A/D92E double mutant also were generated using a baculovirus-High 5 insect cell expression system. Insect cells were cultured for 4 days and the resulting condition medium was applied to an anti-FLAG column. Purified wild-type tryptase ϵ and its mutants were incubated at 37°C (C) or 4°C (D) for the indicated times. Aliquots of the resulting products were then evaluated for their enzymatic activity using H-D-Leu-Thr-Arg-pNA (top rows). Other aliquots were subjected to SDS-PAGE under reducing conditions. In the bottom row of panel C, the resulting gels were stained with Coomassie blue. Molecular-mass standards are indicated on the left. In the lower rows of panel D, immunoblots were prepared and probed with anti–tryptase ϵ or anti-FLAG antibodies. Error bars in this and subsequent figures indicate the mean ± standard error of the mean from a single experiment carried out in triplicate. Experiments were carried out more than 2 times to confirm the data.
Generation of recombinant human tryptase ϵ in COS-7 and insect cells, and spontaneous activation of the recombinant protease. (A) Wild-type pro–tryptase ϵ and its activation-resistant R-1A mutant were expressed in COS-7 cells. After 24 and 48 hours, aliquots of the conditioned medium were subjected to SDS-PAGE and the resulting protein blots were probed with anti–tryptase ϵ antibody (top) or anti-V5 antibody (bottom). (B) Wild-type, COS-7 cell–derived tryptase ϵ (▵, ▴), and its R-1A mutant (▪, □) that had incubated for 24 (▵, □)or 48 (▴, ▪) hours also were evaluated for their enzymatic activity using the tryptase ϵ–susceptible synthetic substrate H-D-Leu-Thr-Arg-pNA. OD indicates optical density. (C-D) Recombinant pro–tryptase ϵ, its C90A mutant, and its C90A/D92E double mutant also were generated using a baculovirus-High 5 insect cell expression system. Insect cells were cultured for 4 days and the resulting condition medium was applied to an anti-FLAG column. Purified wild-type tryptase ϵ and its mutants were incubated at 37°C (C) or 4°C (D) for the indicated times. Aliquots of the resulting products were then evaluated for their enzymatic activity using H-D-Leu-Thr-Arg-pNA (top rows). Other aliquots were subjected to SDS-PAGE under reducing conditions. In the bottom row of panel C, the resulting gels were stained with Coomassie blue. Molecular-mass standards are indicated on the left. In the lower rows of panel D, immunoblots were prepared and probed with anti–tryptase ϵ or anti-FLAG antibodies. Error bars in this and subsequent figures indicate the mean ± standard error of the mean from a single experiment carried out in triplicate. Experiments were carried out more than 2 times to confirm the data.
To confirm that wild-type tryptase ϵ is able to autoactivate, a 20-fold excess of its zymogen was incubated with previously activated tryptase ϵ or its similarly treated C90A/D92E mutant. Whereas the C90A/D92E mutant was rapidly cleaved by wild-type tryptase ϵ, the resulting processed protease was unable to cleave H-D-Leu-Thr-Arg-pNA (Figure 2A, right top and bottom panels). In addition, the processed mutant was unable to activate wild-type pro–tryptase ϵ. In contrast, wild-type pro–tryptase ϵ was quickly converted into its enzymatically active form by previously activated tryptase ϵ (Figure 2A, left top and bottom panels). N-terminal amino acid sequencing and SDS-PAGE immunoblot analysis of the enzymatically active β chain revealed a truncated protein with a single N terminus. Thus, activated tryptase ϵ quickly removes a small portion of its C-terminal sequence. As assessed by matrix assisted laser desorption/ionization-time of flight mass spectrometry, the C-terminal cleavage site resides at Arg246 or Arg248, thereby resulting in the loss of 22 to 24 amino acids. Because none of the loops that form the protease's substrate-binding cleft are adversely affected, the C-terminal processing event does not appear to greatly diminish the protease's enzymatic activity for the colorimetric substrate. The smallest form generated in this experiment (highlighted by the third arrow in Figure 2B) is the mature, biologically active protease. A summary of the processing events is highlighted in Figure 2C. Surprisingly, the ability of the C-9A/C90A/C112A mutant to cleave H-D-Leu-Thr-Arg-pNA was significantly reduced relative to the C90A mutant (P < .01, n = 4). Thus, the physical retention of the conserved propeptide appears to be necessary for optimal enzymatic activity of processed tryptase ϵ.
Substrate specificity of recombinant human tryptase ϵ
Activated tryptase ϵ cleaved H-D-Leu-Thr-Arg-pNA (Figure 2A) but not any of the other tested trypsin- and chymotrypsin-susceptible chromogenic substrates. The extracellular matrix proteins fibronectin, vitronectin, and laminin could not be cleaved by tryptase ϵ under the tested conditions (data not shown). In addition, no prominent peptide formed when tryptase ϵ was incubated with normal human plasma. Based on these data, tryptase ϵ is a highly restricted serine protease.
Activation of purified pro–tryptase ϵ with active tryptase ϵ. (A) Insect cell–expressed pro–tryptase ϵ (left column) or its C90A/D92E mutant (right column) were incubated with spontaneously activated tryptase ϵ in an approximately 20:1 ratio at 37°C for up to 120 minutes. Aliquots of the resulting digests were subjected to SDS-PAGE under reducing conditions. Shown are the Coomassie blue–stained gels (top row). Other aliquots were evaluated for their enzymatic activities using H-D-Leu-Thr-Arg-pNA (bottom row). In a control experiment for the left-column experiments, pro–tryptase ϵ was not activated if incubated with the C90A/D92E mutant that lacks its propeptide (data not shown). (B) An excess of pro–tryptase ϵ was incubated with active tryptase ϵ for 0, 30, or 120 minutes. SDS-PAGE/immunoblots were probed with anti–tryptase ϵ antibody (left panel) or anti-FLAG antibody (right panel). (C) The N-terminal amino acid sequences of protein bands 1, 2, and 3 in panel B are shown, as well as a schematic model of the 3 tryptase ϵ products identified in this study. The black (▪) domain in each right panel is the protease's propeptide, which is covalently linked to the catalytic domain presumably via the Cys-9-Cys112 disulfide bond. The hatched (▨) domain at the protein's C terminus corresponds to the FLAG peptide that is found only in the unprocessed tryptase ϵ zymogen.
Activation of purified pro–tryptase ϵ with active tryptase ϵ. (A) Insect cell–expressed pro–tryptase ϵ (left column) or its C90A/D92E mutant (right column) were incubated with spontaneously activated tryptase ϵ in an approximately 20:1 ratio at 37°C for up to 120 minutes. Aliquots of the resulting digests were subjected to SDS-PAGE under reducing conditions. Shown are the Coomassie blue–stained gels (top row). Other aliquots were evaluated for their enzymatic activities using H-D-Leu-Thr-Arg-pNA (bottom row). In a control experiment for the left-column experiments, pro–tryptase ϵ was not activated if incubated with the C90A/D92E mutant that lacks its propeptide (data not shown). (B) An excess of pro–tryptase ϵ was incubated with active tryptase ϵ for 0, 30, or 120 minutes. SDS-PAGE/immunoblots were probed with anti–tryptase ϵ antibody (left panel) or anti-FLAG antibody (right panel). (C) The N-terminal amino acid sequences of protein bands 1, 2, and 3 in panel B are shown, as well as a schematic model of the 3 tryptase ϵ products identified in this study. The black (▪) domain in each right panel is the protease's propeptide, which is covalently linked to the catalytic domain presumably via the Cys-9-Cys112 disulfide bond. The hatched (▨) domain at the protein's C terminus corresponds to the FLAG peptide that is found only in the unprocessed tryptase ϵ zymogen.
Because many tryptic proteases function as proprotein convertases (eg, Factor D in the complement cascade and Factor XIIa/Hageman Factor in the coagulation cascade), searches of GenBank's protein database were carried out using the 6-mer target sequences Arg-Val-Val-Gly-Gly-Glu and Arg-Ile-Val-Gly-Gly-Glu that reside at the respective endogenous cleavage sites in human and mouse pro–tryptase ϵ in an attempt to identify candidate substrate(s). Those BLASTp searches resulted in the identification of nearly identical sequences in the corresponding activation domains of mouse, rat, and human pro-uPA (Table 1), thereby implicating a possible role for tryptase ϵ in uPA-dependent pathways in the lung and other epithelial cell–enriched tissues. When pro-uPA was incubated with enzymatically active recombinant tryptase ϵ, the former zymogen was rapidly converted into a functional protease that contained disulfide-bonded chains of approximately 33 and approximately 18 kDa (Figure 3A,C). This proteolytic event was not obtained when pro-uPA was incubated with pro–tryptase ϵ or its enzymatically inactive C90A/D92E mutant. For comparison, pro-uPA was not susceptible to recombinant human tryptase β1 (data not shown).
Activation sites of pro-tryptase ϵ, pro-uPA, pro-tPA, and plasminogen
. | Zymogen processing site . |
|---|---|
| . | ↓ . |
| Protease . | propeptide-P1-P1′-P2′-P3′-P4′-P5′-mature domain . |
| Human pro-tryptase ϵ | Arg-Val-Val-Gly-Gly-Glu |
| Rat pro-tryptase ϵ | Arg-Val-Val-Gly-Gly-Glu |
| Mouse pro-tryptase ϵ | Arg-Ile-Val-Gly-Gly-Glu |
| Mouse pro-uPA | Lys-Ile-Val-Gly-Gly-Glu |
| Rat pro-uPA | Lys-Ile-Val-Gly-Gly-Glu |
| Human pro-uPA | Lys-Ile-Ile-Gly-Gly-Glu |
| Human pro-tPA | Arg-Ile-Lys-Gly-Gly-Leu |
| Mouse pro-tPA | Arg-Ile-Lys-Gly-Gly-Leu |
| Rat pro-tPA | Arg-Ile-Lys-Gly-Gly-Leu |
| Human plasminogen | Arg-Val-Val-Gly-Gly-Cys |
| Mouse plasminogen | Arg-Val-Val-Gly-Gly-Cys |
| Rat plasminogen | Arg-Val-Val-Gly-Gly-Cys |
| Danio rerio plasminogen | Arg-Ile-Val-Gly-Gly-Cys |
. | Zymogen processing site . |
|---|---|
| . | ↓ . |
| Protease . | propeptide-P1-P1′-P2′-P3′-P4′-P5′-mature domain . |
| Human pro-tryptase ϵ | Arg-Val-Val-Gly-Gly-Glu |
| Rat pro-tryptase ϵ | Arg-Val-Val-Gly-Gly-Glu |
| Mouse pro-tryptase ϵ | Arg-Ile-Val-Gly-Gly-Glu |
| Mouse pro-uPA | Lys-Ile-Val-Gly-Gly-Glu |
| Rat pro-uPA | Lys-Ile-Val-Gly-Gly-Glu |
| Human pro-uPA | Lys-Ile-Ile-Gly-Gly-Glu |
| Human pro-tPA | Arg-Ile-Lys-Gly-Gly-Leu |
| Mouse pro-tPA | Arg-Ile-Lys-Gly-Gly-Leu |
| Rat pro-tPA | Arg-Ile-Lys-Gly-Gly-Leu |
| Human plasminogen | Arg-Val-Val-Gly-Gly-Cys |
| Mouse plasminogen | Arg-Val-Val-Gly-Gly-Cys |
| Rat plasminogen | Arg-Val-Val-Gly-Gly-Cys |
| Danio rerio plasminogen | Arg-Ile-Val-Gly-Gly-Cys |
N-terminal amino acid sequence analysis of the approximately 33-kDa chain obtained after tryptase ϵ treatment revealed that pro-uPA was cleaved between Lys158 and Ile159, thereby generating the biologically active form of the protease. The amino acid sequence of the N-terminal portion of the 33-kDa fragment is shown in Figure 3A. Because tryptase ϵ was unable to cleave the uPA-susceptible peptide substrate H-Glu-Gly-Arg-pNA, this synthetic substrate could be used to monitor the tryptase ϵ–mediated conversion of pro-uPA to mature uPA. As expected, tryptase ϵ–generated uPA was able to cleave H-Glu-Gly-Arg-pNA effectively (Figure 3B). This activation step occurred in a time-dependent manner (Figure 3C). The estimated Km, kcat, and kcat/Km values for the tryptase ϵ–mediated conversion of pro-uPA to uPA were 28 μM, 6.8 s-1, and 0.24 μM-1 s-1, respectively. A better assessment of tryptase ϵ's specificity for pro-uPA was the subsequent finding that the tryptase ϵ–mediated activation of pro-uPA was not inhibited by the diverse array of protease inhibitors present in normal human plasma (n = 2). In agreement with this finding, the ability of recombinant tryptase ϵ to cleave H-Leu-Thr-Arg-pNA was not inhibited by any plasma protein (n = 3, data not shown). A1AT and SLPI are effective inhibitors of TMT/tryptase γ/PRSS31.31 Even though the amino acid sequences of the 2-chromosome 16p13.3 family members are approximately 50% identical, neither A1AT nor SLPI was able to inhibit tryptase ϵ (Table 2). Finally, neither pro-tPA nor plasminogen could be activated by tryptase ϵ when incubated for 2 hours at 37°C under comparable conditions (Figure 3D). The accumulated data indicate that tryptase ϵ possesses a restricted substrate specificity that differs significantly from its other family members. Moreover, pro-uPA is one of its preferred substrates. The subsequent finding that uPA could not convert pro–tryptase ϵ into its mature form (data not shown) implies that tryptase ϵ acts upstream of uPA in uPA-dependent pathways.
Activation of uPA by tryptase ϵ. A single-chain pro-uPA was incubated in buffer alone (left lane) or buffer containing pro–tryptase ϵ, active wild-type tryptase ϵ, or its inactive C90A/D92A mutant (lanes 2, 3, and 4, respectively). After a 2-hour incubation at 37°C, the treated samples were subjected to SDS-PAGE under reducing conditions; the resulting gels were stained with Coomassie blue. N-terminal amino acid sequence analysis also was performed on the indicated approximately 33-kDa product detected in the third lane (arrow). (B) Aliquots of the resulting reactions were evaluated for the presence of an active protease that cleaves H-Glu-Gly-Arg-pNA. As noted (▴), tryptase ϵ treatment of pro-uPA resulted in the generation of a mature enzyme that readily cleaved the uPA-sensitive substrate. (C) Time-dependent activation of pro-uPA with tryptase ϵ is shown in the left 4 lanes. In lane 5, tryptase ϵ was added to a 20-fold excess of pro-uPA after incubation with plasma (2% final volume) for 30 minutes at room temperature. Lane 6 on the far right is the control, depicting the proteins in plasma before tryptase ϵ treatment. In a second experiment (data not shown), purified, recombinant tryptase ϵ again quickly activated pro-uPA even in the presence of plasma proteins. (D) pro-tPA and plasminogen were incubated in the absence (-) or presence (+) of active tryptase ϵ. After a 2-hour incubation at 37°C, the resulting reaction products were subjected to SDS-PAGE under reducing conditions. The resulting gel was stained with Coomassie blue. The top and bottom arrows point to plasminogen and pro-tPA, respectively. As noted, tryptase ϵ was unable to activate either zymogen under the tested conditions.
Activation of uPA by tryptase ϵ. A single-chain pro-uPA was incubated in buffer alone (left lane) or buffer containing pro–tryptase ϵ, active wild-type tryptase ϵ, or its inactive C90A/D92A mutant (lanes 2, 3, and 4, respectively). After a 2-hour incubation at 37°C, the treated samples were subjected to SDS-PAGE under reducing conditions; the resulting gels were stained with Coomassie blue. N-terminal amino acid sequence analysis also was performed on the indicated approximately 33-kDa product detected in the third lane (arrow). (B) Aliquots of the resulting reactions were evaluated for the presence of an active protease that cleaves H-Glu-Gly-Arg-pNA. As noted (▴), tryptase ϵ treatment of pro-uPA resulted in the generation of a mature enzyme that readily cleaved the uPA-sensitive substrate. (C) Time-dependent activation of pro-uPA with tryptase ϵ is shown in the left 4 lanes. In lane 5, tryptase ϵ was added to a 20-fold excess of pro-uPA after incubation with plasma (2% final volume) for 30 minutes at room temperature. Lane 6 on the far right is the control, depicting the proteins in plasma before tryptase ϵ treatment. In a second experiment (data not shown), purified, recombinant tryptase ϵ again quickly activated pro-uPA even in the presence of plasma proteins. (D) pro-tPA and plasminogen were incubated in the absence (-) or presence (+) of active tryptase ϵ. After a 2-hour incubation at 37°C, the resulting reaction products were subjected to SDS-PAGE under reducing conditions. The resulting gel was stained with Coomassie blue. The top and bottom arrows point to plasminogen and pro-tPA, respectively. As noted, tryptase ϵ was unable to activate either zymogen under the tested conditions.
Effects of varied protease inhibitors on the enzymatic activity of recombinant tryptase ϵ
. | . | % Inhibition . | . | |
|---|---|---|---|---|
| Inhibitor . | Major specificity . | Tryptase ϵ . | Trypsin . | |
| Antipain | Papain and trypsin | 77 | 99 | |
| Bestatin | Amino peptidases | 8 | 13 | |
| Pepstatin | Aspartyl proteases | 0 | 0 | |
| Aprotinin | Serine proteases | 5 | 100 | |
| A1AT | Neutrophil elastase and TMT | 0 | 95 | |
| SLPI | Varied serine proteases | 15 | 90 | |
. | . | % Inhibition . | . | |
|---|---|---|---|---|
| Inhibitor . | Major specificity . | Tryptase ϵ . | Trypsin . | |
| Antipain | Papain and trypsin | 77 | 99 | |
| Bestatin | Amino peptidases | 8 | 13 | |
| Pepstatin | Aspartyl proteases | 0 | 0 | |
| Aprotinin | Serine proteases | 5 | 100 | |
| A1AT | Neutrophil elastase and TMT | 0 | 95 | |
| SLPI | Varied serine proteases | 15 | 90 | |
The protease activity of the recombinant tryptase ϵ and trypsin was determined with synthetic substrate H-D-Leu-Thr-Arg-pNA in the presence of different inhibitors. The enzyme-protease inhibitor ratio used in these experiments was approximately 1:10. The experiment was done in duplicate; shown are the mean values. Similar findings were obtained in a second experiment.
A computational docking approach was then used to understand how tryptase ϵ selectively recognizes the activation sequence in pro-uPA. For docking purposes, we considered only the last 7 residues of the propeptide and the first 6 residues of the catalytic chain that are conserved in mouse, rat, and human uPA. As noted in Figure 4, the electrostatic potential of this 13-residue segment that spans the cleavage site in pro-uPA is predicted to complement that in the substrate-binding cleft of tryptase ϵ. Binding energy studies (data not shown) confirmed that tryptase ϵ prefers pro-uPA over plasminogen. The P5′ residues in the activation domains of pro-tPA and plasminogen are very different from those in pro-uPA and pro–tryptase ϵ (Table 1). The protein modeling data predicted that tryptase ϵ uses Lys20 in its A loop to recognize the conserved acidic amino acid in the processing site of pro-uPA. In agreement with this prediction, the K20A mutant of tryptase ϵ failed to convert pro-uPA to uPA even though the mutant still could cleave the tryptic substrate H-Leu-Thr-Arg-pNA effectively (data not shown).
Surface and ball-and-stick representation of the model of tryptase ϵ binding to pro-uPA. The 13-mer sequence in pro-uPA (residues -7 to +6) recognized by tryptase ϵ is shown as a green/magenta ribbon with 3 of its side chains (R-3, K-1, and E5) in stick representation. The green portion of the ribbon represents the C-terminal end of pro-uPA's propeptide; the magenta portion of the ribbon represents the N-terminal end of the protease's main-chain, catalytic domain. Tryptase ϵ is shown in surface representation and is colored according to its electrostatic potential. The red, blue, and white regions are negatively, positively, and neutrally charged, respectively. The active-site triad residues H41, D92, and S193 are labeled in their approximate locations on the surface of tryptase ϵ. The figure was generated using the MOLMOL program.39
Surface and ball-and-stick representation of the model of tryptase ϵ binding to pro-uPA. The 13-mer sequence in pro-uPA (residues -7 to +6) recognized by tryptase ϵ is shown as a green/magenta ribbon with 3 of its side chains (R-3, K-1, and E5) in stick representation. The green portion of the ribbon represents the C-terminal end of pro-uPA's propeptide; the magenta portion of the ribbon represents the N-terminal end of the protease's main-chain, catalytic domain. Tryptase ϵ is shown in surface representation and is colored according to its electrostatic potential. The red, blue, and white regions are negatively, positively, and neutrally charged, respectively. The active-site triad residues H41, D92, and S193 are labeled in their approximate locations on the surface of tryptase ϵ. The figure was generated using the MOLMOL program.39
Effect of tryptase ϵ on human bronchial smooth-muscle cells
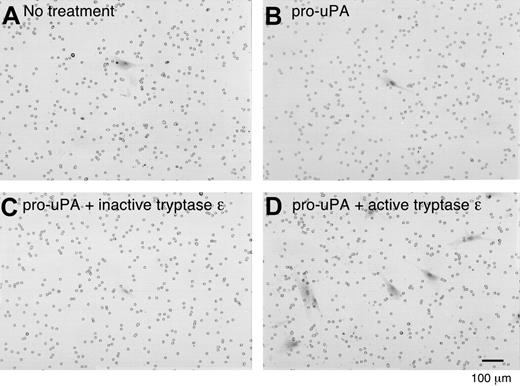
To evaluate whether tryptase ϵ can affect uPA-mediated pathways in a cell system, we next used cultured airway smooth muscle cells whose migration and proliferation can be regulated by uPA and its receptor.40 As assessed by SDS-PAGE immunoblot analysis, pro-uPA is constitutively released into the conditioned medium of cultured smooth muscle cells (Figure 5A). In support of this conclusion, the conditioned medium lacked a protease capable of cleaving the uPA-susceptible substrate H-Glu-Gly-Arg-pNA (Figure 5B). The preferential release of pro-uPA into the conditioned medium is in agreement with that obtained in other smooth muscle cell systems.41 The amount of enzymatically active uPA in the conditioned medium increased significantly when wild-type tryptase ϵ or its active C90A mutant was added to the culture medium but not its inactive C90A/D92E mutant (Figure 5B). In addition, at least some of the tryptase ϵ–generated uPA formed a multimeric complex with endogenous PAI-1 and uPAR (Figure 5A). Thus, in the context of a living cell, human tryptase ϵ is able to convert endogenous pro-uPA to active uPA and active uPA is able to bind to its receptor. In regard to the biologic relevance of the data, human bronchial smooth muscle cells increased their ability to migrate through Matrigel approximately 10-fold when cultured in the presence of enzymatically active tryptase ϵ but not its inactive C90A/D92 mutant (Figure 6).
Discussion
Tryptase ϵ/PRSS22 is a recently cloned member of the chromosome 16p13.3 family of human serine proteases whose expression is normally restricted to epithelial cells.1 It is the likely human ortholog of the Xenopus protease Xepsin,42 a tryptic protease that participates in anteroposterior axis specification in the amphibian's developing epidermis. We now show that tryptase ϵ differs from its other family members in terms of its substrate specificity and ability to autoactivate. We also show that tryptase ϵ is a potent and selective activator of pro-uPA even in the presence of the diverse array of protease inhibitors in blood and lung.
Whether expressed in mammalian or insect cells, exocytosed pro–tryptase ϵ is slowly converted into its mature, enzymatically active protease (Figure 1). Because this spontaneous activation event does not occur with its R-1A and C90A/D92E mutants or with any of the other tryptases we previously expressed in insect cells, the activation event is not the result of a contaminating protease in either expression system. Not only can wild-type pro–tryptase ϵ correctly process itself, Arg-1 plays an essential role in the protease's ability to remove its own propeptide. A small amount of C-terminal processing of tryptase ϵ also occurred in the 2 expression systems after the protease was activated (Figures 1, 2). Rat mast cell protease 2 and human neutrophil cathepsin G, elastase, and proteinase 3 are all cleaved at their C termini downstream of loop 2 in a manner that does not adversely affect their enzymatic activities. It therefore is unlikely that the C-terminal processing of tryptase ϵ is an in vitro artifact caused by the bioengineered C-terminal tags used in the mammalian and insect cell expression systems. More important, the posttranslational event apparently does not adversely affect the protease's enzymatic activity.
Effect of tryptase ϵ on human bronchial smooth-muscle cells. (A) Human bronchial smooth muscle cells were placed in serum-free medium alone (control) or with tryptase ϵ, its C90A mutant, or its C90A/D92E mutant. After an overnight incubation, the conditioned medium was collected, concentrated, and subjected to SDS-PAGE under nonreducing conditions. The resulting protein blots were probed with anti-uPA (left), anti–PAI-1 (middle), and anti-uPAR (right) antibodies. Not shown are the immunoblot data of the residual cell lysates, which in all instances contained substantial amounts of uPAR. Although some uPAR is shed in tryptase ϵ–treated cells, most of the receptor remains cell associated. (B) Samples of the conditioned medium also were evaluated for the presence of enzymatically active uPA using the substrate H-Glu-Gly-Arg-pNA.
Effect of tryptase ϵ on human bronchial smooth-muscle cells. (A) Human bronchial smooth muscle cells were placed in serum-free medium alone (control) or with tryptase ϵ, its C90A mutant, or its C90A/D92E mutant. After an overnight incubation, the conditioned medium was collected, concentrated, and subjected to SDS-PAGE under nonreducing conditions. The resulting protein blots were probed with anti-uPA (left), anti–PAI-1 (middle), and anti-uPAR (right) antibodies. Not shown are the immunoblot data of the residual cell lysates, which in all instances contained substantial amounts of uPAR. Although some uPAR is shed in tryptase ϵ–treated cells, most of the receptor remains cell associated. (B) Samples of the conditioned medium also were evaluated for the presence of enzymatically active uPA using the substrate H-Glu-Gly-Arg-pNA.
Tryptase ϵ/uPA-mediated extravasation of smooth-muscle cells in a Matrigel cell-invasion assay. The ability of normal human bronchial smooth muscle cells to migrate through a basement membrane–like extracellular matrix was evaluated in the presence or absence of tryptase ϵ and uPA. Smooth muscle cells were added to the top wells of each chamber. Nothing else was added in panel A. Pro-uPA alone or with inactive C90A/D92E tryptase ϵ or active tryptase ϵ was added to panels B, C, and D, respectively. FCS-enriched culture medium was added to each bottom chamber as a source of chemotactic factors. Shown are the Masson trichrome+ smooth muscle cells (red) that migrated through each Matrigel-coated membrane. Similar results were obtained in a second experiment. Pro-uPA–expressing smooth muscle cells (SMCs) were able to invade the Matrigel when exposed to approximately 100 nM enzymatically active tryptase ϵ alone (data not shown) but not its inactive C90A/D92 mutant. However, a better response was obtained if a small amount of exogenous pro-uPA was present, presumably because the tryptase ϵ–generated exogenous uPA can overwhelm the amount of PAI-1 present in the 10% FCS and conditioned medium that counteracts the chemotactic response.
Tryptase ϵ/uPA-mediated extravasation of smooth-muscle cells in a Matrigel cell-invasion assay. The ability of normal human bronchial smooth muscle cells to migrate through a basement membrane–like extracellular matrix was evaluated in the presence or absence of tryptase ϵ and uPA. Smooth muscle cells were added to the top wells of each chamber. Nothing else was added in panel A. Pro-uPA alone or with inactive C90A/D92E tryptase ϵ or active tryptase ϵ was added to panels B, C, and D, respectively. FCS-enriched culture medium was added to each bottom chamber as a source of chemotactic factors. Shown are the Masson trichrome+ smooth muscle cells (red) that migrated through each Matrigel-coated membrane. Similar results were obtained in a second experiment. Pro-uPA–expressing smooth muscle cells (SMCs) were able to invade the Matrigel when exposed to approximately 100 nM enzymatically active tryptase ϵ alone (data not shown) but not its inactive C90A/D92 mutant. However, a better response was obtained if a small amount of exogenous pro-uPA was present, presumably because the tryptase ϵ–generated exogenous uPA can overwhelm the amount of PAI-1 present in the 10% FCS and conditioned medium that counteracts the chemotactic response.
Tryptase ϵ possesses an unpaired Cys that is 2 residues upstream of the catalytic Asp in the protease's charge-relay triad. While no other member of the tryptase superfamily possesses a Cys at residue 90, we were intrigued by the fact that serine carboxypeptidases possess a conserved unpaired Cys that is 3 residues downstream of the catalytic Asp.32,33 Because of the importance of unpaired Cys residues in serine carboxypeptidases, we focused our attention on Cys90 in tryptase ϵ. The observation that the C90A mutant of pro–tryptase ϵ underwent a much faster rate of self-activation than the naturally occurring protease (Figure 1D) implies that Cys90 plays a regulatory role in maintaining the protease's latency. Our model of the tryptase ϵ/pro-uPA complex (Figure 4) sheds no light on the role of Cys,90 as it happens to be approximately 7 Å away from the active docked fragment of pro-uPA. The crystal structure of pro–tryptase ϵ should give valuable insight as to how Cys90 controls its latency, but it is presumed that this amino acid somehow controls access to the protease's S1 substrate-binding pocket. What factors and/or mechanisms counteract the Cys-dependent inhibition of pro–tryptase ϵ in vivo remain to be determined, but the nitrosylation or oxidation of Cys90 is a strong possibility.
Recombinant tryptase ϵ can cleave H-D-Leu-Thr-Arg-pNA but not the other tested trypsin-susceptible substrates. Although fibronectin, vitronectin, and laminin are susceptible to a diverse array of neutral proteases, none of these extracellular proteins were cleaved by tryptase ϵ under the conditions used in this study. These data imply that tryptase ϵ possesses a substrate preference very different from that of its other family members. In support of this conclusion is the uniqueness of the amino acid sequences that comprise the loops that form the substrate-binding pocket of tryptase ϵ.1 In contrast to TMT/tryptase γ/PRSS31,31 tryptase ϵ does not recognize the bait regions in those protease inhibitors that reside in the blood and lung. Moreover, no obvious change in the SDS-PAGE pattern of any plasma protein was noted after tryptase ϵ treatment (data not shown), in sharp contrast to what occurs with mMCP-7.24
No other member of the human chromosome 16p13.3 and mouse chromosome 17A3.3 families of serine proteases has been identified to date that can autoactivate. While the ability of tryptase ϵ to autoactivate could be an in vitro phenomenon, this unique feature allowed us to identify one of its physiologic substrates. Many biologic processes in the body are regulated by protease cascades. The fact that tryptase ϵ autoactivates raised the possibility that this serine protease is a convertase that initiates an undefined protease cascade by proteolytically removing the propeptide of a downstream member of the regulated cascade. Tryptase ϵ autoactivates by cleaving the R-1-V1 peptide bond that separates the protease's 17-mer propeptide from its 268-mer catalytic domain (Figure 2; Table 1). The failure of the R-1A mutant (Figure 1) to autoactivate implies that tryptase ϵ recognizes substrates that possess a positively charged amino acid at the P1 site and a hydrophobic amino acid at the P1′ site. A search of GenBank's protein databases using the target peptide sequence Arg-Val-Val-Gly-Gly-Glu that resides at the endogenous cleavage site in the tryptase ϵ zymogen revealed a nearly identical site in the corresponding activation domain of pro-uPA, but not pro-tPA or plasminogen (Table 1). The ability of tryptase ϵ to activate pro-uPA, pro-tPA, and plasminogen was therefore evaluated.
Schematic model of the tryptase ϵ–mediated activation of uPA/uPAR-dependent reactions. In this model, active tryptase ϵ converts pro-uPA to uPA (red symbol) in the epithelium. uPA converts plasminogen to plasmin which, in turn, promotes fibrinolysis and the degradation of extracellular matrices. Some uPA binds to the uPAR (blue symbol) on the surface of the smooth muscle cell and induces varied signaling events in the cell. The uPA/uPAR-activated cells eventually increase their expression of the inhibitory factor PAI-1 (yellow symbol), and the exocytosed serpin binds to the uPA/uPAR complex to dampen the protease-mediated activation response. Some of the multimeric complex is internalized and endocytosed uPA is rapidly destroyed. The remainder is shed in a phospholipase C–dependent manner from the plasma membrane with or without PAI-1. In some cells, uPA can directly bind to the αMβ2.48 Thus, the uPA that is generated by tryptase ϵ also could regulate certain integrin-dependent signaling events. ECM indicates extracellular matrix.
Schematic model of the tryptase ϵ–mediated activation of uPA/uPAR-dependent reactions. In this model, active tryptase ϵ converts pro-uPA to uPA (red symbol) in the epithelium. uPA converts plasminogen to plasmin which, in turn, promotes fibrinolysis and the degradation of extracellular matrices. Some uPA binds to the uPAR (blue symbol) on the surface of the smooth muscle cell and induces varied signaling events in the cell. The uPA/uPAR-activated cells eventually increase their expression of the inhibitory factor PAI-1 (yellow symbol), and the exocytosed serpin binds to the uPA/uPAR complex to dampen the protease-mediated activation response. Some of the multimeric complex is internalized and endocytosed uPA is rapidly destroyed. The remainder is shed in a phospholipase C–dependent manner from the plasma membrane with or without PAI-1. In some cells, uPA can directly bind to the αMβ2.48 Thus, the uPA that is generated by tryptase ϵ also could regulate certain integrin-dependent signaling events. ECM indicates extracellular matrix.
Tryptase ϵ readily converted pro-uPA into active uPA at neutral pH but was unable to activate plasminogen or pro-tPA (Figure 3). The P5′ residues in the activation domains of pro–tryptase ϵ, pro-uPA, pro-tPA, and plasminogen are Glu, Glu, Leu, and Cys, respectively (Table 1). The failure to activate plasminogen or pro-tPA suggested that the P5′ Glu that is present in pro-uPA is essential in substrate recognition. The modeling studies (Figure 4) lend further credence to this hypothesis. The predominantly negatively charged substrate-binding site of tryptase ϵ is predicted to be occupied by the positively charged residues that reside toward the end of the propeptide part of pro-uPA. The negatively charged Glu in pro-uPA complements the positively charged surface patch adjacent to the active site, thereby adding further specificity to tryptase ϵ's recognition of pro-uPA. Computational estimations of the free energy of binding also predicted that tryptase ϵ should recognize and bind pro-uPA much more tightly than plasminogen. The 3D modeling data predicted that tryptase ϵ uses Lys20 to recognize the P5′ Glu residue in pro-uPA. The inability of the K20A mutant to process pro-uPA efficiently supports this prediction and explains why tryptase ϵ cannot activate pro-tPA or plasminogen efficiently. The observation that uPA was unable to activate pro–tryptase ϵ implies that tryptase ϵ acts upstream of uPA in plasmin-dependent pathways.
uPA is a physiologic activator of plasminogen. While plasmin can activate pro-uPA to amplify uPA/plasminogen/plasmin pathways, the fact that the level of uPA is not reduced in the urine of plasminogen-null mice43 implies that an undefined protease often is used to activate uPA. It therefore remains to be determined which naturally occurring protease in the epithelium converts inactive pro-uPA to active uPA to initiate the enzymatic cascade. Because uPA converts plasminogen to plasmin, it is likely that tryptase ϵ indirectly promotes fibrinolysis and the degradation of extracellular matrices. Binding of uPA to its receptor results in cell surface–associated plasmin activity. The ability to concentrate uPA and plasmin at the leading edge of a uPAR-expressing cell is believed to be important in its directional movement through an extracellular matrix.44,45 Migration and accumulation of smooth muscle cells often occur in chronic inflammation of blood vessels and airways,46 and enzymatically active uPA induces the growth and migration of smooth muscle cells.47 The discovery that tryptase ϵ is a potent activator of uPA (Figures 3, 5) and helps smooth muscle cells migrate through a basement membrane–like extracellular matrix (Figure 6) raises the additional possibility that tryptase ϵ participates in airway remodeling and the hypertrophy of smooth muscle cells, as well cancer invasion and/or metastasis. Figure 7 highlights a scenario of what might be occurring in the lung when enzymatically active tryptase ϵ is present.
While it is likely that tryptase ϵ exhibits adverse roles in some disorders of human epithelium, it is important to note that uPA and its receptor are essential for combating life-threatening Cryptococcus neoformans infections in the lung.28 It has been shown that tryptase β1 and its mouse ortholog mMCP-6 are needed for optimally combating Klebsiella pneumoniae infections.23 Because uPA, tryptase ϵ, and tryptase β1 probably work in concert to control bacteria and fungal infections in the lung and other epithelial cell sites, it is likely that tryptase ϵ is a beneficial neutral protease in innate immunity in most instances, like its family member tryptase β1.
Prepublished online as Blood First Edition Paper, February 8, 2005; DOI 10.1182/blood-2003-10-3501.
Supported by grants from the National Health and Medical Research Council of Australia, and by grants DK067835, HL63284, and HL36110 from the National Institutes of Health.
S.Y. and N.M. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal