Abstract
Burkitt lymphoma (BL) is a highly aggressive B-cell neoplasm harboring chromosomal rearrangements of the c-myc oncogene. BL cells frequently resist apoptosis induction by chemotherapeutic agents; however, the mechanism of unresponsiveness has not been elucidated. Here, we show that cytochrome c fails to stimulate apoptosome formation and caspase activation in cytosolic extracts of human BL-derived cell lines, due to insufficient levels of apoptotic protease-activating factor-1 (Apaf-1). Enforced expression of Apaf-1 increased its concentration in the cytosolic compartment, restored cytochrome c-dependent caspase activation, and rendered the prototypic Raji BL cell line sensitive to etoposide- and staurosporine-induced apoptosis. Surprisingly, in nontransfected BL cells, the bulk of Apaf-1 was found to associate with discrete domains in the plasma membrane. Disruption of lipid raft domains or the actin cytoskeleton of Raji cells liberated Apaf-1 and restored sensitivity to cytochrome c–dependent apoptosis, indicating that constitutive Apaf-1 retained its ability to promote caspase activation. Moreover, disruption of lipid rafts sensitized BL cells to apoptosis induced by etoposide. Together, our findings suggest that ectopic (noncytosolic) localization of Apaf-1 may constitute a novel mechanism of chemoresistance in B lymphoma.
Introduction
Resistance to apoptosis is one of the cardinal features of cancer cells1 and contributes to their chemoresistance.2 Most chemotherapeutic drugs activate the intrinsic (ie, mitochondria-dependent) apoptosis pathway, resulting in the release of apoptogenic factors, including cytochrome c, that normally reside in the intermembrane space of these organelles. Subsequently, cytochrome c, Apaf-1 (apoptotic protease-activating factor-1), and pro–caspase-9 form an apoptosome complex in the cytosol. Apoptosome-driven oligomerization of Apaf-1 triggers the activation of pro–caspase-9, and caspase-9, in turn, activates downstream caspases such as pro–caspase-3, resulting in the rapid dismantling of the cell (for a recent review, see Danial and Korsmeyer3 ). Dysfunction of this pathway of caspase activation has been associated with chemoresistance. Loss of Apaf-1 because of hypermethylation of the promotor region was reported in chemoresistant melanomas, and demethylating agents were shown to restore Apaf-1 expression and rescue the apoptosis defect in these cells.4 Similarly, resistance of certain leukemia cell lines to UV light–induced apoptosis was shown to be associated with defective expression of Apaf-1.5,6 Impaired apoptosome activation has also been documented in ovarian carcinoma cell lines but could not be explained by Apaf-1 deletion, degradation, or mutation.7,8 Furthermore, a loss of Apaf-1 expression has been demonstrated in (nonmalignant) human skeletal muscle cytosols, which are refractory to cytochrome c-dependent caspase activation.9 Apoptosome formation is thus a key event in intrinsic apoptosis signaling and, as such, is subject to regulation by numerous cellular proteins.10-12
Burkitt lymphoma (BL) is a highly malignant non-Hodgkin lymphoma that invariably harbors translocations of the c-myc oncogene.13 Other recurrent genetic aberrations, including mutations of the p53 gene and transcriptional silencing of its homolog, p73, have also been reported.14,15 Such aberrations may antagonize cell death signaling pathways and thus contribute to chemoresistance, at least in some forms of BL. Studies in apoptosis-resistant BL cell lines have suggested a link between c-myc activation and alterations in cytosolic proteolysis.16 Other investigators have reported that apoptotic signal transduction in the cytoplasm of Raji, a prototypic BL cell line, is defective; however, the molecular mechanism was not clarified.17 Similarly, a failure of etoposide to activate certain upstream effector(s) of caspase-3 was suggested to contribute to chemoresistance in the BL-derived cell line, BL29.18 Here, we show that a subthreshold level of Apaf-1 in the cytosolic compartment renders BL cells resistant to mitochondria-targeted apoptosis stimuli, including etoposide. Furthermore, apoptosis resistance in the Raji BL cell line could be overcome either by stable overexpression of Apaf-1 or through mobilization of endogenous Apaf-1 from its ectopic localization near or at the plasma membrane of these cells. These results show, for the first time, that plasma membrane sequestration of Apaf-1 may confer chemoresistance to human cancer cells.
Materials and methods
Cell lines
The Jurkat leukemic T-cell line, obtained from the European Cell Culture Collection (Salisbury, United Kingdom), and the BL-derived cell lines, Raji, Ramos, DG75, Rael, Mutu I, and Mutu III (from the Microbiology and Tumor Biology Center, Karolinska Institutet, Stockholm, Sweden), were grown in RPMI-1640 medium (Life Technologies, Paisley, Scotland) supplemented with 10% fetal calf serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 1 mM glutamine.
Induction and quantification of apoptosis
Cells in logarithmic growth phase were treated with 10 μM etoposide (Bristol-Myers, Solna, Sweden) or 1 μM staurosporine (Sigma, St Louis, MO), in the presence or absence of zVAD-fmk (N-benzyloxycarbonyl-Val-Ala-aspartyl-fluoromethyl ketone; 50 μM; Enzyme Systems Products, Dublin, CA) or methyl-β-cyclodextrin (MCD; 100 μM; Sigma), for the indicated time points. Apoptosis was determined by flow cytometric evaluation of DNA content using propidium iodide (PI):RNase A (10 μg/mL PI, from Molecular Probes, Leiden, The Netherlands, and 250 μg/mL RNase A, from Sigma), as previously described.19 Samples were analyzed on a FACScan (Becton Dickinson, San Jose, CA) using the CellQuest software (Becton Dickinson). Data are shown as the percentage of cells displaying hypodiploid DNA content. In some experiments, nuclear apoptotic features were determined by fluorescence microscopy of Hoechst 33342–labeled cells (10 μg/mL; Molecular Probes), as previously described.20 Data are displayed as the percentage of cells with a fragmented/condensed nucleus.
Determination of caspase activities
Caspase-3, caspase-8, and caspase-9 activities were determined by using the fluorogenic peptide substrates, DEVD-AMC (aminomethylcoumarin; 50 μM), IETD-AMC (100 μM), and LEHD-AMC (100 μM) (Biomol, Plymouth Meeting, PA), respectively. Cell lysates and substrates were combined in a standard reaction buffer,21 and real-time measurements of enzyme-catalyzed release of AMC were obtained using a Fluoroscan II plate reader (Labsystems, Stockholm, Sweden) operating with Genesis software (Labsystems). Fluorescence values were converted to picomoles of AMC release by using a standard curve generated with free AMC, and the maximum rate of AMC release (pmol/min) was estimated for each sample; alternatively, the fold-increase of AMC release in treated samples compared with control is depicted.
Western blotting
For Western blotting of total cell lysates, cells were harvested and suspended in 1% Triton X-100–containing extraction buffer. To obtain mitochondrial and cytosolic fractions, cells were resuspended in buffer A (50 mM PIPES (piperazine diethanesulfonic acid)–KOH at pH 7.4, 220 mM mannitol, 68 mM sucrose, 50 mM KCl, 5mM EGTA (ethylene glycol tetraacetic acid), 2 mM MgCl2, 1 mM DTT (dithiothreitol)) supplemented with protease inhibitors (Boehringer Mannheim, Mannheim, Germany). Cell disruption was performed by passing cells through a 20-gauge needle 8 to 10 times. Homogenates were then centrifuged at 12 000g for 5 minutes at 4°C, and supernatant (cytosolic fraction) and pellet (enriched for nuclei and mitochondria) were recovered. Samples (30 μg protein, as determined using the Bradford reagent; BioRad Laboratories, Hercules, CA) were then denatured in Laemmli buffer and resolved on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels, followed by transfer to nitrocellulose membranes. Immunoblotting was performed with antibodies to Apaf-1 (Chemicon, Temecula, CA), caspase-3 (BD Biosciences, San Diego, CA), caspase-9 (BD Biosciences), cytochrome c (BD Biosciences), poly(ADP-ribose) polymerase (PARP; Biomol), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Trevigen, Gaithersburg, MD), and proteins were visualized by enhanced chemiluminescence (BioRad Laboratories). Densitometric analysis of membranes was performed using a Fuji Film Intelligent Darkbox II densitometer and Image Gauge 4.0 software (Fuji Film, Dusseldorf, Germany).
Analysis of mitochondrial membrane potential
Loss of mitochondrial membrane potential (ΔΨm) was detected by using the potential-sensitive fluorescent probe, tetramethylrhodamine ethyl ester (TMRE; Molecular Probes), as described elsewhere.22 Treated or untreated cells were allowed to incubate with TMRE (25 nM) for 15 minutes at 37°C prior to analysis on a FACSscan (Becton Dickinson) operating with CellQuest software. As a positive control, cells were incubated with 100 μM uncoupling agent, carbonyl cyanide 3-chlorophenylhydrazone (CCCP; Molecular Probes).
Cell-free apoptosis reactions
For preparation of S-100 cytosolic extracts, 50 × 106 cells were collected and incubated in ice-cold S-100 buffer (20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid)–KOH at pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA (ethylenediaminetetraacetic acid), 1 mM EGTA, 1 mM DTT) supplemented with protease inhibitors (Boehringer Mannheim). Cells were disrupted using a Dounce homogenizer, and the pellet was centrifuged at 1000g for 10 minutes at 4°C. Supernatants were further centrifuged at 100 000g for 1 hour, and the resulting supernatant (S-100 fraction) was either stored at -80°C or used immediately. For cell-free caspase activation,23 S-100 cytosol (120 μg) was incubated with 1 mM dATP (deoxyadenosine triphosphate) and 4 μM cytochrome c (Sigma) for 30 minutes at 30°C. Aliquots were then collected for assessment of pro-caspase processing by Western blot and caspase activity by fluorogenic assay as detailed in “Determination of caspase activities.” For granzyme B–induced caspase activation, S-100 cytosols were incubated for 30 minutes at 30°C with 2 units human recombinant granzyme B (Calbiochem, Nottingham, United Kingdom).
Assessment of apoptosome formation
S-100 cytosolic extracts (3 μg/μL protein) were prepared as described in “Cell-free apoptosis reactions” and incubated in the presence or absence of dATP (1 mM) and cytochrome c (4 μM) to trigger apoptosome formation. Lysates were then loaded on a Sephacryl S-300 gel filtration column (Amersham Biosciences, Uppsala, Sweden) at a flow rate of 0.5 mL/min, using a BioLogic HR workstation (BioRad). The column was calibrated with a gel filtration protein standard kit (Sigma) ranging from 10 kDa to 2000 kDa. A total of 70 fractions were collected using a Fraction Collector FRAC-100 (Pharmacia, Uppsala, Sweden), and aliquots from each fraction were resolved by SDS-PAGE followed by immunoblotting using antibodies to Apaf-1 and caspase-9.
Transfection and generation of stable cell lines
Apaf-1 pcDNA3.1 (-) plasmid24 was grown in Escherichia coli XL-1 blue strain and purified using the Plasmid DNA MiniPrep kit (Qiagen, Valencia, CA). Raji BL cell lines stably overexpressing Apaf-1 were obtained by electroporation with Apaf-1 pcDNA3.1 (-) using a BioRad Gene Pulser and isolation of stable clones after 3 weeks of selection in geneticin (Life Technologies). Western blotting for Apaf-1 was performed as described in “Western blotting” to assess the transfection efficiency.
RT-PCR and sequencing of Apaf-1
Raji cell RNA was isolated using the RNeasy Mini kit (Qiagen). First-strand DNA was synthesized by reverse transcription–polymerase chain reaction (RT-PCR) using an RT kit (Life Technologies) and a primer targeted toward Apaf-1 mRNA downstream of the stop codon (5′-tactgcacaacaattaagagct-3′). The following 5 primer pairs were used to amplify Apaf-1 cDNA: (1) 5′-tggttgacagctcagagag-3′, 5′-attactgaatctgtaacactc-3′; (2) 5′-gcttttgacagtcagtgtca-3′, 5′-atcagatgagcagggccta-3′; (3) 5′-ccctggattggattaaagca-3′, 5′-cccaaagctttaaggttcca-3′; (4), 5′-ttggctagttgttcagctga-3′, 5′-gtcttctcatcggctgtga-3′; and (5), 5′-ctgtatggcacatccagtt-3′, 5′-agttgatatcagggtttcatta-3′. PCR products were resolved by agarose gel electrophoresis, purified using the QIAQuick Gel Extraction kit (Qiagen), and sequenced with the ABI PRISM Big Dye Primer Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA).
Confocal microscopy
Cells were fixed and permeabilized in 4% formaldehyde and 1% Triton X-100, respectively, and blocked in 2% bovine serum albumin (BSA) before staining with Apaf-1–specific antibodies. Alexa 488–conjugated secondary antibodies were from Molecular Probes. For visualization of mitochondria, cells were incubated with MitoTracker Red (250 nM; Molecular Probes), and for detection of the Golgi apparatus and endoplasmic reticulum, Texas Red–conjugated wheat germ agglutinin (WGA; 5 μg/mL) (Molecular Probe) was used. Localization of nuclei in Raji and Jurkat cells was determined with Hoechst 33342 (10 μg/mL; Molecular Probes), and Alexa 555–conjugated cholera toxin B (CTB; 10 μg/mL; Molecular Probes) was used to locate lipid rafts. All confocal images were captured on a Zeiss Meta LSM510 confocal microscope (Carl Zeiss, Herts, United Kingdom) using a Zeiss 40×/1.3 numerical aperture (NA) oil immersion lens. For some experiments, series of optical sections distanced 4 μm apart were acquired and merged. All image analysis was performed using LSM510 Meta software (Carl Zeiss). For Apaf-1 mobilization studies, cells were incubated with 10 mM MCD or 0.5 μg/mL cytochalasin B (CB; Sigma) for 1 hour prior to immunodetection of Apaf-1 and co-labeling of lipid rafts.
Results
BL cell lines are resistant to etoposide-induced caspase activation
We first tested the response to the commonly used chemotherapeutic drug, etoposide, in a panel of established BL-derived cell lines, including the Raji cell line, previously shown to be resistant to ceramide as well as to various chemotherapeutic agents.17,25 A reduced level of apoptosis was seen in Raji, DG75, Ramos, Rael, Mutu I, and Mutu III cells after 24 hours of treatment; apoptosis-sensitive Jurkat T cells were included as a control (Figure 1A). To ascertain whether apoptosis is abrogated and not merely delayed, cells were also incubated for 48 hours with the apoptogenic drug. These experiments revealed that all BL cell lines with the exception of Mutu I (50% apoptosis in Mutu I under these conditions) were completely resistant to apoptosis induction; in contrast, more than 90% apoptosis was detected in Jurkat cells (data not shown). DEVD-AMC cleavage (indicative of caspase-3 activation) was also significantly reduced in all BL cell lines tested, when compared with Jurkat cells (Figure 1B). Moreover, no activation of caspase-8 or caspase-9 could be detected in BL cells upon etoposide treatment (Figure 1C). To confirm these results, cytosolic extracts were immunoblotted with specific antibodies against caspase-3 and caspase-9. Etoposide-triggered processing of pro–caspase-3 into the p20 fragment and the p17 active subunit was readily detected in Jurkat cells (Figure 1D). Concomitant processing of pro–caspase-9 into its characteristic p35 fragment was also seen; moreover, a p37 fragment of caspase-9, known to be generated by caspase-3, was also evident in Jurkat extracts. However, no processing of pro–caspase-9 was seen in BL cell extracts, with the exception of Rael, in which the p35/p37 bands could be detected (Figure 1D). Processing of pro–caspase-3 was also absent in most BL cell lines, although some evidence of the p20/p17 subunits was consistently seen in Rael, and partial processing to the p17 fragment was observed in Ramos. In addition, processing of PARP to its 85-kDa signature fragment (an indicator of nuclear activation of caspases) was evident in Jurkat cells but could not be detected in BL cells (data not shown). Taken together, these results demonstrate that resistance to etoposide-induced activation of caspases is a common feature of BL cell lines.
BL-derived cell lines are resistant to etoposide-induced caspase activation. (A) BL and Jurkat cell lines were incubated for 24 hours in the absence (□) or presence (▪) of etoposide, and apoptosis was assessed by flow cytometry. (B) Cells treated for 24 hours with etoposide were harvested for measurement of caspase-3–like enzyme activity according to specific cleavage of the fluorescent DEVD-AMC substrate. (C) Raji and Jurkat cells preincubated in the absence (▪) or presence (▨) of the pan-caspase inhibitor, zVAD-fmk, were treated with etoposide for 6 hours, and caspase activities were determined. □ indicates control cells. Data are depicted as fold increases of AMC release. (D) Cell lysates from BL and Jurkat cell lines treated for 12 hours with etoposide were evaluated for processing of pro–caspase-3 and pro–caspase-9 by using specific antibodies. Results shown in panels A-C are mean values ± SDs (n = 3). Data shown in panel D are representative of 3 independent experiments.
BL-derived cell lines are resistant to etoposide-induced caspase activation. (A) BL and Jurkat cell lines were incubated for 24 hours in the absence (□) or presence (▪) of etoposide, and apoptosis was assessed by flow cytometry. (B) Cells treated for 24 hours with etoposide were harvested for measurement of caspase-3–like enzyme activity according to specific cleavage of the fluorescent DEVD-AMC substrate. (C) Raji and Jurkat cells preincubated in the absence (▪) or presence (▨) of the pan-caspase inhibitor, zVAD-fmk, were treated with etoposide for 6 hours, and caspase activities were determined. □ indicates control cells. Data are depicted as fold increases of AMC release. (D) Cell lysates from BL and Jurkat cell lines treated for 12 hours with etoposide were evaluated for processing of pro–caspase-3 and pro–caspase-9 by using specific antibodies. Results shown in panels A-C are mean values ± SDs (n = 3). Data shown in panel D are representative of 3 independent experiments.
Apoptosis is blocked downstream of mitochondria in the Raji BL cell line
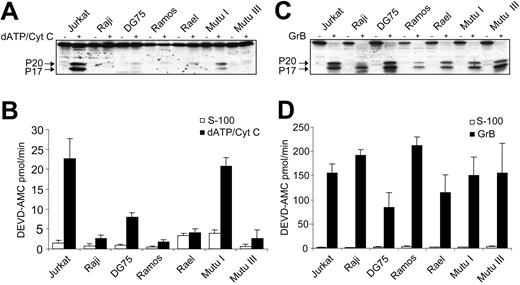
Etoposide triggers mitochondrial release of cytochrome c in cancer cells, resulting in activation of the caspase cascade and induction of apoptosis.26 To further delineate the apoptosis defect in BL cells, we tested whether etoposide treatment of Raji cells could induce mitochondrial alterations. Raji cells were chosen for these experiments because they serve as a prototypic example of a chemoresistant BL-derived cell line and are widely used in lymphoma studies.27 As seen in Figure 2, etoposide triggers a dissipation of the ΔΨm and mitochondrial release of cytochrome c both in Jurkat and Raji cells. Therefore, defective activation of caspase-9 and caspase-3 in the Raji cell line is not due to the absence of cytochrome c release. We then used a cell-free system to further evaluate whether BL-derived cell lines are capable of promoting Apaf-1–dependent activation of caspases. To this end, exogenous cytochrome c was added to S-100 cytosolic extracts derived from the 6 BL cell lines. Jurkat cell extracts served as a positive control. Caspase-3 processing was readily detected in Jurkat cells, whereas no activation of pro–caspase-3 was observed in Raji cells or in any other BL-derived cell line, with the exception of Mutu I (Figure 3A). Similarly, cleavage of DEVD-AMC was absent or reduced in all BL-derived cell lines (except Mutu I) following the addition of exogenous cytochrome c (Figure 3B). In contrast, all BL-derived cell lines responded to recombinant granzyme B–mediated processing and activation of pro–caspase-3 (Figure 3C-D), an event that transpires independently of Apaf-1.28 These data indicate that the response of BL cells to etoposide is blocked downstream of cytochrome c release, yet upstream of caspase-3 activation.
Etoposide triggers mitochondrial perturbations in the Raji BL cell line. Dissipation of mitochondrial membrane potential (A) and release of cytochrome c (B) in etoposide-treated Raji BL cells was evaluated by flow cytometry and immunoblotting of cytosolic extracts versus mitochondrial pellets (30 μg per lane), respectively. CCCP was used as a positive control for membrane depolarization. Jurkat cells were included for comparison in panels A and B. Results are representative of at least 3 independent experiments.
Etoposide triggers mitochondrial perturbations in the Raji BL cell line. Dissipation of mitochondrial membrane potential (A) and release of cytochrome c (B) in etoposide-treated Raji BL cells was evaluated by flow cytometry and immunoblotting of cytosolic extracts versus mitochondrial pellets (30 μg per lane), respectively. CCCP was used as a positive control for membrane depolarization. Jurkat cells were included for comparison in panels A and B. Results are representative of at least 3 independent experiments.
Subthreshold levels of Apaf-1 in the cytosol of human BL cells
Silencing of Apaf-1 has previously been reported in certain melanoma- and leukemia-derived cell lines.4,6 To explore whether chemoresistance in BL cells could be explained by defective Apaf-1 expression, we performed immunoblotting with specific antibodies against Apaf-1 in our panel of BL cells. When cytosolic extracts of Jurkat cells were compared with extracts from apoptosisresistant BL cell lines, a marked reduction of Apaf-1 was observed, most notably in Raji (Figure 4A). However, Apaf-1 was found to be present at comparable levels in total cell extracts of Jurkat and Raji cells (Figure 4B), suggesting that subcellular compartmentalization of Apaf-1 differs between the 2 cell lines. Indeed, approximately 80% of Apaf-1 was present in the cytosol in Jurkat cells, whereas only 30% of total cellular Apaf-1 was recovered in the cytosol of the Raji cell line (Figure 4B). Hence, it appeared that caspase activation in Raji cells might be hampered by a subthreshold level of cytosolic Apaf-1. Of note, our sequence analysis of Apaf-1 mRNA in Raji cells disclosed 2 previously reported mRNA species (corresponding to Apaf-1 isoforms with and without an extra WD40 repeat in the COOH terminal domain)29 ; however, no sequence anomalies were found (data not shown). Hence, the ectopic subcellular localization of Apaf-1 in these cells could not be explained by alternative (mutated or deleted) forms of Apaf-1.
Defective apoptosome activation in the human Raji BL cell line
To test whether a decreased level of cytosolic Apaf-1 precludes apoptosome formation, we used size-exclusion chromatography to fractionate cytosolic extracts from Jurkat and Raji cells. After activation with cytochrome c to induce Apaf-1 oligomerization, or a similar treatment in the absence of cytochrome c as a control, fractions separated by gel filtration chromatography were analyzed for Apaf-1 with the use of specific antibodies. Control samples from both cell lines demonstrated Apaf-1 elution in fractions corresponding to approximately Mr (molecular weight ratio) 150 000, consistent with the monomeric form of this protein (Figure 4C-D). As expected, incubation of Jurkat extracts with exogenous cytochrome c yielded a shift in the Apaf-1 elution profile such that the majority of Apaf-1 appeared in fractions corresponding to Mr 700 000 to 2 000 000 (Figure 4D). No such shift was discernible in Raji cell extracts treated with cytochrome c (Figure 4C), suggesting that Apaf-1 oligomerization is defective in Raji cell cytosolic extracts. Caspase-9 becomes activated in the apoptosome complex30 ; moreover, a diminished amount of caspase-9 in the apoptosome was previously linked to a reduced susceptibility of ovarian cancer cells to cisplatin-triggered apoptosis.8 As seen in Figure 4C, defective apoptosome activation in Raji cells was associated with negligible amounts of active caspase-9 in the Apaf-1–containing fractions. In contrast, after cytochrome c–dependent activation of Jurkat cytosolic extracts, a significant amount of pro–caspase-9 was processed into the active p35/p37 subunits (Figure 4D). Hence, defective cytosolic expression of Apaf-1 in Raji BL cells is associated with the absence of apoptosome formation and, consequently, with a lack of pro–caspase-9 activation.
Overexpression of Apaf-1 restores sensitivity to apoptosis in the Raji BL cell line
To test whether enhanced levels of Apaf-1 in the cytosolic compartment could sensitize BL cells to apoptosis, we generated stable Apaf-1–overexpressing BL cell lines through transfection of the APAF1 gene into Raji cells and subsequent selection with antibiotics. Increased Apaf-1 expression was detected in total cell extracts prepared from the transfectants when compared with extracts from vector controls; furthermore, Apaf-1 expression was increased in the cytosolic fraction of the transfected Raji cells (Figure 5A). Apaf-1–transfected Raji cells were considerably more sensitive to etoposide-triggered apoptosis as compared with mocktransfectants (Figure 5B). Moreover, sensitivity to staurosporine, another apoptotic agent that targets the mitochondrial pathway,31 was also significantly enhanced. In concordance with these findings, both etoposide and staurosporine triggered robust activation of caspase-3 in Apaf-1–transfected cells, whereas their mocktransfected counterparts remained resistant (Figure 5C). As seen in Figure 5D, enforced expression of Apaf-1 also potentiated caspase-3 activation in S-100 extracts in response to exogenous cytochrome c, to a level comparable to that seen in Jurkat cytosols. Raji cells harbor wild-type Apaf-1 (discussed in “Subthreshold levels of Apaf-1 on the cytosol of human BL cells”), and it, therefore, remains to be determined why transfected, but not constitutive, Apaf-1 is expressed in the cytosol of Raji cells. However, this could perhaps be a consequence of a supranormal level of expression of Apaf-1 in the former case. Our data nonetheless demonstrate that re-expression of Apaf-1 in the cytosol renders Raji BL cells susceptible to cytochrome c-dependent caspase activation.
Lack of cytochrome c-dependent activation of pro–caspase-3 in BL cell lines. (A) S-100 cytosols were prepared from the Jurkat and BL cell lines, and exogenous dATP and cytochrome c (dATP/Cyt c) were introduced to initiate caspase activation. Processing of pro–caspase-3 was determined by Western blotting. (B) Measurement of caspase-3–like enzyme activity by DEVD-AMC cleavage in Jurkat and BL cell lines treated as in panel A. □ indicates S-100 cytosols alone; ▪, with exogenous dATP and cytochrome c. Results are depicted as mean values ± SDs (n = 3). (C) Granzyme B (GrB) processing of pro–caspase-3 remains intact in BL-derived cell lines. S-100 cytosols from a panel of BL cell lines were incubated for 30 minutes with recombinant human granzyme B, and samples were assessed for processing of pro–caspase-3 (using specific anti–caspase-3 antibodies) and for caspase-3–like enzyme activity (D), using the fluorogenic DEVD-AMC substrate. Cytosolic extracts from Jurkat cells were included as a positive control. In D, □ indicates S-100 cytosols alone; ▪, with granzyme B. Results in panel D are depicted as mean values ± SDs (n = 3).
Lack of cytochrome c-dependent activation of pro–caspase-3 in BL cell lines. (A) S-100 cytosols were prepared from the Jurkat and BL cell lines, and exogenous dATP and cytochrome c (dATP/Cyt c) were introduced to initiate caspase activation. Processing of pro–caspase-3 was determined by Western blotting. (B) Measurement of caspase-3–like enzyme activity by DEVD-AMC cleavage in Jurkat and BL cell lines treated as in panel A. □ indicates S-100 cytosols alone; ▪, with exogenous dATP and cytochrome c. Results are depicted as mean values ± SDs (n = 3). (C) Granzyme B (GrB) processing of pro–caspase-3 remains intact in BL-derived cell lines. S-100 cytosols from a panel of BL cell lines were incubated for 30 minutes with recombinant human granzyme B, and samples were assessed for processing of pro–caspase-3 (using specific anti–caspase-3 antibodies) and for caspase-3–like enzyme activity (D), using the fluorogenic DEVD-AMC substrate. Cytosolic extracts from Jurkat cells were included as a positive control. In D, □ indicates S-100 cytosols alone; ▪, with granzyme B. Results in panel D are depicted as mean values ± SDs (n = 3).
Defective cytosolic expression of Apaf-1 and lack of apoptosome oligomerization in BL cells. (A) Cytosolic Apaf-1 expression in Jurkat and BL cell lines. Protein (30 μg) was loaded in each lane. (B) Apaf-1 expression in total cell lysates versus cytosolic extracts. The ratio of cytosolic-total Apaf-1 was determined by densitometry. GAPDH staining is shown in panels A and B as confirmation of equal loading of protein. Results are representative of at least 3 independent experiments. (C) Defective apoptosome activation. S-100 lysates from Raji (C) and Jurkat (D) cells were incubated for 30 minutes in the presence or absence of cytochrome c and dATP and then fractionated on a Sephacryl S-300 column. Equal amounts of each fraction were resolved by SDS-PAGE, and Apaf-1 and caspase-9 were detected by immunoblotting by using specific antibodies. The pro-form (p46) of caspase-9 and the 2 subunits (p37 and p35) are indicated with arrows. The sizes of selected standards are shown at the top of each panel.
Defective cytosolic expression of Apaf-1 and lack of apoptosome oligomerization in BL cells. (A) Cytosolic Apaf-1 expression in Jurkat and BL cell lines. Protein (30 μg) was loaded in each lane. (B) Apaf-1 expression in total cell lysates versus cytosolic extracts. The ratio of cytosolic-total Apaf-1 was determined by densitometry. GAPDH staining is shown in panels A and B as confirmation of equal loading of protein. Results are representative of at least 3 independent experiments. (C) Defective apoptosome activation. S-100 lysates from Raji (C) and Jurkat (D) cells were incubated for 30 minutes in the presence or absence of cytochrome c and dATP and then fractionated on a Sephacryl S-300 column. Equal amounts of each fraction were resolved by SDS-PAGE, and Apaf-1 and caspase-9 were detected by immunoblotting by using specific antibodies. The pro-form (p46) of caspase-9 and the 2 subunits (p37 and p35) are indicated with arrows. The sizes of selected standards are shown at the top of each panel.
Plasma membrane sequestration of Apaf-1 in chemoresistant BL cells
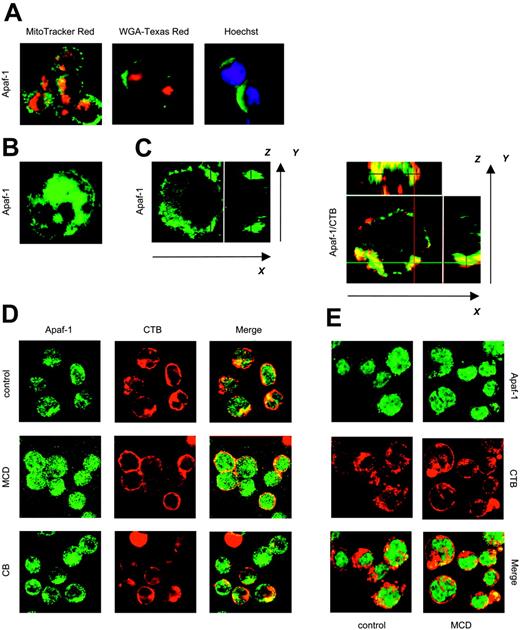
Apaf-1 has been reported to reside exclusively in the cytosol,32 although other studies have suggested that the subcellular localization of Apaf-1 is cell type dependent; indeed, overexpression of Bcl-2 in a murine B lymphoma cell line was shown to trigger redistribution of Apaf-1 from the Golgi apparatus to a perinuclear compartment.33 To investigate the subcellular distribution of Apaf-1 in Raji cells, we performed colocalization studies by using specific markers of the nucleus, Golgi apparatus, and mitochondria. Confocal microscopic analyses did not provide any evidence of Apaf-1 localization in these subcellular compartments in Raji cells (Figure 6A) or in Jurkat cells (data not shown). Moreover, no alterations in the distribution of endogenous Apaf-1 were observed upon etoposide treatment of these cell lines (data not shown). However, while Apaf-1 in Jurkat cells exhibited a diffuse cytoplasmic distribution (Figure 6B), Apaf-1 in Raji cells appeared to be closely associated with the plasma membrane. In fact, confocal analysis of different x-y planes along the z-axis of individual cells suggested the aggregation of Apaf-1 in discrete membrane domains (Figure 6C). Lipid rafts are subdomains of the plasma membrane that are enriched in sphingolipids in optimal admixture with cholesterol.34 To test the hypothesis that Apaf-1 is associated with lipid rafts, we labeled Raji cells with CTB, a well-known marker of such microdomains. Costaining of cells with antibodies specific for Apaf-1 revealed a substantial overlap between the lipid raft signal and Apaf-1 (Figure 6D, top; see Figure 6C for z-axis projections). No such overlap was observed in Jurkat cells (Figure 6E). To obtain further evidence for lipid raft association, Raji cells were treated with the cholesterol-depleting agent, MCD, that is known to disrupt lipid rafts and caveolae.34 As seen in Figure 6D (middle), Apaf-1 was dispersed in the cytoplasm upon MCD treatment, and the colocalization with CTB became less evident. The pattern of Apaf-1 distribution in Jurkat cells, however, remained unchanged (Figure 6E). Importantly, S-100 cytosolic extracts prepared from MCD-treated Raji cells supported cytochrome c–dependent activation of caspase-3, while control extracts failed to do so (Figure 7A). MCD treatment of Jurkat cells, however, did not significantly affect apoptosome-driven caspase activation.
Studies have shown that lipid raft domains interact with (and are anchored to) the underlying actin cytoskeleton.35 We, therefore, assessed whether manipulation of the cytoskeleton might affect Apaf-1 distribution in the Raji BL cell line. To this end, cells were treated with CB, a fungal metabolite that prevents actin polymerization and promotes disruption of the microfilament network. CB triggered a redistribution of Apaf-1 in Raji cells, similar to that observed in MCD-treated cells (Figure 6D, bottom). Moreover, cytosolic extracts from CB-treated Raji cells were also able to promote cytochrome c–dependent caspase activation (Figure 7A). Again, no significant effects were observed in Jurkat cells. Together, these results suggest that Apaf-1 is sequestered in discrete domains at or near the plasma membrane of the Raji BL cell line and indicate that Apaf-1, upon mobilization from these membrane domains, is capable of supporting cytochrome c-dependent caspase activation in the cytosol.
Stable overexpression of Apaf-1 sensitizes BL cells to cytochrome c-dependent apoptosis. (A) Apaf-1 expression in total cell lysates versus cytosolic extracts obtained from APAF1- and mock-transfected Raji cells (Raji-vector). Protein (30 μg) was loaded in each lane. Reprobing of membranes with anti-GAPDH antibodies was performed to confirm equal loading of protein. (B) Stable APAF1-(□) or mock-transfected (▪) Raji cells were treated with staurosporine or etoposide for 72 hours, and apoptosis was determined by flow cytometry. (C) Jurkat cells and APAF1- and mock-transfected Raji cells were harvested after 24 hours of treatment with etoposide (▪) or staurosporine (▨), and DEVD-AMC cleavage was determined. □ indicates control cells. (D) dATP and cytochrome c were added (▪) or not (□) to S-100 cytosols from APAF1- and mock-transfected Raji cells to initiate Apaf-1–dependent caspase-3 activation. S-100 extracts from Jurkat cells were used as a positive control. Results in panels B-D are shown as mean values ± SDs (n = 3).
Stable overexpression of Apaf-1 sensitizes BL cells to cytochrome c-dependent apoptosis. (A) Apaf-1 expression in total cell lysates versus cytosolic extracts obtained from APAF1- and mock-transfected Raji cells (Raji-vector). Protein (30 μg) was loaded in each lane. Reprobing of membranes with anti-GAPDH antibodies was performed to confirm equal loading of protein. (B) Stable APAF1-(□) or mock-transfected (▪) Raji cells were treated with staurosporine or etoposide for 72 hours, and apoptosis was determined by flow cytometry. (C) Jurkat cells and APAF1- and mock-transfected Raji cells were harvested after 24 hours of treatment with etoposide (▪) or staurosporine (▨), and DEVD-AMC cleavage was determined. □ indicates control cells. (D) dATP and cytochrome c were added (▪) or not (□) to S-100 cytosols from APAF1- and mock-transfected Raji cells to initiate Apaf-1–dependent caspase-3 activation. S-100 extracts from Jurkat cells were used as a positive control. Results in panels B-D are shown as mean values ± SDs (n = 3).
Plasma membrane sequestration of Apaf-1 in BL cells: mobilization by lipid raft–disrupting agents. (A) Raji cells were fixed and stained with anti–Apaf-1 antibodies followed by Alexa 488–conjugated secondary antibodies (green). Simultaneous staining of mitochondria (MitoTracker Red), Golgi (WGA-Texas Red), and nuclei (Hoechst 33342; blue) was performed, and merged confocal images are shown. The same pattern of Apaf-1 distribution was observed in 95 of 100 cells. (B) Merged image of 10 consecutive cross-sections of 1 representative Jurkat cell labeled with Apaf-1–specific antibodies. (C) Representative cross-sections of Apaf-1–labeled Raji cells are displayed in the large panels, whereas several optical sections of each cell were merged in the small panels to generate images in the x-z and y-z axes, respectively. Note the overlay between Apaf-1 (green) and the lipid raft marker, CTB (red). (D) Cytosplasmic redistribution of Apaf-1 upon disruption of the actin cytoskeleton or lipid rafts. Raji cells were incubated for 1 hour in medium alone or in the presence of MCD or CB. Cells were subsequently fixed and stained for Apaf-1 (green) and for lipid rafts, using Alexa 555–conjugated CTB (red). (E) Jurkat cells are labeled as in panel D. No apparent changes in the pattern of Apaf-1 distribution were observed upon incubation with MCD. Original magnification of all panels was × 40.
Plasma membrane sequestration of Apaf-1 in BL cells: mobilization by lipid raft–disrupting agents. (A) Raji cells were fixed and stained with anti–Apaf-1 antibodies followed by Alexa 488–conjugated secondary antibodies (green). Simultaneous staining of mitochondria (MitoTracker Red), Golgi (WGA-Texas Red), and nuclei (Hoechst 33342; blue) was performed, and merged confocal images are shown. The same pattern of Apaf-1 distribution was observed in 95 of 100 cells. (B) Merged image of 10 consecutive cross-sections of 1 representative Jurkat cell labeled with Apaf-1–specific antibodies. (C) Representative cross-sections of Apaf-1–labeled Raji cells are displayed in the large panels, whereas several optical sections of each cell were merged in the small panels to generate images in the x-z and y-z axes, respectively. Note the overlay between Apaf-1 (green) and the lipid raft marker, CTB (red). (D) Cytosplasmic redistribution of Apaf-1 upon disruption of the actin cytoskeleton or lipid rafts. Raji cells were incubated for 1 hour in medium alone or in the presence of MCD or CB. Cells were subsequently fixed and stained for Apaf-1 (green) and for lipid rafts, using Alexa 555–conjugated CTB (red). (E) Jurkat cells are labeled as in panel D. No apparent changes in the pattern of Apaf-1 distribution were observed upon incubation with MCD. Original magnification of all panels was × 40.
Reversal of chemoresistance in human BL cells upon liberation of Apaf-1
Finally, we were interested to determine whether mobilization of constitutive Apaf-1 could restore sensitivity of BL cells to chemotherapeutic agents. To test this, Raji cells were pretreated with a low dose of MCD, insufficient by itself to trigger cell death, and then subjected to etoposide treatment. As seen in Figure 7B, MCD sensitized the Raji BL cell line to etoposide-induced apoptosis, in a time-dependent manner. Similar results were obtained in the chemoresistant BL-derived cell line, DG75 (Figure 7C). The Mutu I cell line, however, in which partial postmitochondrial caspase activation is evident (Figure 3), was not sensitized further upon preincubation with MCD (data not shown). While we cannot exclude other, concurrent effects of MCD-mediated lipid raft disruption in BL cells, these results are consistent with the view that MCD treatment triggers Apaf-1 relocalization to the cytosolic compartment, thus allowing for etoposide-stimulated, apoptosome-driven apoptosis of otherwise resistant cancer cells (Figure 7D).
Discussion
BL is a highly aggressive form of B lymphoma that remains problematic in areas where it is endemic and that has increased dramatically worldwide as a result of its association with the acquired immunodeficiency syndrome.36 In the present study, we show that BL-derived cell lines are deficient in apoptosome-dependent activation of caspases downstream of mitochondria. However, pro–caspase-3 itself was not defective since recombinant granzyme B induced prominent caspase-3 activation in all BL cell lines tested. These findings suggested that the block conferring chemoresistance resides downstream of cytochrome c, yet upstream of caspase-3. Indeed, our data obtained in the Raji model show that the defective activation of caspases is linked to a lack of expression of the adaptor molecule, Apaf-1, in the cytosolic compartment. Transfection of Apaf-1 to resistant Raji cells yielded elevated levels of Apaf-1 in the cytosol and conferred sensitivity to apoptosis, thus providing direct evidence that Apaf-1 is an important determinant of cytochrome c-dependent apoptosis in these cells. Deletion or inactivation of Apaf-1 has been documented previously in melanoma and leukemia cell lines.4,5 The present study, however, is the first to identify defective Apaf-1 localization as a potential mechanism of chemoresistance in human cancer cells. Of note, some BL cell lines included in our study were found to express Apaf-1 in the cytosol, yet remained refractory to cytochrome c–dependent caspase activation, and it is thus conceivable that other postmitochondrial defects may contribute to chemoresistance in these cells. For comparison, Hodgkin lymphoma–derived B-cell lines were recently shown to harbor elevated levels of XIAP (X-linked inhibitor of apoptosis protein), resulting in an impairment of cytochrome c–dependent caspase activation.37 Studies are currently under way to investigate alternative mechanisms of chemoresistance in BL-derived cells.
Sensitization of BL cells to etoposide upon incubation with a lipid raft–disrupting agent. (A) S-100 cytosols from Raji cells pretreated with CB or MCD as detailed in the legend to Figure 6 were incubated for 30 minutes in the absence (▪) or presence (□) of cytochrome c and dATP, and caspase-3–like enzyme activity was determined by using the DEVD-AMC fluorogenic substrate. Data shown are mean values ± SDs (n = 4). (B) Raji BL cells were preincubated in the presence or absence of MCD for 1 hour prior to treatment with etoposide, and apoptosis was determined at the indicated time points by fluorescence microscopic evaluation of Hoechst 33342–labeled cells. Data are shown as mean values ± SDs (n = 3). (C) DG75 BL cells preincubated with MCD as in panel B, and then treated for 48 hours with etoposide prior to assessment of apoptosis. Data are shown as mean values ± SDs (n = 3). (D) Schematic model showing the apoptosis defect in BL-derived Raji cells, and the putative mechanism whereby lipid raft disruption sensitizes these cells to etoposide-induced killing. MCD indicates methyl-β-cyclodextrin.
Sensitization of BL cells to etoposide upon incubation with a lipid raft–disrupting agent. (A) S-100 cytosols from Raji cells pretreated with CB or MCD as detailed in the legend to Figure 6 were incubated for 30 minutes in the absence (▪) or presence (□) of cytochrome c and dATP, and caspase-3–like enzyme activity was determined by using the DEVD-AMC fluorogenic substrate. Data shown are mean values ± SDs (n = 4). (B) Raji BL cells were preincubated in the presence or absence of MCD for 1 hour prior to treatment with etoposide, and apoptosis was determined at the indicated time points by fluorescence microscopic evaluation of Hoechst 33342–labeled cells. Data are shown as mean values ± SDs (n = 3). (C) DG75 BL cells preincubated with MCD as in panel B, and then treated for 48 hours with etoposide prior to assessment of apoptosis. Data are shown as mean values ± SDs (n = 3). (D) Schematic model showing the apoptosis defect in BL-derived Raji cells, and the putative mechanism whereby lipid raft disruption sensitizes these cells to etoposide-induced killing. MCD indicates methyl-β-cyclodextrin.
Previous studies have shown that Bad, a proapoptotic Bcl-2 family member, is attached to lipid rafts in proliferating cells and redistributes to mitochondria during apoptosis.38 Similarly, proapoptotic Bim associates with distinct components of the cytoskeleton in viable cells and translocates to other organelles upon induction of apoptosis.39 Furthermore, translocation from mitochondria to nuclear membranes of CED-4, a nematode homolog of Apaf-1, precedes caspase activation and is thought to drive the cell death process in this model.40 Together, these studies, in conjunction with the current data, underscore the fundamental importance of correct compartmentalization of the cell death machinery and suggest that BL cells may subvert this control mechanism to resist apoptosis induction. Further studies are warranted to determine whether Apaf-1 sequestration is seen in cell lines derived from other types of human cancers and to determine whether plasma membrane localization occurs in normal cells or is restricted to cancer cells. Indeed, although Apaf-1 is commonly reported to be a cytosolic protein,32 subcellular localization of Apaf-1 has not been exhaustively investigated in normal cells to date. However, Zou et al24 have shown, in their pioneering experiments, that Apaf-1 participates in cytochrome c–dependent activation of cytosolic caspases in a cell-free system, thus lending further support to the notion that cytosolic expression of APAF-1 is a prerequisite for apoptosome-driven cell death. More recent studies have confirmed that direct targeting of the apoptosome in the cytosolic compartment is a viable approach to selectively kill cancer cells.11,41
Several isoforms of Apaf-1 have been described in mammalian cells, some of which exhibit a decreased ability to support caspase activation.29 Moreover, proteolytic degradation of Apaf-1 by caspases has been suggested as a means of disrupting Apaf-1–dependent apoptosis signaling.42 Human cancer cells thus appear to have adopted a number of different strategies, including caspase-mediated degradation, gene silencing, and ectopic subcellular localization of Apaf-1, to circumvent apoptosome-driven apoptosis. An important finding in the present study is that the resistant Raji BL cell line expressed wild-type Apaf-1 that retained its capacity to support caspase activation upon liberation from the plasma membrane. Hence, treatment of these cells with a lipid raft–disrupting agent was found to restore susceptibility to chemotherapy-induced apoptosis. It is theoretically possible that a fraction of Apaf-1 is located in cholesterol-rich vesicles in close proximity of the plasma membrane, rather than in the membrane itself. Notwithstanding such considerations, our results provide evidence for the sequestration of Apaf-1 in a noncytosolic compartment, at least in Raji cells, and suggest that the disruption of this ectopic localization renders cells susceptible to apoptosis. Novel treatment modalities that promote Apaf-1 mobilization might, therefore, serve to sensitize chemoresistant tumors lacking cytosolic expression of this key molecule. Future studies should assess whether mislocalization of Apaf-1 plays a role in BL in vivo and whether targeting of Apaf-1 can reverse chemoresistance also in the clinical setting.
Prepublished online as Blood First Edition Paper, February 3, 2005; DOI 10.1182/blood-2004-10-4075.
Supported by the Swedish Research Council, the Swedish Children's Cancer Foundation, the Jeansson Foundation, and Karolinska Institutet.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Ingemar Ernberg (Karolinska Institutet) and Xiaodong Wang (University of Texas Southwestern Medical Center, Dallas, TX) for the generous provision of BL cell lines and Apaf-1 cDNA, respectively. We also thank Gunilla Karlsson, Susanne Ahlberg, Mikhail Baryshev, and Yvonne Hofmann (Karolinska Institutet) for helpful comments and technical assistance.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal