Abstract
Hypoxia-inducible factor 1 (HIF-1) activates transcription of genes encoding angiogenic growth factors, which are secreted by hypoxic cells and stimulate endothelial cells, leading to angiogenesis. To determine whether HIF-1 also mediates cell-autonomous responses to hypoxia, we have compared gene expression profiles in arterial endothelial cells cultured under nonhypoxic versus hypoxic conditions and in nonhypoxic cells infected with adenovirus encoding beta-galactosidase versus a constitutively active form of HIF-1α (AdCA5). There were 245 gene probes that showed at least 1.5-fold increase in expression in response to hypoxia and in response to AdCA5; 325 gene probes showed at least 1.5-fold decrease in expression in response to hypoxia and in response to AdCA5. The largest category of genes down-regulated by both hypoxia and AdCA5 encoded proteins involved in cell growth/proliferation. Many genes up-regulated by both hypoxia and AdCA5 encoded cytokines/growth factors, receptors, and other signaling proteins. Transcription factors accounted for the largest group of HIF-1–regulated genes, indicating that HIF-1 controls a network of transcriptional responses to hypoxia in endothelial cells. Infection of endothelial cells with AdCA5 under nonhypoxic conditions was sufficient to induce increased basement membrane invasion and tube formation similar to the responses induced by hypoxia, indicating that HIF-1 mediates cell-autonomous activation of endothelial cells.
Introduction
The ability to sense and respond to changes in O2 concentration is a fundamental property of all nucleated cells. The regulation of gene transcription by hypoxia-inducible factor 1 (HIF-1) represents the most well-defined molecular mechanism for maintaining O2 homeostasis in metazoans. HIF-1 is a heterodimeric protein composed of HIF-1α and HIF-1β subunits that each contain basic helix-loop-helix and PAS domains that mediate heterodimerization and DNA binding.1,2 Whereas HIF-1β is constitutively expressed, HIF-1α expression increases exponentially as O2 concentration declines.3 In order to respond rapidly to hypoxia, cells devote considerable energy to the continuous synthesis and degradation of HIF-1α under nonhypoxic conditions. Under hypoxic conditions, the degradation of HIF-1α is inhibited, resulting in accumulation of the protein, dimerization with HIF-1β, binding to hypoxia response elements (HREs) within target genes, and activation of transcription via recruitment of the coactivators p300 and CBP (for review, see Poellinger and Johnson4 ).
The degradation of HIF-1α is controlled by binding of the von Hippel-Lindau protein (VHL), which is the recognition component of an E3 ubiquitin-protein ligase that targets HIF-1α for proteasomal degradation.5-8 VHL binding is dependent upon the hydroxylation of proline-402 and/or proline-564 of HIF-1α.9-11 The prolyl hydroxylases (PHDs 1-3) that are responsible for this modification use O2 as a substrate with a Michaelis-Menten constant (Km) that is slightly above atmospheric concentration, such that O2 is rate-limiting for enzymatic activity under physiologic conditions,12-15 providing a mechanism by which changes in O2 concentration can be directly transduced into changes in gene expression.
In mice, complete deficiency of HIF-1α results in embryonic lethality at midgestation that is associated with dramatic vascular regression due to extensive endothelial cell (EC) death.16-18 Conditional knock-out mice lacking HIF-1α expression in neural cells have marked cerebral atrophy associated with vascular regression.19 Vascularization of tumor xenografts derived from HIF-1α–null mouse embryonic stem cells is also severely impaired.18,20 In humans, HIF-1 has been implicated in protective or pathogenic responses in ischemic heart disease, cancer, stroke, and chronic lung disease,21,22 which are the major causes of mortality in the US population. Vascular involvement contributes to the pathogenesis of all of these diseases.23,24
HIF-1 plays a critical role in angiogenesis by activating transcription of genes encoding angiogenic growth factors including vascular endothelial growth factor (VEGF), angiopoietin 1 (ANGPT1) and ANGPT2, placental growth factor (PGF), and platelet-derived growth factor B (PDGFB).25,26 HIF-1 directly activates transcription of the VEGF gene by binding to an HRE located approximately 1 kb 5′ to the gene,27 whereas it is not known whether regulation of ANGPT1, ANGPT2, PGF, and PDGFB is direct or indirect. The regulation of these genes is remarkably cell-type specific. ANGPT2 expression is induced by hypoxia in arterial ECs, repressed in arterial smooth muscle cells, and unchanged in cardiac fibroblasts and myocytes, whereas VEGF is induced by hypoxia in all 4 cell types.25 These results underscore the importance of determining the subset of HIF-1 target genes that is regulated by hypoxia in a particular cell type. Remarkably, infection of all 4 cell types under nonhypoxic conditions with AdCA5, an adenovirus encoding a mutant form of HIF-1α that is resistant to O2-dependent degradation, induces the same pattern of angiogenic factor gene expression as that observed in hypoxic cells.25
In addition to genes encoding angiogenic growth factors, more than 70 other HIF-1 target genes have been identified using various criteria, such as the identification of an HRE, loss of expression in HIF-1α–null cells, or gain of expression in VHL-null cells (for review, see Semenza28 ). Each of these approaches has limitations: identification of HREs is labor intensive since they may be located anywhere within the gene or its flanking sequences and can be demonstrated functionally only in reporter assays; HIF-1α–null cells are thus-far limited to mouse embryonic stem cells and fibroblasts; and not all of the effects of VHL loss-of-function are HIF-1 dependent.29,30 To address these issues, we have developed an experimental strategy that is designed to identify HIF-1–dependent gene expression induced in primary cells exposed to hypoxia. We have applied this approach to the analysis of ECs because of their central role in angiogenesis and their potential as therapeutic targets in cancer and ischemic cardiovascular disease.23
Materials and methods
Cell culture
Human pulmonary artery ECs were obtained cryopreserved at third passage (Clonetics/Cambrex BioScience, Walkersville, MD). Cells were cultured on uncoated polystyrene dishes in endothelial basal medium supplemented with Clonetics EGM-2 Bullet Kit containing fetal bovine serum, nonessential amino acids, hydrocortisone, epidermal growth factor, VEGF, basic fibroblast growth factor, insulin-like growth factor 1, ascorbic acid, heparin, and gentamicin/amphotericin B. The cultures were maintained at 37°C in a humidified 5% CO2 incubator. Experiments were performed with 3 completely independent isolates of pulmonary artery ECs following a 1:6 split on to 10-cm dishes at passage 6.
Hypoxic exposure
Tissue culture plates were placed in a modular incubator chamber (Billups-Rothenberg, Del Mar, CA) and flushed at 2 psi for 3 minutes with a gas mixture of 1% O2, 5% CO2, and balance N2. The chamber was sealed and placed in a 37°C incubator for 24 hours.
Adenoviral exposure
AdCA5 is a replication-defective adenovirus that encodes green fluorescent protein (GFP) and a form of HIF-1α that is constitutively active as a result of a deletion (amino acids 392-520) and 2 substitutions (Pro567Thr and Pro658Gln), as previously described.25 Cells were exposed to AdCA5 or AdLacZ (obtained from the NHLBI PEGT Vector Core Facility, University of Pittsburgh) at 50 plaque-forming units (pfu) per cell based upon the total cell number from a replicate plate. Under these conditions, approximately 100% of cells were infected based upon GFP expression as determined by fluorescence microscopy (AdCA5) or X-gal staining (AdLacZ) at 24 hours after infection (data not shown).
RNA isolation
After 24-hour exposure to hypoxia or adenovirus, ECs were rinsed twice with ice-cold phosphate-buffered saline (PBS). Total RNA was extracted in 5 mL TRIzol (Invitrogen, Frederick, MD) and then Dnase I–treated and purified using RNeasy (Qiagen, Valencia, CA). The RNA was quantified by spectrophotometry, and its integrity was confirmed by agarose gel electrophoresis and ethidium bromide staining.
Microarray hybridization and data analysis
cDNA was synthesized from each RNA preparation using SuperScript System (Invitrogen) and used as template for the preparation of biotin-labeled cRNA using the BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics, Farmingdale, NY). Biotin-labeled cRNA was hybridized to the Human Genome U133A Array (Affymetrix, Santa Clara, CA), washed, stained with phycoerythrin-streptavidin, and laser scanned according to the manufacturer's instructions. The array contained 22 283 human gene probe sets, each of which (hereafter designated a “gene probe” or “probe”) consisted of 11 probe pairs corresponding to a single mRNA transcript. Data were saved as raw image files and converted into probe set data using Microarray Suite 5.0 (Affymetrix). Annotation by Unigene database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene) number, gene symbol, and gene description was performed using DAVID31 (http://david.niaid.nih.gov./david/beta/index.htm) and Affymetrix databases. Statistical analysis of the data was performed by t test using Excel (Microsoft, Seattle, WA). Mean data for fold change in gene expression and t test were log2-transformed and plotted.32 The primary data sets are available at http://www.hopkins-genomics.org/expression.html.
Quantitative real-time reverse-transcription–polymerase chain reaction (RT-PCR)
Primers were designed using Beacon Designer 2.1 software, and cDNA was prepared using the iScript cDNA Synthesis Kit (BioRad, Hercules, CA). Real-time PCR was performed using iQ SYBR Green Supermix and the iCycler Real-Time PCR Detection System (BioRad). For each primer pair (sequences available upon request), annealing temperature was optimized by gradient PCR. The fold change in expression of each target mRNA relative to 18S rRNA was calculated based on the threshold cycle (Ct) as 2–Δ(ΔCt), where ΔCt = Cttarget – Ct18S and Δ(ΔCt) =ΔCt1% – ΔCt20% or ΔCtAdCA5 – ΔCtAdLacZ.
Invasion and tube formation assays
Matrigel (10 mg/mL; BD Biosciences, San Jose, CA) was diluted to a final concentration of 500 μg/mL with basal medium. Millicell-PCF 12-μm filter inserts (Millipore, Billerica, MA) were placed in 24-well plates, and 100 μL of the diluted Matrigel was pipetted on top of each filter. The plates were dried overnight in a nonhumidified incubator at 37°C. ECs (5 × 104) in 200 μL basal medium were plated on each Matrigel-coated filter in a modified Boyden chamber with complete medium in the lower chamber and incubated for 24 hours. Cells on the lower surface of the filter were scraped with a rubber policeman into the medium from the lower chamber, pelleted, resuspended in 50 μL medium, and counted using a hemocytometer. Each of the 4 experimental conditions was performed in triplicate. To analyze tube formation, cells were plated on Matrigel and photographed with a Spot RT digital camera (Diagnostic Instruments, Sterling Heights, MI) mounted on an Olympus Bx60 microscope (Olympus America, Melville, NY).
Results
Microarray analysis of gene expression in human arterial endothelial cells exposed to hypoxia or AdCA5
Primary human pulmonary arterial ECs were subjected to 4 different culture conditions. To evaluate the effect of hypoxia on gene expression, one plate of cells was exposed to 20% O2 (standard nonhypoxic tissue culture conditions of 95% air/5% CO2) and a second plate was exposed to 1% O2 (with 5% CO2/94% N2). To evaluate the effect of HIF-1 on gene expression, 2 plates were exposed, under nonhypoxic conditions, to 50 pfu of adenovirus encoding the constitutively expressed form of HIF-1α (AdCA5) or Escherichia coli β-galactosidase (AdLacZ). After 24 hours, cells were harvested for isolation of total RNA, which was used to synthesize cDNA and labeled cRNA for hybridization to microarrays that contained 22 283 gene probes.
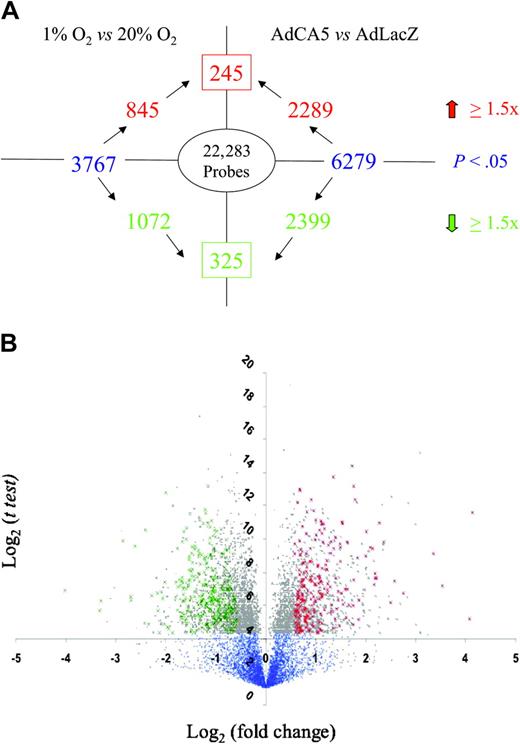
The experimental protocol was performed 3 times using independent primary cell cultures. For each gene probe, the data were subjected to statistical analysis by t test to identify those probes for which a significant difference (P < .05) in mean hybridization intensity was observed between conditions in a given pairwise comparison (ie, 1% vs 20% O2 or AdCA5 vs AdLacZ). Among those probes demonstrating a significant difference between conditions, a threshold of 1.5-fold increase or decrease was selected because this cutoff captured many, but not all, previously identified HIF-1 target genes. However, t tests alone are not statistically robust because of the problem of multiple comparisons. We therefore performed a more stringent analysis by identifying gene probes whose expression varied in a similar manner in response to both hypoxia and to AdCA5 (Figure 1). We identified 245 gene probes with increased expression (Table 1 and Supplementary Table 1) and 325 gene probes with decreased expression (Table 2 and Supplementary Table 2) in response to both hypoxia and AdCA5. Based upon these results, a minimum of 570 (2.6%) of 22 283 of all gene probes was regulated by hypoxia in a HIF-1–dependent manner in these primary cultures of arterial ECs.
Identification of genes regulated by hypoxia and HIF-1. (A) Experimental design and summary of results. The number inside the oval indicates the total number of gene probes on the microarray. Text in blue indicates the total number of gene probes showing a statistically significant difference between experimental conditions (1% O2 vs 20% O2; AdCA5 vs AdLacZ). The number in the box indicates the total number of gene probes with a statistically significant 1.5-fold or higher increase (red) or decrease (green) in expression in response to both 1% O2 and AdCA5. (B) Volcano plot of 1% O2 versus 20% O2 data. For each gene probe, the log2 (mean fold change in gene expression) is plotted on the x-axis and the log2 (t test) is plotted on the y-axis. Blue dots represent gene probes with P > .05 by t test. Gray dots represent gene probes with P < .05. Green crosses indicate the 325 gene probes with a statistically significant 1.5-fold or higher decrease in expression in response to both hypoxia and AdCA5. Red crosses indicate the 245 gene probes with a statistically significant 1.5-fold or higher increase in expression in response to both hypoxia and AdCA5.
Identification of genes regulated by hypoxia and HIF-1. (A) Experimental design and summary of results. The number inside the oval indicates the total number of gene probes on the microarray. Text in blue indicates the total number of gene probes showing a statistically significant difference between experimental conditions (1% O2 vs 20% O2; AdCA5 vs AdLacZ). The number in the box indicates the total number of gene probes with a statistically significant 1.5-fold or higher increase (red) or decrease (green) in expression in response to both 1% O2 and AdCA5. (B) Volcano plot of 1% O2 versus 20% O2 data. For each gene probe, the log2 (mean fold change in gene expression) is plotted on the x-axis and the log2 (t test) is plotted on the y-axis. Blue dots represent gene probes with P > .05 by t test. Gray dots represent gene probes with P < .05. Green crosses indicate the 325 gene probes with a statistically significant 1.5-fold or higher decrease in expression in response to both hypoxia and AdCA5. Red crosses indicate the 245 gene probes with a statistically significant 1.5-fold or higher increase in expression in response to both hypoxia and AdCA5.
Selected genes induced by both hypoxia and AdCA5
Unigene . | Gene name . | Symbol . | Hypoxia . | AdCA5 . |
|---|---|---|---|---|
| Oxidoreductases | ||||
| Hs.302085 | Prostaglandin I2 (prostacyclin) synthase | PTGIS | 11.5 | 16.4 |
| Hs.302085 | Prostaglandin I2 (prostacyclin) synthase | PTGIS | 3.1 | 5.0 |
| Hs.443906 | Prolyl hydroxylase domain-containing protein 3 | EGLN3 | 4.6 | 5.0 |
| Hs.318567 | N-myc downstream regulated gene 1 | NDRG1 | 2.2 | 3.7 |
| Hs.88474 | Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | PTGS1 | 2.3 | 2.8 |
| Hs.302085 | Prostaglandin I2 (prostacyclin) synthase | PTGIS | 2.5 | 2.3 |
| Hs.88474 | Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | PTGS1 | 1.6 | 2.2 |
| Hs.130946 | Prolyl hydroxylase domain-containing protein 2 | EGLN1 | 2.4 | 2.1 |
| Hs.352733 | Peptidylglycine alpha-amidating monooxygenase | PAM | 1.7 | 1.9 |
| Hs.10949 | ERO1-like | ERO1L | 1.7 | 1.8 |
| Hs.179526 | Thioredoxin interacting protein | TXNIP | 2.6 | 1.7 |
| Hs.352733 | Peptidylglycine alpha-amidating monooxygenase | PAM | 1.6 | 1.7 |
| Hs.352733 | Peptidylglycine alpha-amidating monooxygenase | PAM | 1.8 | 1.7 |
| Hs.83354 | Lysyl oxidase-like 2 | LOXL2 | 1.7 | 1.6 |
| Collagens/modifying enzymes | ||||
| Hs.102267 | Lysyl oxidase | LOX | 2.7 | 7.1 |
| Hs.102267 | Lysyl oxidase | LOX | 2.4 | 6.7 |
| Hs.104772 | Procollagen proline 4-hydroxylase, alpha 2 | P4HA2 | 2.0 | 4.6 |
| Hs.232115 | Collagen, type 1, alpha 2 | COL1A2 | 4.5 | 4.1 |
| Hs.41270 | Procollagen lysine hydroxylase 2 | PLOD2 | 2.2 | 3.5 |
| Hs.433695 | Collagen, type V, alpha 1 | COL5A1 | 2.3 | 3.0 |
| Hs.76768 | Procollagen proline 4-hydroxylase, alpha 1 | PRHA1 | 3.4 | 2.6 |
| Hs.433695 | Collagen, type V, alpha 1 | COL5A1 | 2.4 | 2.6 |
| Hs.41270 | Procollagen lysine hydroxylase 2 | PLOD2 | 1.8 | 2.6 |
| Hs.413175 | Collagen, type XVIII, alpha 1 | COL18A1 | 1.8 | 2.0 |
| Hs.149809 | Collagen, type IX, alpha 1 | COL9A1 | 1.7 | 1.8 |
| Hs.437173 | Collagen, type IV, alpha 1 | COL4A1 | 1.6 | 1.7 |
| Hs.407912 | Collagen, type IV, alpha 2 | COL4A2 | 1.6 | 1.7 |
| Hs.75093 | Procollagen lysine hydroxylase | PLOD | 2.0 | 1.5 |
| Hs.413175 | Collagen, type XVIII, alpha 1 | COL18A1 | 1.6 | 1.5 |
| Growth factors/cytokines | ||||
| Hs.9613 | Angiopoietin-like 4 | ANGPTL4 | 17.5 | 14.9 |
| Hs.25590 | Stanniocalcin 1 | STC1 | 3.1 | 12.2 |
| Hs.9613 | Inhibin, beta A (activin A, activin AB alpha) | INHBA | 10.2 | 11.6 |
| Hs.73793 | Vascular endothelial growth factor | VEGF | 6.7 | 8.8 |
| Hs.279497 | Inhibin, beta E | INHBE | 2.2 | 4.8 |
| Hs.155223 | Stanniocalcin 2 | STC2 | 2.3 | 4.3 |
| Hs.511899 | Endothelin 1 | EDN1 | 1.9 | 4.0 |
| Hs.2171 | Growth differentiation factor 10 | GDF10 | 1.5 | 3.3 |
| Hs.450230 | Insulin-like growth factor binding protein 3 | IGFBP3 | 2.5 | 3.2 |
| Hs.80420 | Chemokine (C-X3-C motif) ligand 1 | CX3CL1 | 2.1 | 2.9 |
| Hs.79141 | Vascular endothelial growth factor C | VEGFC | 2.8 | 2.6 |
| Hs.368996 | Relaxin 1 | RLN1 | 1.5 | 2.5 |
| Hs.1976 | Platelet-derived growth factor B polypeptide | PDGFB | 1.9 | 2.5 |
| Hs.73793 | Vascular endothelial growth factor | VEGF | 2.5 | 2.4 |
| Hs.73793 | Vascular endothelial growth factor | VEGF | 1.9 | 2.2 |
| Hs.252820 | Placental growth factor | PGF | 1.9 | 2.1 |
| Hs.441047 | Adrenomedullin | ADM | 4.2 | 2.1 |
| Hs.1976 | Platelet-derived growth factor B polypeptide | PDGFB | 2.3 | 1.7 |
| Receptors | ||||
| Hs.439141 | Gamma-aminobutyric acid (GABA) A receptor, pi | GABRP | 3.4 | 8.1 |
| Hs.231853 | Chemokine orphan receptor 1 | CMKOR1 | 1.7 | 6.4 |
| Hs.370422 | Very low density lipoprotein receptor | VLDLR | 2.0 | 5.9 |
| Hs.198288 | Protein tyrosine phosphatase, receptor type, R | PTPRR | 6.0 | 5.6 |
| Hs.166206 | Cubilin (intrinsic factor-cobalamin receptor) | CUBN | 2.0 | 4.5 |
| Hs.421986 | Chemokine (C-X-C motif) receptor 4 | CXCR4 | 3.1 | 2.9 |
| Hs.197029 | Adenosine A2a receptor | ADORA2A | 2.3 | 2.8 |
| Hs.438864 | Natriuretic peptide receptor A/guanylate cyclase A | NPR1 | 1.8 | 2.7 |
| Hs.438669 | Insulin receptor | INSR | 2.3 | 2.5 |
| Hs.434375 | Protein tyrosine phosphatase, receptor type, B | PTPRB | 2.1 | 2.5 |
| Hs.371974 | Activin A receptor, type IB | ACVR1B | 2.0 | 2.4 |
| Hs.198288 | Protein tyrosine phosphatase, receptor type, R | PTPRR | 5.1 | 2.3 |
| Hs.434375 | Protein tyrosine phosphatase, receptor type, B | PTPRB | 2.6 | 2.2 |
| Hs.17109 | Integral membrane protein 2A | ITM2A | 1.8 | 1.8 |
| Hs.127826 | Erythropoietin receptor | EPOR | 1.7 | 1.8 |
| Hs.334131 | Cadherin 2, type 1, N-cadherin (neuronal) | CDH2 | 1.6 | 1.8 |
| Hs.51233 | TNF receptor superfamily, member 10b | TNFRSF10B | 1.6 | 1.7 |
| Hs.279899 | TNF receptor superfamily, member 14 | TNFRSF14 | 1.5 | 1.7 |
| Hs.170129 | Opsin 3 (encephalopsin, panopsin) | OPN3 | 1.5 | 1.6 |
| Hs.23581 | Leptin receptor | LEPR | 2.6 | 1.6 |
| Hs.75216 | Protein tyrosine phosphatase, receptor type, F | PTPRF | 1.7 | 1.5 |
| Hs.512235 | Inositol 1,4,5-triphosphate receptor, type 2 | ITPR2 | 1.8 | 1.5 |
| Hs.83341 | AXL receptor tyrosine kinase | AXL | 1.5 | 1.5 |
| Hs.23581 | Leptin receptor | LEPR | 1.6 | 1.5 |
| Hs.211569 | G protein—coupled receptor kinase 5 | GRK5 | 1.5 | 1.5 |
| Other signal transduction | ||||
| Hs.106070 | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) | CDKN1C | 3.6 | 17.2 |
| Hs.512298 | Phospholipase C, gamma 2 | PLCG2 | 2.1 | 4.8 |
| Hs.200598 | AMP-activated protein kinase family member 5 | ARK5 | 1.5 | 4.5 |
| Hs.287460 | PDZ domain containing 3 | PDZK3 | 4.4 | 4.2 |
| Hs.106070 | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) | CDKN1C | 1.6 | 3.9 |
| Hs.213840 | Ectonucleotide pyrophosphatase/phosphodiesterase 1 | ENPP1 | 2.6 | 3.5 |
| Hs.458389 | SPRY domain-containing SOCS box protein SSB-1 | SSB1 | 1.6 | 3.0 |
| Hs.298654 | Dual specificity phosphatase 6 | DUSP6 | 2.3 | 2.9 |
| Hs.44038 | Pellino homolog 2 | PELI2 | 1.9 | 2.8 |
| Hs.301664 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 2 | RPS6KA2 | 1.7 | 2.7 |
| Hs.10862 | Adenylate kinase 3 | AK3 | 5.7 | 2.6 |
| Hs.15099 | Rho-related BTB domain containing 1 | RHOBTB1 | 1.6 | 2.6 |
| Hs.288196 | Calcium/calmodulin-dependent serine protein kinase | CASK | 1.5 | 2.6 |
| Hs.173380 | CK2 interacting protein 1 | CKIP-1 | 1.5 | 2.3 |
| Hs.82294 | Regulator of G-protein signaling 3 | RGS3 | 1.9 | 2.3 |
| Hs.112378 | LIM and senescent cell antigen-like domains 1 | LIMS1 | 1.6 | 2.2 |
| Hs.10862 | Adenylate kinase 3 | AK3 | 3.8 | 2.1 |
| Hs.80905 | Ras association (RalGDS/AF-6) domain family 2 | RASSF2 | 1.5 | 1.8 |
| Hs.299883 | FAD104 | FAD104 | 1.6 | 1.8 |
| Hs.446403 | WAS protein family, member 2 | WASF2 | 1.6 | 1.8 |
| Hs.110571 | Growth arrest and DNA-damage-inducible, beta | GADD45B | 2.3 | 1.7 |
| Hs.13291 | Cyclin G2 | CCNG2 | 1.5 | 1.7 |
| Hs.129836 | Cyclin-dependent kinase (CDC2-like) 11 | CDK11 | 1.5 | 1.7 |
| Hs.367689 | Triple functional domain (PTPRF interacting) | TRIO | 1.5 | 1.7 |
| Hs.301664 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 2 | RPS6KA2 | 1.7 | 1.6 |
| Hs.9651 | Related RAS viral (r-ras) oncogene homolog | RRAS | 2.1 | 1.6 |
| Hs.301242 | Myocytic induction/differentiation originator | MIDOR1 | 1.9 | 1.5 |
| Hs.497972 | FK506 binding protein 9, 63 kDa | FKBP9 | 1.5 | 1.5 |
| Transcription factors | ||||
| Hs.460889 | V-maf oncogene homolog F | MAFF | 2.1 | 5.0 |
| Hs.387667 | Peroxisome proliferative activated receptor, gamma | PPARG | 16.8 | 3.9 |
| Hs.232068 | Transcription factor 8 | TCF8 | 1.7 | 3.3 |
| Hs.321707 | Jumonji domain containing 1 | JMJD1 | 2.1 | 3.1 |
| Hs.511938 | Zinc finger and BTB domain containing 1 | ZBTB1 | 1.7 | 2.9 |
| Hs.420830 | Hypoxia-inducible factor 3, alpha subunit | HIF3A | 1.6 | 2.7 |
| Hs.357901 | SRY (sex-determining region Y)-box 4 | SOX4 | 1.7 | 2.3 |
| Hs.171825 | Basic helix-loop-helix domain containing, class B, 2 | BHLHB2 | 3.4 | 2.3 |
| Hs.324051 | RelA-associated inhibitor | RAI | 2.8 | 2.1 |
| Hs.321164 | Neuronal PAS domain protein 2 | NPAS2 | 1.6 | 2.1 |
| Hs.436100 | Notch homolog 4 | NOTCH4 | 2.6 | 2.1 |
| Hs.118630 | MAX interacting protein 1 | MXI1 | 3.0 | 2.0 |
| Hs.415033 | Myocyte enhancer factor 2A | MEF2A | 1.8 | 2.0 |
| Hs.406491 | Transducin-like enhancer of split 1 homolog | TLE1 | 1.5 | 1.9 |
| Hs.75063 | HIV-1 enhancer binding protein 2 | HIVEP2 | 1.8 | 1.9 |
| Hs.318517 | Transcription factor 7-like 1 | TCF7L1 | 1.8 | 1.9 |
| Hs.448341 | Zinc finger protein 292 | ZNF292 | 1.9 | 1.8 |
| Hs.55967 | Short stature homeobox 2 | SHOX2 | 1.7 | 1.8 |
| Hs.357901 | SRY box 4 | SOX4 | 1.5 | 1.8 |
| Hs.179526 | Thioredoxin interacting protein | TXNIP | 2.6 | 1.7 |
| Hs.250666 | Hairy and enhancer of split 1 | HES1 | 2.2 | 1.7 |
| Hs.30209 | Zinc fingers and homeoboxes 2 | ZHX2 | 2.3 | 1.6 |
| Hs.155024 | B-cell CLL/lymphoma 6 (zinc finger protein 51) | BCL6 | 1.7 | 1.6 |
| Hs.511950 | Sirtuin (sir2 homolog) 3 | SIRT3 | 1.7 | 1.6 |
| Hs.250666 | Hairy and enhancer of split 1 | HES1 | 2.0 | 1.6 |
| Hs.153863 | MAD, mothers against decapentaplegic homolog 6 | MADH6 | 1.9 | 1.6 |
| Hs.171825 | Basic helix-loop-helix domain containing, class B, 2 | BHLHB2 | 2.2 | 1.5 |
| Hs.77810 | Nuclear factor of activated T cells 4 | NFATC4 | 1.9 | 1.5 |
| Hs.59943 | cAMP responsive element binding protein 3-like 2 | CREB3L2 | 1.5 | 1.5 |
Unigene . | Gene name . | Symbol . | Hypoxia . | AdCA5 . |
|---|---|---|---|---|
| Oxidoreductases | ||||
| Hs.302085 | Prostaglandin I2 (prostacyclin) synthase | PTGIS | 11.5 | 16.4 |
| Hs.302085 | Prostaglandin I2 (prostacyclin) synthase | PTGIS | 3.1 | 5.0 |
| Hs.443906 | Prolyl hydroxylase domain-containing protein 3 | EGLN3 | 4.6 | 5.0 |
| Hs.318567 | N-myc downstream regulated gene 1 | NDRG1 | 2.2 | 3.7 |
| Hs.88474 | Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | PTGS1 | 2.3 | 2.8 |
| Hs.302085 | Prostaglandin I2 (prostacyclin) synthase | PTGIS | 2.5 | 2.3 |
| Hs.88474 | Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | PTGS1 | 1.6 | 2.2 |
| Hs.130946 | Prolyl hydroxylase domain-containing protein 2 | EGLN1 | 2.4 | 2.1 |
| Hs.352733 | Peptidylglycine alpha-amidating monooxygenase | PAM | 1.7 | 1.9 |
| Hs.10949 | ERO1-like | ERO1L | 1.7 | 1.8 |
| Hs.179526 | Thioredoxin interacting protein | TXNIP | 2.6 | 1.7 |
| Hs.352733 | Peptidylglycine alpha-amidating monooxygenase | PAM | 1.6 | 1.7 |
| Hs.352733 | Peptidylglycine alpha-amidating monooxygenase | PAM | 1.8 | 1.7 |
| Hs.83354 | Lysyl oxidase-like 2 | LOXL2 | 1.7 | 1.6 |
| Collagens/modifying enzymes | ||||
| Hs.102267 | Lysyl oxidase | LOX | 2.7 | 7.1 |
| Hs.102267 | Lysyl oxidase | LOX | 2.4 | 6.7 |
| Hs.104772 | Procollagen proline 4-hydroxylase, alpha 2 | P4HA2 | 2.0 | 4.6 |
| Hs.232115 | Collagen, type 1, alpha 2 | COL1A2 | 4.5 | 4.1 |
| Hs.41270 | Procollagen lysine hydroxylase 2 | PLOD2 | 2.2 | 3.5 |
| Hs.433695 | Collagen, type V, alpha 1 | COL5A1 | 2.3 | 3.0 |
| Hs.76768 | Procollagen proline 4-hydroxylase, alpha 1 | PRHA1 | 3.4 | 2.6 |
| Hs.433695 | Collagen, type V, alpha 1 | COL5A1 | 2.4 | 2.6 |
| Hs.41270 | Procollagen lysine hydroxylase 2 | PLOD2 | 1.8 | 2.6 |
| Hs.413175 | Collagen, type XVIII, alpha 1 | COL18A1 | 1.8 | 2.0 |
| Hs.149809 | Collagen, type IX, alpha 1 | COL9A1 | 1.7 | 1.8 |
| Hs.437173 | Collagen, type IV, alpha 1 | COL4A1 | 1.6 | 1.7 |
| Hs.407912 | Collagen, type IV, alpha 2 | COL4A2 | 1.6 | 1.7 |
| Hs.75093 | Procollagen lysine hydroxylase | PLOD | 2.0 | 1.5 |
| Hs.413175 | Collagen, type XVIII, alpha 1 | COL18A1 | 1.6 | 1.5 |
| Growth factors/cytokines | ||||
| Hs.9613 | Angiopoietin-like 4 | ANGPTL4 | 17.5 | 14.9 |
| Hs.25590 | Stanniocalcin 1 | STC1 | 3.1 | 12.2 |
| Hs.9613 | Inhibin, beta A (activin A, activin AB alpha) | INHBA | 10.2 | 11.6 |
| Hs.73793 | Vascular endothelial growth factor | VEGF | 6.7 | 8.8 |
| Hs.279497 | Inhibin, beta E | INHBE | 2.2 | 4.8 |
| Hs.155223 | Stanniocalcin 2 | STC2 | 2.3 | 4.3 |
| Hs.511899 | Endothelin 1 | EDN1 | 1.9 | 4.0 |
| Hs.2171 | Growth differentiation factor 10 | GDF10 | 1.5 | 3.3 |
| Hs.450230 | Insulin-like growth factor binding protein 3 | IGFBP3 | 2.5 | 3.2 |
| Hs.80420 | Chemokine (C-X3-C motif) ligand 1 | CX3CL1 | 2.1 | 2.9 |
| Hs.79141 | Vascular endothelial growth factor C | VEGFC | 2.8 | 2.6 |
| Hs.368996 | Relaxin 1 | RLN1 | 1.5 | 2.5 |
| Hs.1976 | Platelet-derived growth factor B polypeptide | PDGFB | 1.9 | 2.5 |
| Hs.73793 | Vascular endothelial growth factor | VEGF | 2.5 | 2.4 |
| Hs.73793 | Vascular endothelial growth factor | VEGF | 1.9 | 2.2 |
| Hs.252820 | Placental growth factor | PGF | 1.9 | 2.1 |
| Hs.441047 | Adrenomedullin | ADM | 4.2 | 2.1 |
| Hs.1976 | Platelet-derived growth factor B polypeptide | PDGFB | 2.3 | 1.7 |
| Receptors | ||||
| Hs.439141 | Gamma-aminobutyric acid (GABA) A receptor, pi | GABRP | 3.4 | 8.1 |
| Hs.231853 | Chemokine orphan receptor 1 | CMKOR1 | 1.7 | 6.4 |
| Hs.370422 | Very low density lipoprotein receptor | VLDLR | 2.0 | 5.9 |
| Hs.198288 | Protein tyrosine phosphatase, receptor type, R | PTPRR | 6.0 | 5.6 |
| Hs.166206 | Cubilin (intrinsic factor-cobalamin receptor) | CUBN | 2.0 | 4.5 |
| Hs.421986 | Chemokine (C-X-C motif) receptor 4 | CXCR4 | 3.1 | 2.9 |
| Hs.197029 | Adenosine A2a receptor | ADORA2A | 2.3 | 2.8 |
| Hs.438864 | Natriuretic peptide receptor A/guanylate cyclase A | NPR1 | 1.8 | 2.7 |
| Hs.438669 | Insulin receptor | INSR | 2.3 | 2.5 |
| Hs.434375 | Protein tyrosine phosphatase, receptor type, B | PTPRB | 2.1 | 2.5 |
| Hs.371974 | Activin A receptor, type IB | ACVR1B | 2.0 | 2.4 |
| Hs.198288 | Protein tyrosine phosphatase, receptor type, R | PTPRR | 5.1 | 2.3 |
| Hs.434375 | Protein tyrosine phosphatase, receptor type, B | PTPRB | 2.6 | 2.2 |
| Hs.17109 | Integral membrane protein 2A | ITM2A | 1.8 | 1.8 |
| Hs.127826 | Erythropoietin receptor | EPOR | 1.7 | 1.8 |
| Hs.334131 | Cadherin 2, type 1, N-cadherin (neuronal) | CDH2 | 1.6 | 1.8 |
| Hs.51233 | TNF receptor superfamily, member 10b | TNFRSF10B | 1.6 | 1.7 |
| Hs.279899 | TNF receptor superfamily, member 14 | TNFRSF14 | 1.5 | 1.7 |
| Hs.170129 | Opsin 3 (encephalopsin, panopsin) | OPN3 | 1.5 | 1.6 |
| Hs.23581 | Leptin receptor | LEPR | 2.6 | 1.6 |
| Hs.75216 | Protein tyrosine phosphatase, receptor type, F | PTPRF | 1.7 | 1.5 |
| Hs.512235 | Inositol 1,4,5-triphosphate receptor, type 2 | ITPR2 | 1.8 | 1.5 |
| Hs.83341 | AXL receptor tyrosine kinase | AXL | 1.5 | 1.5 |
| Hs.23581 | Leptin receptor | LEPR | 1.6 | 1.5 |
| Hs.211569 | G protein—coupled receptor kinase 5 | GRK5 | 1.5 | 1.5 |
| Other signal transduction | ||||
| Hs.106070 | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) | CDKN1C | 3.6 | 17.2 |
| Hs.512298 | Phospholipase C, gamma 2 | PLCG2 | 2.1 | 4.8 |
| Hs.200598 | AMP-activated protein kinase family member 5 | ARK5 | 1.5 | 4.5 |
| Hs.287460 | PDZ domain containing 3 | PDZK3 | 4.4 | 4.2 |
| Hs.106070 | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) | CDKN1C | 1.6 | 3.9 |
| Hs.213840 | Ectonucleotide pyrophosphatase/phosphodiesterase 1 | ENPP1 | 2.6 | 3.5 |
| Hs.458389 | SPRY domain-containing SOCS box protein SSB-1 | SSB1 | 1.6 | 3.0 |
| Hs.298654 | Dual specificity phosphatase 6 | DUSP6 | 2.3 | 2.9 |
| Hs.44038 | Pellino homolog 2 | PELI2 | 1.9 | 2.8 |
| Hs.301664 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 2 | RPS6KA2 | 1.7 | 2.7 |
| Hs.10862 | Adenylate kinase 3 | AK3 | 5.7 | 2.6 |
| Hs.15099 | Rho-related BTB domain containing 1 | RHOBTB1 | 1.6 | 2.6 |
| Hs.288196 | Calcium/calmodulin-dependent serine protein kinase | CASK | 1.5 | 2.6 |
| Hs.173380 | CK2 interacting protein 1 | CKIP-1 | 1.5 | 2.3 |
| Hs.82294 | Regulator of G-protein signaling 3 | RGS3 | 1.9 | 2.3 |
| Hs.112378 | LIM and senescent cell antigen-like domains 1 | LIMS1 | 1.6 | 2.2 |
| Hs.10862 | Adenylate kinase 3 | AK3 | 3.8 | 2.1 |
| Hs.80905 | Ras association (RalGDS/AF-6) domain family 2 | RASSF2 | 1.5 | 1.8 |
| Hs.299883 | FAD104 | FAD104 | 1.6 | 1.8 |
| Hs.446403 | WAS protein family, member 2 | WASF2 | 1.6 | 1.8 |
| Hs.110571 | Growth arrest and DNA-damage-inducible, beta | GADD45B | 2.3 | 1.7 |
| Hs.13291 | Cyclin G2 | CCNG2 | 1.5 | 1.7 |
| Hs.129836 | Cyclin-dependent kinase (CDC2-like) 11 | CDK11 | 1.5 | 1.7 |
| Hs.367689 | Triple functional domain (PTPRF interacting) | TRIO | 1.5 | 1.7 |
| Hs.301664 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 2 | RPS6KA2 | 1.7 | 1.6 |
| Hs.9651 | Related RAS viral (r-ras) oncogene homolog | RRAS | 2.1 | 1.6 |
| Hs.301242 | Myocytic induction/differentiation originator | MIDOR1 | 1.9 | 1.5 |
| Hs.497972 | FK506 binding protein 9, 63 kDa | FKBP9 | 1.5 | 1.5 |
| Transcription factors | ||||
| Hs.460889 | V-maf oncogene homolog F | MAFF | 2.1 | 5.0 |
| Hs.387667 | Peroxisome proliferative activated receptor, gamma | PPARG | 16.8 | 3.9 |
| Hs.232068 | Transcription factor 8 | TCF8 | 1.7 | 3.3 |
| Hs.321707 | Jumonji domain containing 1 | JMJD1 | 2.1 | 3.1 |
| Hs.511938 | Zinc finger and BTB domain containing 1 | ZBTB1 | 1.7 | 2.9 |
| Hs.420830 | Hypoxia-inducible factor 3, alpha subunit | HIF3A | 1.6 | 2.7 |
| Hs.357901 | SRY (sex-determining region Y)-box 4 | SOX4 | 1.7 | 2.3 |
| Hs.171825 | Basic helix-loop-helix domain containing, class B, 2 | BHLHB2 | 3.4 | 2.3 |
| Hs.324051 | RelA-associated inhibitor | RAI | 2.8 | 2.1 |
| Hs.321164 | Neuronal PAS domain protein 2 | NPAS2 | 1.6 | 2.1 |
| Hs.436100 | Notch homolog 4 | NOTCH4 | 2.6 | 2.1 |
| Hs.118630 | MAX interacting protein 1 | MXI1 | 3.0 | 2.0 |
| Hs.415033 | Myocyte enhancer factor 2A | MEF2A | 1.8 | 2.0 |
| Hs.406491 | Transducin-like enhancer of split 1 homolog | TLE1 | 1.5 | 1.9 |
| Hs.75063 | HIV-1 enhancer binding protein 2 | HIVEP2 | 1.8 | 1.9 |
| Hs.318517 | Transcription factor 7-like 1 | TCF7L1 | 1.8 | 1.9 |
| Hs.448341 | Zinc finger protein 292 | ZNF292 | 1.9 | 1.8 |
| Hs.55967 | Short stature homeobox 2 | SHOX2 | 1.7 | 1.8 |
| Hs.357901 | SRY box 4 | SOX4 | 1.5 | 1.8 |
| Hs.179526 | Thioredoxin interacting protein | TXNIP | 2.6 | 1.7 |
| Hs.250666 | Hairy and enhancer of split 1 | HES1 | 2.2 | 1.7 |
| Hs.30209 | Zinc fingers and homeoboxes 2 | ZHX2 | 2.3 | 1.6 |
| Hs.155024 | B-cell CLL/lymphoma 6 (zinc finger protein 51) | BCL6 | 1.7 | 1.6 |
| Hs.511950 | Sirtuin (sir2 homolog) 3 | SIRT3 | 1.7 | 1.6 |
| Hs.250666 | Hairy and enhancer of split 1 | HES1 | 2.0 | 1.6 |
| Hs.153863 | MAD, mothers against decapentaplegic homolog 6 | MADH6 | 1.9 | 1.6 |
| Hs.171825 | Basic helix-loop-helix domain containing, class B, 2 | BHLHB2 | 2.2 | 1.5 |
| Hs.77810 | Nuclear factor of activated T cells 4 | NFATC4 | 1.9 | 1.5 |
| Hs.59943 | cAMP responsive element binding protein 3-like 2 | CREB3L2 | 1.5 | 1.5 |
Selected genes repressed by both hypoxia and AdCA5
Unigene . | Gene name . | Symbol . | Hypoxia . | AdCA5 . |
|---|---|---|---|---|
| CDC/cyclins | ||||
| Hs.408658 | Cyclin E2 | CCNE2 | - 1.7 | - 4.1 |
| Hs.244723 | Cyclin E1 | CCNE1 | - 1.9 | - 3.9 |
| Hs.1973 | Cyclin F | CCNF | - 2.1 | - 3.0 |
| Hs.408658 | Cyclin E2 | CCNE2 | - 2.4 | - 2.8 |
| Hs.334562 | Cell division cycle 2 | CDC2 | - 1.5 | - 2.2 |
| Hs.405958 | Cell division cycle 6 | CDC6 | - 2.7 | - 2.1 |
| Hs.334562 | Cell division cycle 2 | CDC2 | - 1.8 | - 2.1 |
| Hs.28853 | Cell division cycle 7 | CDC7 | - 1.6 | - 2.1 |
| Hs.334562 | Cell division cycle 2 | CDC2 | - 1.8 | - 1.9 |
| MCM proteins | ||||
| Hs.460184 | Minichromosome maintenance deficient 4 | MCM4 | - 2.4 | - 3.1 |
| Hs.460184 | Minichromosome maintenance deficient 4 | MCM4 | - 2.2 | - 3.1 |
| Hs.57101 | Minichromosome maintenance deficient 2 | MCM2 | - 2.2 | - 3.1 |
| Hs.77171 | Minichromosome maintenance deficient 5 | MCM5 | - 2.4 | - 2.7 |
| Hs.444118 | Minichromosome maintenance deficient 6 | MCM6 | - 1.9 | - 2.4 |
| Hs.198363 | Minichromosome maintenance deficient 10 | MCM10 | - 2.2 | - 2.3 |
| Hs.179565 | Minichromosome maintenance deficient 3 | MCM3 | - 1.5 | - 2.2 |
| Hs.460184 | Minichromosome maintenance deficient 4 | MCM4 | - 1.8 | - 2.1 |
| Hs.460184 | Minichromosome maintenance deficient 4 | MCM4 | - 2.0 | - 1.8 |
| DNA/RNA polymerases | ||||
| Hs.437186 | Polymerase (RNA) III polypeptide K | POLR3K | - 2.1 | - 4.3 |
| Hs.441072 | Polymerase (RNA) II polypeptide L | RNAPOL2 | - 2.0 | - 3.3 |
| Hs.99185 | Polymerase (DNA directed), epsilon 2 | POLE2 | - 2.0 | - 2.6 |
| Hs.282387 | Polymerase (RNA) III (32kD) | RPC32 | - 1.9 | - 1.9 |
| Hs.306791 | Polymerase (DNA directed), delta 2 | POLD2 | - 1.5 | - 1.7 |
| Hs.290921 | Polymerase (DNA directed), gamma | POLG | - 1.7 | - 1.6 |
| Replication factors | ||||
| Hs.79018 | Chromatin assembly factor 1, subunit A | CHAF1A | - 6.0 | - 10.0 |
| Hs.433180 | DNA replication complex GINS protein PSF2 | PFS2 | - 2.2 | - 3.4 |
| Hs.194665 | DNA replication helicase 2-like | DNA2L | - 1.7 | - 3.3 |
| Hs.443227 | Replication factor C5 | RFC5 | - 1.9 | - 3.2 |
| Hs.115474 | Replication factor C (activator 1) 3 | RFC3 | - 1.9 | - 2.9 |
| Hs.443227 | Replication factor C5 | RFC5 | - 1.6 | - 2.3 |
| Hs.49760 | Origin recognition complex, subunit 6-like | ORC6L | - 1.9 | - 1.9 |
| Hs.139226 | Replication factor C (activator 1) 2 | RFC2 | - 1.6 | - 1.6 |
| Other cell cycle | ||||
| Hs.23348 | S-phase kinase-associated protein 2 | SKP2 | - 2.4 | - 3.1 |
| Hs.79078 | MAD2 mitotic arrest deficient-like 1 | MAD2L1 | - 1.7 | - 2.9 |
| Hs.511945 | Block of proliferation 1 | BOP1 | - 1.7 | - 2.8 |
| Hs.42650 | ZW10 interactor | ZWINT | - 2.0 | - 2.5 |
| Hs.445084 | M-phase phosphoprotein 9 | MPHOSPH9 | - 1.7 | - 2.5 |
| Hs.75337 | Nucleolar and coiled-body phosphoprotein 1 | NOLC1 | - 1.6 | - 2.4 |
| Hs.374491 | Proliferation-associated 2G4 | PA2G4 | - 1.6 | - 2.1 |
| Hs.152759 | Activator of S-phase kinase | ASK | - 2.7 | - 2.0 |
| Hs.344037 | Protein regulator of cytokinesis 1 | PRC1 | - 2.0 | - 1.6 |
| Hs.433008 | NIMA (never in mitosis a)-related kinase 4 | NEK4 | - 1.5 | - 1.5 |
| Ribonucleotide metabolism | ||||
| Hs.226390 | Ribonucleotide reductase M2 polypeptide | RRM2 | - 2.4 | - 7.2 |
| Hs.464813 | Dihydrofolate reductase | DHFR | - 2.0 | - 4.5 |
| Hs.251871 | CTP synthase | CTPS | - 1.8 | - 3.2 |
| Hs.226390 | Ribonucleotide reductase M2 polypeptide | RRM2 | - 1.8 | - 2.4 |
| Hs.56 | Phosphoribosyl pyrophosphate synthetase 1 | PRPS1 | - 1.6 | - 2.3 |
| Hs.2057 | Uridine monophosphate synthetase | UMPS | - 1.6 | - 2.3 |
| Hs.464813 | Dihydrofolate reductase | DHFR | - 1.5 | - 2.3 |
| Hs.90280 | 5-aminoimidazole-4-carboxamide ribonucleotide transformylase | ATIC | - 1.5 | - 2.2 |
| Hs.177766 | Poly(ADP-ribose) polymerase | ADPRT | - 1.7 | - 2.0 |
| Hs.215766 | GTP-binding protein | GTPBP4 | - 1.5 | - 2.0 |
| Hs.409412 | ADP-ribosyltransferase (NAD +; poly(ADP-ribose) polymerase)-like 2 | ADPRTL2 | - 1.5 | - 1.8 |
| Hs.2057 | Uridine monophosphate synthetase | UMPS | - 1.5 | - 1.7 |
| Hs.383396 | Ribonucleotide reductase M1 polypeptide | RRM1 | - 1.6 | - 1.5 |
| Ribosomal/mitochondrial biogenesis | ||||
| Hs.411125 | Mitochondrial ribosomal protein S12 | MRPS12 | - 1.7 | - 9.9 |
| Hs.435643 | Ts translation elongation factor, mitochondrial | TSFM | - 3.7 | - 6.5 |
| Hs.511949 | Ribosomal RNA processing 4 | RRP4 | - 2.0 | - 4.4 |
| Hs.71827 | Ribosome biogenesis regulator homolog | RRS1 | - 1.9 | - 3.1 |
| Hs.132748 | Ribosomal protein L39 | RPL39 | -1.8 | -2.5 |
| Hs.256583 | Interleukin enhancer binding protein 3 | ILF3 | -2.0 | -2.3 |
| Hs.511949 | Ribosomal RNA processing 4 | RRP4 | -1.7 | -2.3 |
| Hs.101414 | Zinc finger protein 500 | ZNF500 | -1.6 | -2.3 |
| Hs.431307 | Mitochondrial ribosomal protein L40 | MRPL40 | -1.6 | -2.1 |
| Hs.376064 | Nucleolar protein (KKED repeat) | NOL5A | -1.9 | -2.0 |
| Hs.376681 | Mitofusin 2 | MFN2 | -1.7 | -1.7 |
| Hs.424264 | Mitochondrial ribosomal protein L46 | MRPL46 | -1.5 | -1.5 |
| RNA binding/metabolism | ||||
| Hs.443960 | DEAD/H box polypeptide 11 | DDX11 | -4.1 | -3.9 |
| Hs.434901 | Small nuclear ribonucleoprotein polypeptide A′ | SNRPA1 | -1.6 | -3.0 |
| Hs.166463 | Heterogeneous nuclear ribonucleoprotein U | HNRPU | -1.7 | -2.9 |
| Hs.153768 | RNA, U3 small nucleolar interacting protein 2 | RNU3IP2 | -1.7 | -2.4 |
| Hs.363492 | DEAD box polypeptide 18 | DDX18 | -1.5 | -2.4 |
| Hs.434901 | Small nuclear ribonucleoprotein polypeptide A′ | SNRPA1 | -1.9 | -2.1 |
| Hs.416994 | POP7 (processing of precursor) homolog | RPP20 | -1.8 | -2.1 |
| Hs.30174 | Small nuclear RNA activating complex 5 | SNAPC5 | -1.7 | -2.1 |
| Hs.372673 | Heterogeneous nuclear ribonucleoprotein D-like | HNRPDL | -1.6 | -2.1 |
| Hs.130098 | DEAD box polypeptide 23 | DDX23 | -1.5 | -2.1 |
| Hs.7174 | DEAH (Asp-Glu-Ala-His) box polypeptide 35 | DHX35 | -2.1 | -2.0 |
| Hs.73965 | Splicing factor, arginine/serine-rich 2 | SFRS2 | -1.6 | -2.0 |
| Hs.438726 | Heterogeneous nuclear ribonucleoprotein D | HNRPD | -2.2 | -1.9 |
| Hs.434901 | Small nuclear ribonucleoprotein polypeptide A′ | SNRPA1 | -1.9 | -1.9 |
| Hs.438726 | DEAD box polypeptide 21 | DDX21 | -1.7 | -1.9 |
| Hs.372673 | Heterogeneous nuclear ribonucleoprotein D-like | HNRPDL | -1.6 | -1.7 |
| Hs.86948 | Small nuclear ribonucleoprotein D1 polypeptide | SNRPD1 | -1.6 | -1.7 |
| Hs.356549 | Small nuclear ribonucleoprotein D3 polypeptide | SNRPD3 | -1.5 | -1.7 |
| Hs.511756 | Ribonuclease P1 | RNASEP1 | -1.6 | -1.6 |
| Hs.139120 | Ribonuclease P (30 kDa) | RPP30 | -1.6 | -1.6 |
| Ubiquitin/proteasome | ||||
| Hs.152978 | Proteasome activator subunit 3 | PSME3 | -1.6 | -2.9 |
| Hs.396393 | Ubiquitin-conjugating enzyme E2S | UBE2S | -1.6 | -2.9 |
| Hs.25223 | Ubiquitin carboxyl-terminal hydrolase L5 | UCHL5 | -1.6 | -2.1 |
| Hs.443379 | Proteasome 26S subunit, non-ATPase, 11 | PSMD11 | -1.5 | -2.1 |
| Hs.152978 | Proteasome activator subunit 3 | PSME3 | -1.6 | -1.8 |
| Hs.119563 | Proteasome activator subunit 4 | PSME4 | -1.6 | -1.8 |
| Hs.12820 | Ubiquitin specific protease 39 | USP39 | -1.6 | -1.6 |
| Hs.35086 | Ubiquitin specific protease 1 | USP1 | -1.5 | -1.5 |
| Transcription factors | ||||
| Hs.446451 | Myeloid/lymphoid leukemia; translocated to, 10 | MLLT10 | -2.2 | -5.1 |
| Hs.54089 | BRCA1 associated RING domain 1 | BARD1 | -1.6 | -3.6 |
| Hs.194143 | Breast cancer 1, early onset | BRCA1 | -2.7 | -3.2 |
| Hs.181128 | ELK1, member of ETS oncogene family | ELK1 | -2.4 | -3.0 |
| Hs.75133 | Transcription factor A, mitochondrial | TFAM | -1.6 | -3.0 |
| Hs.448398 | MYC-associated zinc finger protein | MAZ | -1.6 | -2.8 |
| Hs.194143 | Breast cancer 1, early onset | BRCA1 | -2.0 | -2.4 |
| Hs.106415 | Peroxisome proliferative activated receptor, delta | PPARD | -2.1 | -2.3 |
| Hs.408528 | Retinoblastoma 1 | RB1 | -1.8 | -2.3 |
| Hs.436187 | Thyroid hormone receptor interactor 13 | TRIP13 | -1.9 | -2.1 |
| Hs.75133 | Transcription factor 6-like 1 | TF6M | -1.6 | -1.7 |
| Hs.75133 | Transcription factor A, mitochondrial | TFAM | -1.6 | -1.7 |
| Hs.410900 | Inhibitor of DNA binding 1 | ID1 | -1.6 | -1.5 |
Unigene . | Gene name . | Symbol . | Hypoxia . | AdCA5 . |
|---|---|---|---|---|
| CDC/cyclins | ||||
| Hs.408658 | Cyclin E2 | CCNE2 | - 1.7 | - 4.1 |
| Hs.244723 | Cyclin E1 | CCNE1 | - 1.9 | - 3.9 |
| Hs.1973 | Cyclin F | CCNF | - 2.1 | - 3.0 |
| Hs.408658 | Cyclin E2 | CCNE2 | - 2.4 | - 2.8 |
| Hs.334562 | Cell division cycle 2 | CDC2 | - 1.5 | - 2.2 |
| Hs.405958 | Cell division cycle 6 | CDC6 | - 2.7 | - 2.1 |
| Hs.334562 | Cell division cycle 2 | CDC2 | - 1.8 | - 2.1 |
| Hs.28853 | Cell division cycle 7 | CDC7 | - 1.6 | - 2.1 |
| Hs.334562 | Cell division cycle 2 | CDC2 | - 1.8 | - 1.9 |
| MCM proteins | ||||
| Hs.460184 | Minichromosome maintenance deficient 4 | MCM4 | - 2.4 | - 3.1 |
| Hs.460184 | Minichromosome maintenance deficient 4 | MCM4 | - 2.2 | - 3.1 |
| Hs.57101 | Minichromosome maintenance deficient 2 | MCM2 | - 2.2 | - 3.1 |
| Hs.77171 | Minichromosome maintenance deficient 5 | MCM5 | - 2.4 | - 2.7 |
| Hs.444118 | Minichromosome maintenance deficient 6 | MCM6 | - 1.9 | - 2.4 |
| Hs.198363 | Minichromosome maintenance deficient 10 | MCM10 | - 2.2 | - 2.3 |
| Hs.179565 | Minichromosome maintenance deficient 3 | MCM3 | - 1.5 | - 2.2 |
| Hs.460184 | Minichromosome maintenance deficient 4 | MCM4 | - 1.8 | - 2.1 |
| Hs.460184 | Minichromosome maintenance deficient 4 | MCM4 | - 2.0 | - 1.8 |
| DNA/RNA polymerases | ||||
| Hs.437186 | Polymerase (RNA) III polypeptide K | POLR3K | - 2.1 | - 4.3 |
| Hs.441072 | Polymerase (RNA) II polypeptide L | RNAPOL2 | - 2.0 | - 3.3 |
| Hs.99185 | Polymerase (DNA directed), epsilon 2 | POLE2 | - 2.0 | - 2.6 |
| Hs.282387 | Polymerase (RNA) III (32kD) | RPC32 | - 1.9 | - 1.9 |
| Hs.306791 | Polymerase (DNA directed), delta 2 | POLD2 | - 1.5 | - 1.7 |
| Hs.290921 | Polymerase (DNA directed), gamma | POLG | - 1.7 | - 1.6 |
| Replication factors | ||||
| Hs.79018 | Chromatin assembly factor 1, subunit A | CHAF1A | - 6.0 | - 10.0 |
| Hs.433180 | DNA replication complex GINS protein PSF2 | PFS2 | - 2.2 | - 3.4 |
| Hs.194665 | DNA replication helicase 2-like | DNA2L | - 1.7 | - 3.3 |
| Hs.443227 | Replication factor C5 | RFC5 | - 1.9 | - 3.2 |
| Hs.115474 | Replication factor C (activator 1) 3 | RFC3 | - 1.9 | - 2.9 |
| Hs.443227 | Replication factor C5 | RFC5 | - 1.6 | - 2.3 |
| Hs.49760 | Origin recognition complex, subunit 6-like | ORC6L | - 1.9 | - 1.9 |
| Hs.139226 | Replication factor C (activator 1) 2 | RFC2 | - 1.6 | - 1.6 |
| Other cell cycle | ||||
| Hs.23348 | S-phase kinase-associated protein 2 | SKP2 | - 2.4 | - 3.1 |
| Hs.79078 | MAD2 mitotic arrest deficient-like 1 | MAD2L1 | - 1.7 | - 2.9 |
| Hs.511945 | Block of proliferation 1 | BOP1 | - 1.7 | - 2.8 |
| Hs.42650 | ZW10 interactor | ZWINT | - 2.0 | - 2.5 |
| Hs.445084 | M-phase phosphoprotein 9 | MPHOSPH9 | - 1.7 | - 2.5 |
| Hs.75337 | Nucleolar and coiled-body phosphoprotein 1 | NOLC1 | - 1.6 | - 2.4 |
| Hs.374491 | Proliferation-associated 2G4 | PA2G4 | - 1.6 | - 2.1 |
| Hs.152759 | Activator of S-phase kinase | ASK | - 2.7 | - 2.0 |
| Hs.344037 | Protein regulator of cytokinesis 1 | PRC1 | - 2.0 | - 1.6 |
| Hs.433008 | NIMA (never in mitosis a)-related kinase 4 | NEK4 | - 1.5 | - 1.5 |
| Ribonucleotide metabolism | ||||
| Hs.226390 | Ribonucleotide reductase M2 polypeptide | RRM2 | - 2.4 | - 7.2 |
| Hs.464813 | Dihydrofolate reductase | DHFR | - 2.0 | - 4.5 |
| Hs.251871 | CTP synthase | CTPS | - 1.8 | - 3.2 |
| Hs.226390 | Ribonucleotide reductase M2 polypeptide | RRM2 | - 1.8 | - 2.4 |
| Hs.56 | Phosphoribosyl pyrophosphate synthetase 1 | PRPS1 | - 1.6 | - 2.3 |
| Hs.2057 | Uridine monophosphate synthetase | UMPS | - 1.6 | - 2.3 |
| Hs.464813 | Dihydrofolate reductase | DHFR | - 1.5 | - 2.3 |
| Hs.90280 | 5-aminoimidazole-4-carboxamide ribonucleotide transformylase | ATIC | - 1.5 | - 2.2 |
| Hs.177766 | Poly(ADP-ribose) polymerase | ADPRT | - 1.7 | - 2.0 |
| Hs.215766 | GTP-binding protein | GTPBP4 | - 1.5 | - 2.0 |
| Hs.409412 | ADP-ribosyltransferase (NAD +; poly(ADP-ribose) polymerase)-like 2 | ADPRTL2 | - 1.5 | - 1.8 |
| Hs.2057 | Uridine monophosphate synthetase | UMPS | - 1.5 | - 1.7 |
| Hs.383396 | Ribonucleotide reductase M1 polypeptide | RRM1 | - 1.6 | - 1.5 |
| Ribosomal/mitochondrial biogenesis | ||||
| Hs.411125 | Mitochondrial ribosomal protein S12 | MRPS12 | - 1.7 | - 9.9 |
| Hs.435643 | Ts translation elongation factor, mitochondrial | TSFM | - 3.7 | - 6.5 |
| Hs.511949 | Ribosomal RNA processing 4 | RRP4 | - 2.0 | - 4.4 |
| Hs.71827 | Ribosome biogenesis regulator homolog | RRS1 | - 1.9 | - 3.1 |
| Hs.132748 | Ribosomal protein L39 | RPL39 | -1.8 | -2.5 |
| Hs.256583 | Interleukin enhancer binding protein 3 | ILF3 | -2.0 | -2.3 |
| Hs.511949 | Ribosomal RNA processing 4 | RRP4 | -1.7 | -2.3 |
| Hs.101414 | Zinc finger protein 500 | ZNF500 | -1.6 | -2.3 |
| Hs.431307 | Mitochondrial ribosomal protein L40 | MRPL40 | -1.6 | -2.1 |
| Hs.376064 | Nucleolar protein (KKED repeat) | NOL5A | -1.9 | -2.0 |
| Hs.376681 | Mitofusin 2 | MFN2 | -1.7 | -1.7 |
| Hs.424264 | Mitochondrial ribosomal protein L46 | MRPL46 | -1.5 | -1.5 |
| RNA binding/metabolism | ||||
| Hs.443960 | DEAD/H box polypeptide 11 | DDX11 | -4.1 | -3.9 |
| Hs.434901 | Small nuclear ribonucleoprotein polypeptide A′ | SNRPA1 | -1.6 | -3.0 |
| Hs.166463 | Heterogeneous nuclear ribonucleoprotein U | HNRPU | -1.7 | -2.9 |
| Hs.153768 | RNA, U3 small nucleolar interacting protein 2 | RNU3IP2 | -1.7 | -2.4 |
| Hs.363492 | DEAD box polypeptide 18 | DDX18 | -1.5 | -2.4 |
| Hs.434901 | Small nuclear ribonucleoprotein polypeptide A′ | SNRPA1 | -1.9 | -2.1 |
| Hs.416994 | POP7 (processing of precursor) homolog | RPP20 | -1.8 | -2.1 |
| Hs.30174 | Small nuclear RNA activating complex 5 | SNAPC5 | -1.7 | -2.1 |
| Hs.372673 | Heterogeneous nuclear ribonucleoprotein D-like | HNRPDL | -1.6 | -2.1 |
| Hs.130098 | DEAD box polypeptide 23 | DDX23 | -1.5 | -2.1 |
| Hs.7174 | DEAH (Asp-Glu-Ala-His) box polypeptide 35 | DHX35 | -2.1 | -2.0 |
| Hs.73965 | Splicing factor, arginine/serine-rich 2 | SFRS2 | -1.6 | -2.0 |
| Hs.438726 | Heterogeneous nuclear ribonucleoprotein D | HNRPD | -2.2 | -1.9 |
| Hs.434901 | Small nuclear ribonucleoprotein polypeptide A′ | SNRPA1 | -1.9 | -1.9 |
| Hs.438726 | DEAD box polypeptide 21 | DDX21 | -1.7 | -1.9 |
| Hs.372673 | Heterogeneous nuclear ribonucleoprotein D-like | HNRPDL | -1.6 | -1.7 |
| Hs.86948 | Small nuclear ribonucleoprotein D1 polypeptide | SNRPD1 | -1.6 | -1.7 |
| Hs.356549 | Small nuclear ribonucleoprotein D3 polypeptide | SNRPD3 | -1.5 | -1.7 |
| Hs.511756 | Ribonuclease P1 | RNASEP1 | -1.6 | -1.6 |
| Hs.139120 | Ribonuclease P (30 kDa) | RPP30 | -1.6 | -1.6 |
| Ubiquitin/proteasome | ||||
| Hs.152978 | Proteasome activator subunit 3 | PSME3 | -1.6 | -2.9 |
| Hs.396393 | Ubiquitin-conjugating enzyme E2S | UBE2S | -1.6 | -2.9 |
| Hs.25223 | Ubiquitin carboxyl-terminal hydrolase L5 | UCHL5 | -1.6 | -2.1 |
| Hs.443379 | Proteasome 26S subunit, non-ATPase, 11 | PSMD11 | -1.5 | -2.1 |
| Hs.152978 | Proteasome activator subunit 3 | PSME3 | -1.6 | -1.8 |
| Hs.119563 | Proteasome activator subunit 4 | PSME4 | -1.6 | -1.8 |
| Hs.12820 | Ubiquitin specific protease 39 | USP39 | -1.6 | -1.6 |
| Hs.35086 | Ubiquitin specific protease 1 | USP1 | -1.5 | -1.5 |
| Transcription factors | ||||
| Hs.446451 | Myeloid/lymphoid leukemia; translocated to, 10 | MLLT10 | -2.2 | -5.1 |
| Hs.54089 | BRCA1 associated RING domain 1 | BARD1 | -1.6 | -3.6 |
| Hs.194143 | Breast cancer 1, early onset | BRCA1 | -2.7 | -3.2 |
| Hs.181128 | ELK1, member of ETS oncogene family | ELK1 | -2.4 | -3.0 |
| Hs.75133 | Transcription factor A, mitochondrial | TFAM | -1.6 | -3.0 |
| Hs.448398 | MYC-associated zinc finger protein | MAZ | -1.6 | -2.8 |
| Hs.194143 | Breast cancer 1, early onset | BRCA1 | -2.0 | -2.4 |
| Hs.106415 | Peroxisome proliferative activated receptor, delta | PPARD | -2.1 | -2.3 |
| Hs.408528 | Retinoblastoma 1 | RB1 | -1.8 | -2.3 |
| Hs.436187 | Thyroid hormone receptor interactor 13 | TRIP13 | -1.9 | -2.1 |
| Hs.75133 | Transcription factor 6-like 1 | TF6M | -1.6 | -1.7 |
| Hs.75133 | Transcription factor A, mitochondrial | TFAM | -1.6 | -1.7 |
| Hs.410900 | Inhibitor of DNA binding 1 | ID1 | -1.6 | -1.5 |
Major functional categories of genes induced by hypoxia and AdCA5
Analysis of gene expression that was induced by hypoxia and AdCA5 revealed several large categories of gene products. More than half of all induced gene probes (128/245) could be placed into 1 of 6 categories: oxidoreductases, collagens/modifying enzymes, cytokines/growth factors, receptors, other signal transduction proteins, and transcription factors (Table 1).
Genes encoding oxidoreductases accounted for 14 probes corresponding to 9 unique genes. One of the most highly induced genes was PTGIS (3 probes), which encodes prostaglandin (PG) I2 (PGI2; prostacyclin) synthase. The expression of PTGS (2 probes), which encodes cyclooxygenase 1, was also induced by both hypoxia and AdCA5. PTGS catalyzes the synthesis of PGG2 and PGH2 from arachidonic acid, whereas PTGIS converts PGH2 to PGI2. Thus, HIF-1 coordinately regulates the expression of 2 enzymes in the prostaglandin synthetic pathway in ECs. The 2 other members of the oxidoreductase group are the EGLN1 and EGLN3 genes, which encode the HIF-1α prolyl hydroxylases PHD2 and PHD3, respectively. Induction of these genes provides a feedback mechanism for down-regulating HIF-1α expression.33
Genes encoding collagens and their modifying enzymes accounted for 15 probes (11 unique genes), including COL1A2, COL4A1, COL4A2, COL5A1 (2 probes), COL9A1, and COL18A1 (2 probes), as well as procollagen prolyl hydroxylases (P4HA1, P4HA2), lysyl oxidase (LOX [2 probes]), and lysyl hydroxylases (PLOD, PLOD2 [2 probes]). Thus, HIF-1 coordinately regulates multiple hypoxia-induced changes in collagen biosynthesis by ECs.
Genes encoding cytokines and growth factors accounted for 18 probes and 15 genes, including the known HIF-1 target genes VEGF (3 probes), EDN1, IGFBP3, PDGFB (2 probes), PGF, ADM, and ANGPTL4, which was the gene probe that was most highly induced by hypoxia (17.5-fold). Novel HIF-1 target genes encoding secreted proteins that were identified include CX3CL1, GDF10, INHBA, INHBE, VEGFC, RLN1, STC1, and STC2, the latter of which was recently implicated as a HIF-1 target gene.34
A large group of HIF-1 target genes in arterial ECs encodes receptors (25 probes, 22 genes). Included in this group are genes for receptor tyrosine kinases (AXL, INSR), G protein–coupled receptors (ADORA2A, CMKOR1, GRK5, OPN3), cytokine receptors (CXCR4, EPOR, LEPR, TNFRSF10B, TNFRSF14), a receptor guanylate cyclase (NPR1), and genes for the receptor-type protein tyrosine phosphatases PTPRB (2 probes), PTPRF, and PTPRR (2 probes).
Other signal transduction proteins compose another large group (28 probes, 25 genes) of hypoxia-inducible genes that are regulated by HIF-1. These genes encode multiple serine-threonine kinases (ARK5, CASK, CDK11, RPS6AK2 [2 probes], TRIO) and G-protein signaling molecules (RASSF2, RGS3, RHOBTB1, RRAS).
The largest category of genes induced by hypoxia and AdCA5 encodes transcription factors (29 probes, 26 genes). Included among these is the known HIF-1 target gene BHLHB2 (2 probes), which encodes the transcriptional repressor DEC1/Stra13.30 However, the other 25 transcription factor genes identified are novel HIF-1 targets. The implications of this important finding will be addressed in “Discussion.”
To validate the microarray data, aliquots from the same RNA preparations were analyzed by quantitative real-time RT-PCR (qPCR), using primers specific for EDN1, PTGIS, and VEGF. The qPCR analysis confirmed that expression of all of these genes was increased more than 1.5-fold in ECs exposed to hypoxia or AdCA5 (Figure 2A). Moreover, the rank order of fold induction (PTGIS > VEGF > EDN1) was identical in the microarray and qPCR analyses. In order to investigate whether the gene expression induced by hypoxia was mediated directly by HIF-1 or was secondary to the expression of a HIF-1–regulated transcription factor, we performed a time-course analysis. EPOR, PTGIS, and VEGFC mRNA levels were all increased more than 1.5-fold after only 8 hours of hypoxic exposure (Figure 2B). These results were similar to those for CXCR4 and VEGF mRNAs, which are the products of known HIF-1 target genes,27,35 suggesting that the EPOR, PTGIS, and VEGFC genes are also directly regulated by HIF-1. Further studies are required to identify functional HREs containing HIF-1 binding sites in these genes.
Real-time RT-PCR analysis of gene expression induced in response to hypoxia or AdCA5. (A) Aliquots of the same 12 RNA preparations used for microarray hybridization (3 different EC primary cultures, 4 experimental conditions) were analyzed by quantitative real-time RT-PCR. For each pair of experimental conditions (ie, 1% O2 vs 20% O2 [H] and AdCA5 vs AdLacZ [C]) the fold induction of VEGF, PTGIS, and EDN1 mRNA expression was calculated (“Materials and methods”). The mean and standard error for the 3 independent data sets are shown. (B) ECs were exposed to hypoxia (1% O2) for 0 to 60 hours; total RNA was isolated; and the levels of CXCR4, EPOR, PTGIS, VEGF, and VEGFC mRNA were determined.
Real-time RT-PCR analysis of gene expression induced in response to hypoxia or AdCA5. (A) Aliquots of the same 12 RNA preparations used for microarray hybridization (3 different EC primary cultures, 4 experimental conditions) were analyzed by quantitative real-time RT-PCR. For each pair of experimental conditions (ie, 1% O2 vs 20% O2 [H] and AdCA5 vs AdLacZ [C]) the fold induction of VEGF, PTGIS, and EDN1 mRNA expression was calculated (“Materials and methods”). The mean and standard error for the 3 independent data sets are shown. (B) ECs were exposed to hypoxia (1% O2) for 0 to 60 hours; total RNA was isolated; and the levels of CXCR4, EPOR, PTGIS, VEGF, and VEGFC mRNA were determined.
Major functional categories of genes repressed by hypoxia and AdCA5
Among the 325 gene probes that showed decreased expression in cells exposed to hypoxia or AdCA5, 67 (21%) encoded proteins involved in cell growth/proliferation. Included among these were gene probes encoding the following: cyclins E1, E2 (2 probes), and F; CDC2 (3 probes), CDC6, and CDC7; MCM2, MCM3, MCM4 (4 probes), MCM5, MCM6, and MCM10; replication factors RFC2, RFC3, and RFC5 (2 probes); DNA polymerases δ2, ϵ2, and γ; RNA polymerase II polypeptides K and L; RNA polymerase III 32-kDa subunit; enzymes involved in ribonucleotide metabolism; and proteins involved in mitochondrial and ribosomal biogenesis (Table 2). CHAF1A, encoding chromatin assembly factor 1 subunit A, was the named gene that was most highly repressed by hypoxia (6.0-fold) and by AdCA5 (10.0-fold).
Another large group of genes repressed by hypoxia and AdCA5 (20 probes, 17 genes) encodes proteins involved in RNA binding/metabolism. There are 8 other probes (7 genes) that encode proteins involved in protein ubiquitination or proteasomal degradation. Multiple genes encoding transcription factors were also repressed by both hypoxia and AdCA5 (13 probes, 11 genes).
CDC2, CCNE1, MPHOSP9, POLD, POLG, PRC1, RRM2, and NEK4 gene expression was analyzed by qPCR. For each of these 8 genes, expression was decreased more than 1.5-fold in ECs exposed to hypoxia or AdCA5 for 24 hours (Figure 3).
Real-time RT-PCR analysis of gene expression repressed in response to hypoxia or AdCA5. Aliquots of the same RNA preparations used for microarray hybridization were analyzed by quantitative real-time RT-PCR. For each pair of experimental conditions (ie, 1% O2 vs 20% O2 [H] and AdCA5 vs AdLacZ [C]), the fold change in the levels of CCNE1, MPHOSP9, POLD, POLG, CDC2, PRC1, RRM2, and NEK4 mRNA was calculated. The mean and standard error for the 3 independent data sets are shown.
Real-time RT-PCR analysis of gene expression repressed in response to hypoxia or AdCA5. Aliquots of the same RNA preparations used for microarray hybridization were analyzed by quantitative real-time RT-PCR. For each pair of experimental conditions (ie, 1% O2 vs 20% O2 [H] and AdCA5 vs AdLacZ [C]), the fold change in the levels of CCNE1, MPHOSP9, POLD, POLG, CDC2, PRC1, RRM2, and NEK4 mRNA was calculated. The mean and standard error for the 3 independent data sets are shown.
Effects of hypoxia and AdCA5 on endothelial cell biology
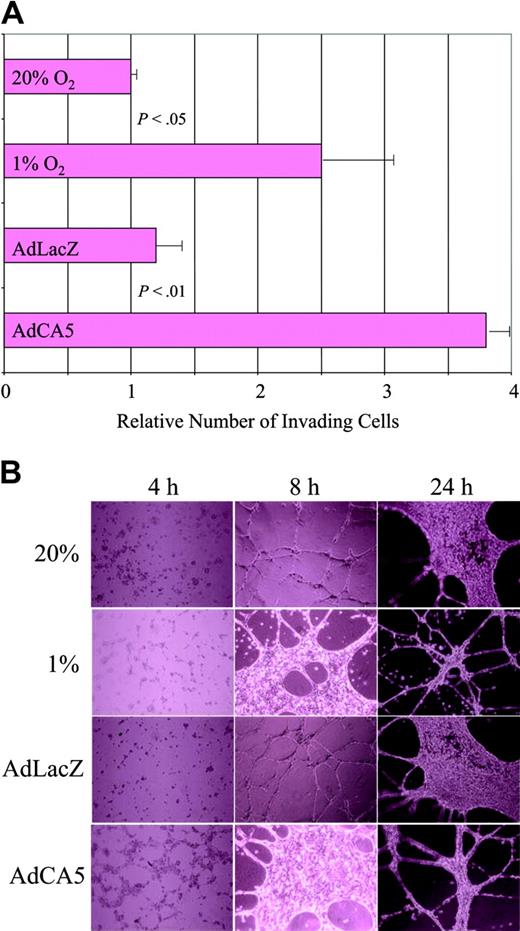
The gene expression data suggest that exposure of ECs to hypoxia or AdCA5 induces major functional changes in these cells. To test this hypothesis, we performed 2 assays of EC activation. First, we plated ECs on Matrigel (an experimental basement membrane) overlying a porous filter, and incubated the cells for 24 hours under the same 4 conditions used for microarray analysis, except that the cells were exposed to only 5 pfu of AdCA5 or AdLacZ. Exposure of cells to hypoxia or AdCA5 resulted in a statistically significant increase in the number of cells that digested the Matrigel and migrated through the pores to the underside of the filter (Figure 4A). These results are consistent with previous studies demonstrating increased Matrigel invasion by hypoxic human umbilical vein ECs.36
Effect of hypoxia and AdCA5 on invasion and tube formation by endothelial cells. (A) Matrigel invasion by ECs exposed to hypoxia or AdCA5. ECs were incubated for 24 hours in the presence or absence of adenovirus (5 pfu/cell), transferred to a Matrigel-coated membrane in a Boyden chamber, and exposed to 20% or 1% O2 for 24 hours, and the number of cells that had invaded through the Matrigel to the underside of the filter was counted. Three independent experiments were performed and the results for each condition were normalized to the result obtained for untreated cells (20% O2). P values were determined from the mean and standard deviation (shown) using Student t test. (B) Tube formation by ECs exposed to hypoxia or AdCA5. ECs were plated on Matrigel in the absence or presence (5 pfu/cell) of adenovirus (AdLacZ or AdCA5) that was added to the medium at the time of plating. Adenovirus-infected cells and one set of uninfected cells were incubated under nonhypoxic conditions (20% O2), whereas another set of uninfected cells was incubated under hypoxic conditions (1% O2). The cultures were analyzed 4, 8, and 24 hours after plating by phase-contrast microscopy. Each condition was performed in triplicate in 2 independent experiments and representative fields are shown.
Effect of hypoxia and AdCA5 on invasion and tube formation by endothelial cells. (A) Matrigel invasion by ECs exposed to hypoxia or AdCA5. ECs were incubated for 24 hours in the presence or absence of adenovirus (5 pfu/cell), transferred to a Matrigel-coated membrane in a Boyden chamber, and exposed to 20% or 1% O2 for 24 hours, and the number of cells that had invaded through the Matrigel to the underside of the filter was counted. Three independent experiments were performed and the results for each condition were normalized to the result obtained for untreated cells (20% O2). P values were determined from the mean and standard deviation (shown) using Student t test. (B) Tube formation by ECs exposed to hypoxia or AdCA5. ECs were plated on Matrigel in the absence or presence (5 pfu/cell) of adenovirus (AdLacZ or AdCA5) that was added to the medium at the time of plating. Adenovirus-infected cells and one set of uninfected cells were incubated under nonhypoxic conditions (20% O2), whereas another set of uninfected cells was incubated under hypoxic conditions (1% O2). The cultures were analyzed 4, 8, and 24 hours after plating by phase-contrast microscopy. Each condition was performed in triplicate in 2 independent experiments and representative fields are shown.
Hypoxia has been shown to stimulate capillary-like tube formation by human skin microvascular ECs cultured on fibrin matrices.37 We analyzed the ability of ECs to form tubelike networks on Matrigel. When incubated under nonhypoxic conditions, ECs formed cell-cell contacts within 4 hours, tubelike extensions by 8 hours, followed by a phase of proliferation that was evident at 24 hours, and then more extensive tube formation (Figure 4B). When cells were subjected to hypoxia or AdCA5 infection, the process was accelerated such that the EC morphology at 8 hours was similar to that of control or AdLacZ-infected ECs at 24 hours. The effect of AdCA5 was striking because the ECs were infected with only 5 pfu/cell, and expression of adenoviral gene products did not peak until 24 hours (data not shown). Thus, exposure of ECs to hypoxia or AdCA5 ex vivo results in biologic responses associated with angiogenic activation in vivo.
Discussion
Previous studies have demonstrated that HIF-1 plays a critical role in angiogenesis by regulating the transcription of multiple growth factors, which are produced by hypoxic parenchymal cells in tissues and activate the angiogenic program in ECs, leading to increased tissue perfusion.23,25,26 Our present studies indicate that HIF-1 plays an equally profound role as a mediator of EC-autonomous responses to hypoxia. Overexpression of a constitutively active form of HIF-1α under nonhypoxic conditions is sufficient to induce changes in EC biology that are remarkably similar to those induced by hypoxia (Figure 4). Infection of ECs with an adenovirus encoding a chimeric HIF-1α/VP-16 protein has also been shown to promote invasion and tube formation,38 results that were interpreted within the well-established paradigm of HIF-1 as an activator of angiogenic growth factor gene expression, whereas our data indicate that there are additional cell-autonomous mechanisms by which HIF-1 may mediate activation of ECs.
The large numbers of HIF-1–regulated genes encoding cytokines/growth factors, receptor tyrosine kinases, G protein–coupled receptors, and associated signaling proteins (Table 1) provide a broad molecular basis for EC activation. Among a group of genes whose expression was induced in human umbilical vein ECs undergoing differentiation into tubelike structures ex vivo39 are several genes that were also induced by hypoxia in pulmonary artery ECs, including AXL, COL4A1, CXCR4, PAM, PGF, and STC1. The majority of the 15 hypoxia-induced and HIF-1–regulated genes encoding cytokines and growth factors have established roles in vascular biology. One notable novel finding is the regulation of VEGFC by HIF-1 in hypoxic ECs. VEGF-C stimulates lymphangiogenesis, which may promote drainage of extravasated fluid resulting from the permeability effects of hypoxia-induced VEGF on ECs.40 Among the genes encoding receptors, the identification of EPOR as the product of a HIF-1 target gene is notable because of the recent demonstration that its ligand, erythropoietin, stimulates the mobilization and differentiation of bone marrow–derived endothelial progenitor cells.41,42
In addition to cell autonomous effects, gene products induced by hypoxia in ECs have more global effects on vascular biology. Prostaglandins produced in ECs via the activity of PTGS1 and PTGIS play an important role in modulating vascular smooth muscle cell tone.43 ECs also play a major role in extracellular matrix production and the number of HIF-1–regulated genes encoding collagens and collagen-modifying enzymes is striking. HIF-1 also regulates the extracellular matrix proteoglycans chondroitin sulfate and keratan sulfate by activating expression of the CHSY1 and CHST1 genes encoding chondroitin synthase 1 and keratan sulfate sulfotransferase 1, respectively (Supplementary Table 1). In addition, HIF-1 activates the expression of GPC1, encoding glypican 1, an integral membrane protein to which heparan sulfate proteoglycans are anchored.
Our data indicate that more than 2% of all human genes are regulated by HIF-1 in ECs. These data underestimate the number of HIF-1–regulated genes in this cell type for several reasons. First, only one type of EC (pulmonary arterial) was cultured under only one set of conditions (substratum, growth medium, cell density) and assayed at only one time point. For example, the experiments were performed with confluent cultures of ECs, and, under these conditions, hypoxia or AdCA5 induced a growth arrest response as manifested by the repression of genes encoding cell cycle proteins (Table 2). These results delineate the molecular basis for the previous demonstration that HIF-1α is required for hypoxia-induced cell cycle arrest.44 In contrast, when cultured at low density, EC proliferation is stimulated by hypoxia.45 Second, multiple target genes were excluded based upon statistical criteria (significant t test using the minimal sample size). For example, although 3 VEGF probes met the inclusion criteria, 1 additional probe was induced 4.1- and 6.2-fold by hypoxia and AdCA5, respectively, but the adenoviral data had a P > .05 and thus the probe was excluded. Third, multiple target genes were excluded based upon the arbitrary threshold value. For example, a probe for the known HIF-1 target gene LDHA showed statistically significant increases of 1.3- and 1.6-fold in response to hypoxia and AdCA5, respectively. If the threshold is lowered to 1.2-fold, the numbers of gene probes showing a significant increase or decrease rise to 442 and 687, respectively, or 5.1% of all gene probes on the array.
The finding that less than half of the hypoxia-induced genes were also AdCA5 induced does not imply that HIF-1 regulates only a minority of O2-regulated genes, because even a smaller fraction of the genes induced by AdCA5 were also induced by hypoxia. Rather, these results are a reflection of the stringent study design, which allowed us to obtain data with high specificity (ie, few false positives, as demonstrated in Figures 2, 3) at the expense of reduced sensitivity for the reasons described above. Based upon these considerations, the total number of HIF-1–regulated genes in ECs may exceed 5% of all human genes. Comparison of these data with previously published microarray analyses29,30,39,46-54 indicates that the battery of genes regulated by hypoxia varies greatly from one cell type to another. Thus, the total number of human genes subject to regulation by HIF-1 in one cell type or another appears to be considerable.
What accounts for the large number of HIF-1–regulated genes identified by microarray analysis? Some of the 570 probes correspond to genes, such as ADM, BNIP3, CXCR4, EDN1, and VEGF, which are known direct targets of HIF-1 that contain HREs with HIF-1 binding sites. Cross-species bioinformatic analyses of genomic DNA and chromatin immunoprecipitation assays34,55 represent 2 methods that can be applied to the data set reported here in an effort to identify novel direct (primary) targets of regulation by HIF-1 in ECs. It is striking that one of the largest functional categories of genes regulated by HIF-1 encodes transcription factors, indicating that HIF-1 is at the top of a hierarchy of O2-regulated gene expression within ECs. In addition to the 29 probes (26 genes) encoding transcription factors in the hypoxia/AdCA5-induced data set (Table 1), 13 probes (11 genes) encoding transcription factors were present in the hypoxia/AdCA5-repressed data set (Table 2). Thus, some of the genes identified in this study may represent secondary HIF-1 targets, which are directly regulated by transcription factors that are encoded by primary HIF-1 target genes. In this case, during the 24-hour exposure to hypoxia or AdCA5, HIF-1α expression would lead to transcription of primary HIF-1 target genes, translation of the resulting mRNAs into functional transcription factors that would then regulate expression of secondary target genes. However, all of the genes for which time-course analysis was performed manifested increased expression as early as 8 hours after onset of hypoxia (Figure 2B), suggesting that they are primary HIF-1 target genes. Since 24 hours is unlikely to be sufficient for maximal effects on secondary targets, additional time points are required to fully delineate the cascade of gene expression that is initiated by HIF-1. In contrast to the cytokines/growth factors, most of the transcription factors that were identified as regulated by HIF-1 in ECs have not been previously studied in the context of EC/vascular biology. The response of ECs to thrombin also has been shown to involve dramatic alterations in transcriptional networks,24 which are mediated by transcription factors that are distinct from those we have identified as involved in the response to hypoxia.
The number of gene probes (5258) that showed a 1.5-fold or higher change in expression in response to AdCA5 is considerably greater than the number (1917) that showed a 1.5-fold or higher change in expression in response to hypoxia (Figure 1). There are at least 2 possible explanations for this result. First, the levels of HIF-1α protein may have been greater in the AdCA5-exposed cells. Second, recent data indicate that many mRNAs synthesized in response to hypoxia are not translated until the cells are reoxygenated.56 If any HIF-1–regulated transcription factor mRNAs are included in this category, then expression of their target genes would be affected in nonhypoxic cells exposed to AdCA5 but not in hypoxic cells. A network/systems biology approach is clearly needed to further define the global impact of HIF-1 as a regulator of EC gene expression, vascular biology, and oxygen homeostasis.
Previous studies have demonstrated that HIF-1 controls the expression of key angiogenic factors, which are secreted by other cell types and activate ECs. The present study is the most comprehensive analysis of gene expression regulated by hypoxia and HIF-1 in human arterial ECs. The cellular assays demonstrate that HIF-1 induces autonomous EC activation, as manifested by extracellular matrix invasion and tube formation. Taken together, these molecular and cellular data provide novel insights into the composition and function of the HIF-1–regulated transcriptome in ECs.
Prepublished online as Blood First Edition Paper, September 16, 2004; DOI 10.1182/blood-2004-07-2958.
Supported by National Heart, Lung, and Blood Institute (NHLBI) grant R01-HL55338 to G.L.S.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Susan Schoonover and Andrea Gambotto (University of Pittsburgh) for adenovirus production; Jeffrey Rade (Johns Hopkins University) for assistance with microscopy; Shannon Berg-Dixon and Mia Pearson for technical assistance; and Rafael Irizarry, Shwu Fan Ma, and Jonathan Pevsner (Johns Hopkins University) for helpful discussions.

![Figure 2. Real-time RT-PCR analysis of gene expression induced in response to hypoxia or AdCA5. (A) Aliquots of the same 12 RNA preparations used for microarray hybridization (3 different EC primary cultures, 4 experimental conditions) were analyzed by quantitative real-time RT-PCR. For each pair of experimental conditions (ie, 1% O2 vs 20% O2 [H] and AdCA5 vs AdLacZ [C]) the fold induction of VEGF, PTGIS, and EDN1 mRNA expression was calculated (“Materials and methods”). The mean and standard error for the 3 independent data sets are shown. (B) ECs were exposed to hypoxia (1% O2) for 0 to 60 hours; total RNA was isolated; and the levels of CXCR4, EPOR, PTGIS, VEGF, and VEGFC mRNA were determined.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/2/10.1182_blood-2004-07-2958/6/m_zh80020572780002.jpeg?Expires=1767707593&Signature=44gscW179V-wzxg3DaGXDnXls6bE4x6v5~A0Pylymh~xu5jZACPWBZBma63NerBef~~0h7bIQwEzDpay9O8W3Pd9KgMWq3ApyM89cPrRwj2CN5X4ealc8C4iHuFyVKBHv7i-QkVpDfLtg3emgnC~8~yAwHzbdrWlbJWh95oXDjLirkZbig-etAxkMgZVXGk-HUk57Gfbim8hnbxOt~TIGLuqv9OGPxWeK-1uoRkzUO7yD5Sj5drMOgHI3me5MgwHpbZdHnVn~UnwF4wf~p2qhYeC1zTbFkK89O1jdb-czSgzPcLNtAtbtTWxMVWaKBDE8Yad22J0bac-DRk144gL1A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Real-time RT-PCR analysis of gene expression repressed in response to hypoxia or AdCA5. Aliquots of the same RNA preparations used for microarray hybridization were analyzed by quantitative real-time RT-PCR. For each pair of experimental conditions (ie, 1% O2 vs 20% O2 [H] and AdCA5 vs AdLacZ [C]), the fold change in the levels of CCNE1, MPHOSP9, POLD, POLG, CDC2, PRC1, RRM2, and NEK4 mRNA was calculated. The mean and standard error for the 3 independent data sets are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/2/10.1182_blood-2004-07-2958/6/m_zh80020572780003.jpeg?Expires=1767707593&Signature=lfnSGCcsZe8Z8qqhNx-1Yqg4KRV7Pxbz0U8jt4fRS24QyLqMgNYqta02arxJTdOU1ANx61aCm-osq9LVPomRD2BFowOpDCmqRcrcJIH-4WXFwYd8KgrT-RPdpprxPctq09HEGWobezVFeqfu6sPnTzccGcjlRboCAegHx~Edrc3Y2ZoR8Iy83tU3NX4-9qh0QMQRbUFVw6Aqv8JA317EvwK0gG4kqo6k~CDrdbZgNlMsJj9KoraQ~ZcmTKB-sPRww5BRvheY1PKJXodOVAZkJ6Gxpvp9e37AeVH9JzdfL~eyCcRHjYqOcXtHe4yhCH76TCMJQv8YmpXwG8m2IC2zJA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal