Abstract
Engagement of the B-cell antigen receptor (BCR) activates kinases of the Src and Syk families and signaling complexes assembled by adaptor proteins, which dictate B-cell fate and function. The adaptor 3BP2/SH3BP2, an Abl Src homology domain 3 (SH3)–binding and Syk-kinases interacting protein, exhibits positive regulatory roles in T, natural killer (NK), and basophilic cells. However, its involvement in BCR signaling is completely unknown. Here we show that 3BP2 is tyrosine phosphorylated following BCR aggregation on B lymphoma cells, and that 3BP2 is a substrate for Syk and Fyn, but not Btk. To further explore the function of 3BP2 in B cells, we screened a yeast 2-hybrid B-lymphocyte library and found 3BP2 as a binding partner of Vav proteins. The interaction between 3BP2 and Vav proteins involved both constitutive and inducible mechanisms. 3BP2 also interacted with other components of the BCR signaling pathway, including Syk and phospholipase C γ (PLC-γ). Furthermore, overexpression and RNAi blocking experiments showed that 3BP2 regulated BCR-mediated activation of nuclear factor of activated T cells (NFATs). Finally, evidence was provided that 3BP2 functionally cooperates with Vav proteins and Rho GTPases to activate NFATs. Our results show that 3BP2 may regulate BCR-mediated gene activation through Vav proteins.
Introduction
B-cell antigen receptor (BCR) engagement triggers multiple signal transduction events regulating B-cell development and function.1 The BCR signals through conserved immunoreceptor tyrosine-based activation motifs (ITAMs) found within the cytoplasmic tails of its subunits. Ligation by antigen results in the activation of associated cytoplasmic protein tyrosine kinases (PTKs), including Syk, the Src-family PTKs Lyn and Fyn, and Btk.2,3 The activated Lyn and Fyn kinases phosphorylate the ITAMs within the immunoglobulin alpha (Ig-α) and Ig-β chains, resulting in the recruitment and activation of Syk. As a result, molecular scaffolds composed of adaptor proteins and key enzymes, such as phospholipase C γ (PLC-γ), phosphatidylinositol 3-kinase (PI3K), and guanine nucleotide exchange factors (GEFs) Vav1 and Vav2, are assembled and activated at the plasma membrane by Src and/or Syk PTKs. These scaffolds transduce signals to the cytoplasm and nucleus to activate gene expression and metabolic changes involved in B-cell homeostasis.
One critical pathway stimulated by BCR triggers intracytoplasmic calcium movements leading to the activation of transcription factors, such as nuclear factor of activated T cells (NFATs). These nuclear factors coordinate the expression of cytokines and cell-cycle proteins involved in B-cell growth and apoptosis. Calcium signals are triggered by activated PLC-γ that produces inositol-1,4,5-triphosphate (IP3), followed by an increase of cytoplasmic calcium.4 The regulation of PLC-γ activity involves the membrane translocation and activation of Btk following PI3K-mediated production of phosphatidylinositol-3,4,5-triphosphate (PIP3).5,6 Vav proteins also participate to the regulation of calcium signals and NFAT activation in lymphocytes. Vav proteins are activators of Rho family GTPases that control cytoskeletal reorganization and gene activation.7,8 Analysis of Vav1- and Vav2-deficient mice showed the importance of these proteins in BCR signaling during development, maturation, and proliferation of B cells.9-11 The major defect in vav1–/– × vav2–/– B cells stimulated through antigen receptor appears to be a defect in calcium release. Although the basis for this defect is still unclear, it likely involves defects in PI3K, PLC-γ, and Tec-family kinases activation, since this phenomenon has been observed in Vav1-deficient T and mast cells.12,13 Moreover, Vav3 has been shown to regulate PI3K activation in the DT40 chicken B-cell line.14
The assembly of signaling complexes is regulated by adaptor proteins that mediate protein-protein or protein-lipid interactions.15 In B cells, one such adaptor is the Syk substrate BLNK/SLP-65, which connects BCR to PLC-γ activation by prodiving docking sites for Btk and PLC-γ2.16,17 In a screen for Syk-kinase interacting proteins, we previously identified 3BP2,18 a cytoplasmic adaptor originally identified as an Abl Src homology domain 3 (SH3) binding protein.19 3BP2 (also known as SH3BP2) is preferentially expressed in hematopoietic tissues,18,20 and positively regulates the activity of the transcription factors NFAT and AP-1 in T cells through calcineurin and Ras-dependent pathways.18 A positive regulatory role of 3BP2 in NK cell–mediated cytotoxicity21 and in the degranulation of rat basophilic leukemia cells22 has also been described. Several cell signaling partners of 3BP2 have been described including Fyn, Zap-70, LAT, and PLC-γ in T cells,18 Vav1 and PLC-γ in NK cells,21 and LAT and Lyn in basophilic cells.22,23 A regulation of 3BP2 adaptor function by the chaperone protein 14-3-3 has also been reported.24 Finally, mutations in the 3bp2/sh3bp2 gene were identified to cause a human bone disease called cherubism.25 Altogether, these observations suggest that 3BP2 plays a crucial role in PTK-dependent signaling pathways during hematopoietic cell differentiation and function.
In this study, we investigated the role of 3BP2 in BCR signaling. Using 3BP2 as a bait in a yeast 2-hybrid system, we found that 3BP2 interacted with members of the Vav family of Rho GTPases activators. A combination of biochemical techniques, overexpression, and RNAi blocking experiments showed that 3BP2 cooperated with Vav proteins to control a Rho GTPases-dependent pathway regulating NFAT activation in B cells. Together, our results provide the first evidence that 3BP2 is an active and important component of BCR signaling.
Materials and methods
Reagents and antibodies
Culture media, oligonucleotides, and enzymes were from Invitrogen (The Netherlands). Chemicals were obtained from Sigma (St Louis, MO). Polyclonal antibodies against 3BP2 were prepared following rabbit immunization with the glutathione S transferase (GST) 3BP2 SH2 domain fusion protein.18 Anti-hemagglutinine (HA; 12CA5) monoclonal antibody (mAb) was from Roche Diagnostics (Basel, Switzerland). The antibodies specific for the myc epitope (9E10), LexA, Vav1, and c-Cbl were from Upstate Biotechnology (Lake Placid, NY). The antibodies against Syk, ERK2, PLC-γ1, and GST were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-phosphotyrosine mAbs were purified from culture supernatants of the corresponding hybridoma by protein A–Sepharose chromatography. The anti–human Vav2 mAb was a gift from J. Downward (London, United Kingdom). F(ab′)2 fragments of anti–human IgM were from Dako (Glostrup, Denmark).
Plasmids
Plasmid construction, cloning, and DNA sequencing were carried out according to standard protocols. The yeast expression plasmids pLex9, pACTII, and the HA-tagged 3BP2 constructs in the expression vector pMT3, and 3BP2 constructs in the yeast expression vector pLex9, were described before.18,24 The NFAT luciferase reporter construct was from G. Crabtree (Stanford, CA). Plasmids encoding myc-tagged Vav1 and Vav2, GST-Vav2 SH3-SH2-SH3, GST-3BP2 SH2, and HA-Rac1 were previously described.18,26-28
Cell culture and transfection
The human B (BJAB, RPMI8866, Daudi, and Raji) and T (Jurkat) cell lines were obtained from American Type Culture Collection (ATCC; Manassas, VA). Cells were grown in RPMI 1640 medium (Invitrogen), supplemented with 10% fetal calf serum (FCS; Dutcher, Brumath, France), 2 mM glutamine, 1 mM sodium pyruvate, 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), nonessential amino acid solution, and 100 U/mL penicillin and streptomycin. Cells grown in a logarithmic growth phase were transfected with the indicated amounts of plasmids by electroporation using the Gene Pulser (Bio-Rad Laboratories, Mississauga, ON, Canada). Simian COS-1 were cultured in Dulbecco modified Eagle medium (Invitrogen) with 10% FCS. Cells (1 × 107) were plated the day before transfection. Transfections were then performed by calcium phosphate precipitation.
Yeast 2-hybrid system
Growth and transformation of the yeast strain L40 were performed as described before.26 Briefly, a human B-lymphocyte cDNA library encoded by the yeast 2-hybrid vector pACTII was transformed into yeast cells that were previously transformed by the pLex9 3BP2 bait vector. Transformed cells were plated on synthetic drop-out medium lacking tryptophan, leucine, histidine, and uracil, and grown for 4 days at 30°C. Colonies were then assayed for β-galactosidase activity using a filter assay.26 After a second round of screening performed with a LexA-lamin plasmid as a negative control, library plasmid cDNAs were rescued and sequenced.
GST pull-down assays
GST fusion proteins were expressed in BL21 bacteria with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 hours at 37°C. Proteins were affinity purified using glutathione-Sepharose beads. Purified proteins were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by Coomassie staining. For GST precipitations, cells were lysed at 1 × 108 cells/mL in ice-cold lysis buffer (1% Nonidet P-40 in 150 mM NaCl, 50 mM HEPES [pH 7.4], 5 mM NaF, 5 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 10 μg/mL aprotinin, 10 μg/mL leupeptin, 1 mM phenylmethylsulfonyl fluoride [PMSF]) for 30 minutes on ice. Lysates were clarified by centrifugation at 15 000g for 15 minutes at 4°C, and incubated with 10 μg of the indicated GST fusion protein and glutathione-Sepharose beads (Sigma) for 3 hours at 4°C. After extensive washing in lysis buffer, bound proteins were detected by immunoblotting as indicated below.
Immunoprecipitation and immunoblotting
Cells were lysed at 1 × 108 cells/mL in ice-cold lysis buffer for 30 minutes on ice. In some experiments, cells were lysed in membrane rafts-solubilizing buffer (1% octylglucoside in 140 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA [ethylenediaminetetraacetic acid], 1 mM NaF, 1 mM sodium orthovanadate, 10% glycerol, 10 μg/mL aprotinin, 10 μg/mL leupeptin, 1 mM PMSF for the same period of time. Cleared lysates were then incubated for 3 hours at 4°C with the indicated antibodies and one hour with protein G–Sepharose beads (Sigma). Pellets were then washed 3 times with ice-cold lysis buffer and resuspended in SDS sample buffer. Eluted immunoprecipitates or whole-cell lysates were separated by SDS-PAGE, and analyzed by immunoblotting. SDS-PAGE–resolved samples were transferred to nitrocellulose membranes, and probed with primary and horseradish peroxidase (HRP)–conjugated secondary antibodies followed by enhanced chemiluminescence detection (ECL).
Reporter assays
Cells were transiently transfected with NFAT reporter plasmid along with the indicated expression plasmids, and a control Renilla luciferase plasmid vector (pRL-TK) for normalization of transfection efficiency (Promega, Madison, WI). After 18 hours, cells were washed and stimulated or not with anti-IgM (2 μg/mL), or a combination of phorbol 12-myristate 13-acetate (PMA) and ionomycin for 6 hours at 37°C. Luciferase activities were determined using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Normalized firefly luciferase activities (NFATs) were expressed as the fold increase relative to the basal activity seen in unstimulated mock-transfected cells.
RNA interference
Synthetic siRNA oligonucleotides were synthesized and purified by Dharmacon (Lafayette, CO). siRNA against human 3BP2 was 5′-AAGAGUAGCACCUCUGCCUCC-3′ (si3BP2). The sequence of the control siRNA against green fluorescent protein (GFP) was 5′-AAGGCUACGUCCAGGAGCGCACC-3′ (siGFP). A 5′-fluorescein-conjugated luciferase siRNA (5′-CGUACGCGGAAUACUUCGU-3′) was used in some experiments for sorting cell populations transfected with siRNA. The efficiency of 3BP2 siRNA was examined in Jurkat (107) or Raji (5 × 107) cells electroporated with 2 μg 3BP2 siRNA or control siRNA in the presence of 5′ fluorescein isothiocyanate (FITC)–conjugated luciferase siRNA. After 24 hours, cells were sorted by fluorescence-activated cell sorting (FACS), lysed, and 3BP2 expression was determined by immunoblotting. For determination of NFAT activity in 3BP2 knock-down cells, Raji or Jurkat cells (2 × 107) were electroporated with 2 μg siRNA, 15 μg NFAT reporter, and 3 μg Renilla luciferase plasmid. After 18 hours, cells were washed and stimulated for 6 hours at 37°C. Triplicate of luciferase activities were measured and expressed as described under “Reporter assays.”
Determination of Rac1 activity
Rac1 activity was determined by a modification of the pull-down assay described before.29 Briefly, cells (2 × 107) were transfected with 5 μg HA-tagged Rac1 expression plasmid28 along with 20 μg of either 3BP2 or Vav1 expression plasmids. At 24 hours after transfection, cells were lysed in Rac pull-down assay buffer (0.5% Triton X-100 in 150 mM NaCl, 25 mM Tris [pH 7.5], 5 mM MgCl2, 0.5 mM EGTA [ethylene glycol-bis(beta-aminoethyl ether)-N,N,N′,N′-tetraacetic acid], 4% glycerol, 1 mM β-glycerophosphate, 2 mM sodium orthovanadate, 5 mM dithiothreitol [DTT], 1 mM PMSF) for 30 minutes at 4°C. Clear lysates were incubated with 10 μg GST-PBD fusion protein expressing the Rac1-binding domain of p21-activated kinase 3 (Pak3) and glutathione-Sepharose beads for 45 minutes at 4°C. After extensive washings in lysis buffer, bound Rac1 proteins were detected by anti-HA immunoblotting. Prior to the incubation with beads, 50 μL aliquots were removed from samples to control the expression of total HA-Rac1 proteins.
Results
3BP2 is phosphorylated following BCR stimulation
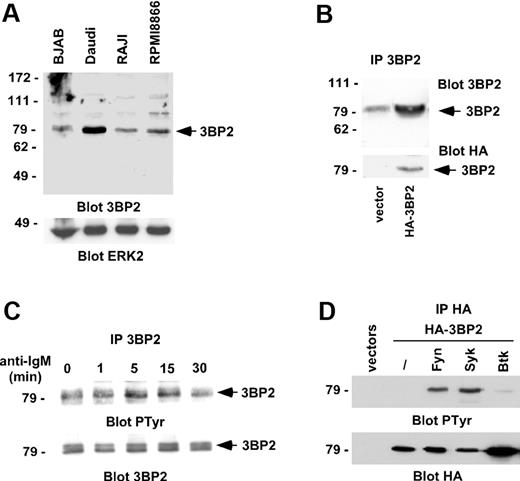
To examine the function of 3BP2 in B cells, we first examined its expression in different B-cell lines. 3BP2 was abundantly expressed in the Burkitt lymphoma cell line, Daudi, and to a lesser extend in BJAB, Raji, and RPMI8866 cells (Figure 1A). The specificity of the polyclonal antibodies against 3BP2 was examined by blotting 3BP2 immunoprecipitates from lysates of Raji cells transfected with HA-tagged 3BP2 cDNA (Figure 1B). We then examined the tyrosine phosphorylation of 3BP2 following BCR engagement. 3BP2 immunoprecipitates from lysates of Daudi cells stimulated with anti-IgM antibodies were analyzed by antiphosphotyrosine immunoblotting. Figure 1C shows that 3BP2 was tyrosine phosphorylated after BCR engagement. As an attempt to identify the kinase or kinases involved in this process, HA-tagged 3BP2 was cotransfected with plasmids encoding Fyn, Syk, or Btk kinases. Western blot analysis of anti-HA immunoprecipitates shows that 3BP2 is tyrosine phosphorylated by Fyn and Syk, but not by Btk in intact cells (Figure 1D), suggesting that 3BP2 may be a substrate of Src- and Syk-, but not Tec-, family kinases in BCR signaling.
3BP2 is phosphorylated by BCR engagement. (A) Lysates of various B-cell lines were analyzed by Western blot for 3BP2 expression. ERK2 expression was used as a control to quantify protein levels in each lane. (B) Daudi cells were transfected with a plasmid encoding HA-tagged 3BP2 or empty vector. HA-immunoprecipitates were then probed with anti-3BP2 or anti-HA antibodies. The membrane was reprobed with anti-HA antibody to control 3BP2 protein expression. (C) BCR cross-linking results in 3BP2 tyrosine phosphorylation. Daudi cells were stimulated for 5 minutes with anti-IgM antibody (2 μg/mL). Immunoprecipitates were prepared with anti-3BP2 antibody and probed with antibodies to phosphotyrosine and 3BP2. (D) COS-1 cells were transiently cotransfected with expression plasmids encoding HA-tagged 3BP2 and Fyn, Syk, or Btk constructs. HA-immunoprecipitates were probed with antiphosphotyrosine antibody. The membrane was reprobed with anti-HA antibody to control 3BP2 protein expression.
3BP2 is phosphorylated by BCR engagement. (A) Lysates of various B-cell lines were analyzed by Western blot for 3BP2 expression. ERK2 expression was used as a control to quantify protein levels in each lane. (B) Daudi cells were transfected with a plasmid encoding HA-tagged 3BP2 or empty vector. HA-immunoprecipitates were then probed with anti-3BP2 or anti-HA antibodies. The membrane was reprobed with anti-HA antibody to control 3BP2 protein expression. (C) BCR cross-linking results in 3BP2 tyrosine phosphorylation. Daudi cells were stimulated for 5 minutes with anti-IgM antibody (2 μg/mL). Immunoprecipitates were prepared with anti-3BP2 antibody and probed with antibodies to phosphotyrosine and 3BP2. (D) COS-1 cells were transiently cotransfected with expression plasmids encoding HA-tagged 3BP2 and Fyn, Syk, or Btk constructs. HA-immunoprecipitates were probed with antiphosphotyrosine antibody. The membrane was reprobed with anti-HA antibody to control 3BP2 protein expression.
Identification of Vav family members as 3BP2 interacting proteins
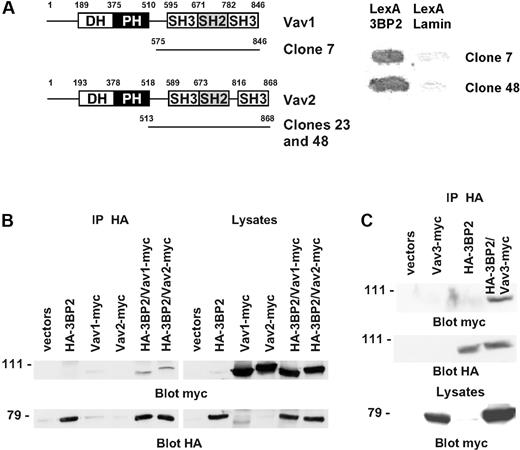
To identify novel partners of 3BP2 in B cells, we employed the yeast 2-hybrid interaction analysis. 3BP2, fused to the DNA-binding domain of LexA, was used to screen a human B-lymphocyte library. Of the clones selected for sequence analysis, one Vav1 and 2 Vav2 clones were identified by comparison with the nucleotide database at the National Library of Medicine using the BLAST algorithm (Figure 2A). To confirm the interaction between 3BP2 and Vav proteins in mammalian cells, COS-1 cells were transiently cotransfected with a combination of plasmids encoding HA-tagged 3BP2, myc-tagged Vav1 and Vav2, or empty vectors. Anti-3BP2 immunoprecipitates were analyzed by Western blot using anti-myc antibody. The membranes were then stripped and 3BP2 expression was controlled by blotting with anti-HA antibody. As shown in Figure 2B, 3BP2 coimmunoprecipitated both with Vav1 and Vav2. Using a similar approach, an association between 3BP2 and Vav3 was also detected (Figure 2C). These data indicate that a constitutive complex between 3BP2 and Vav proteins exists both in yeast and in mammalian cells.
Interaction of 3BP2 with Vav family members. (A) A yeast 2-hybrid screen of a human lymphocyte library using LexA-3BP2 as bait was performed. Shown are the accession number of the different Vav cDNAs isolated during this screen, and the number of independent clones for each cDNA. (B) COS-1 cells were transiently transfected with expression plasmids encoding HA-tagged 3BP2, myc-tagged Vav1, myc-tagged Vav2, or combinations of these plasmids as indicated. Immunoprecipitates were prepared with anti-HA antibody and probed with anti-myc antibody. The membranes were then reprobed with anti-HA antibody to control 3BP2 protein expression. (C) A combination of HA-tagged 3BP2 and myc-tagged Vav3 were expressed in COS-1 cells and coimmunoprecipitations were performed.
Interaction of 3BP2 with Vav family members. (A) A yeast 2-hybrid screen of a human lymphocyte library using LexA-3BP2 as bait was performed. Shown are the accession number of the different Vav cDNAs isolated during this screen, and the number of independent clones for each cDNA. (B) COS-1 cells were transiently transfected with expression plasmids encoding HA-tagged 3BP2, myc-tagged Vav1, myc-tagged Vav2, or combinations of these plasmids as indicated. Immunoprecipitates were prepared with anti-HA antibody and probed with anti-myc antibody. The membranes were then reprobed with anti-HA antibody to control 3BP2 protein expression. (C) A combination of HA-tagged 3BP2 and myc-tagged Vav3 were expressed in COS-1 cells and coimmunoprecipitations were performed.
3BP2 associates Vav1 and other signaling molecules following BCR engagement
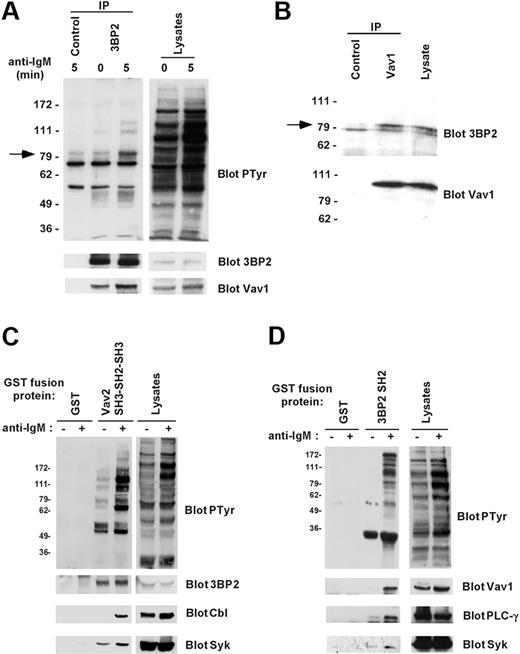
To examine the association between endogenous 3BP2 and Vav proteins in B cells, 3BP2 was immunoprecipitated from lysates of Daudi cells stimulated or not with anti-IgM antibodies. Antiphosphotyrosine blotting analysis showed that following BCR stimulation different phosphoproteins coprecipitated with 3BP2 in resting and anti-IgM–stimulated cells (Figure 3A). By reprobing membranes with antibodies against known tyrosine-phosphorylated proteins with similar molecular mass, we determined that one of these 3BP2-associated phosphoproteins was Vav1. The interaction between 3BP2 and Vav1 was constitutive and increased following B-cell activation. A similar association of 3BP2 with Vav2 was observed in Daudi cells (data not shown). The specificity of these interactions was controlled on immunoprecipitates performed with irrelevant antibodies (Figure 3A). To rule out the possibility that the 3BP2/Vav1 interaction may be indirect due to association of phospho-3BP2 with membrane lipid rafts, unstimulated Daudi cells were solubilized in a buffer containing 1% octylglucoside, and coimmunoprecipitations were performed. As shown in Figure 3B, 3BP2 specifically coprecipitated with Vav1 in detergent conditions under which lipid rafts are dissociated. Similar results were obtained in other B-cell lines including Raji and Ramos (data not shown). To further characterize 3BP2/Vav interaction in B lymphocytes, in vitro GST pull-down assays were next performed. When GST Vav2 SH3-SH2-SH3—but not GST alone—was incubated with lysates from resting or activated Daudi cells, a constitutive binding of 3BP2 was detected, in addition to an inducible binding to Syk and c-Cbl (Figure 3C). A similar approach using GST 3BP2 SH2 showed that the SH2 domain of 3BP2 bound several tyrosine phosphorylated proteins from lysates of BCR-stimulated Daudi B cells (Figure 3D). Bound proteins included important fractions of total Vav1 and PLC-γ, and a significant fraction of Syk. These interactions were not detected on GST precipitates. Together, these data indicate that 3BP2 binds critical molecules of BCR signaling, including Vav proteins.
Binding of 3BP2 to Vav1 in B cells. (A) Interaction between endogenous 3BP2 and Vav1 in B cells. Daudi cells were activated for 0 or 5 minutes with anti-IgM antibody and lysed in NP-40 lysis buffer supplemented with 0.1% SDS. Immunoprecipitates were prepared with anti-3BP2 or irrelevant (control) antibodies and probed with antiphosphotyrosine antibody. The membranes were then stripped and reprobed with antibodies against 3BP2 and Vav1. (B) Interaction between 3BP2 and Vav1 in lipid raft-dissociating conditions. Unstimulated Daudi cells were lysed in 1% octylglucoside lysis buffer, and immunoprecipitates were prepared with anti-Vav1 or irrelevant (control) antibodies and probed with anti-3BP2 antibody. The membranes were then stripped and reprobed with antibodies to Vav1. (C-D) In vitro interaction between 3BP2 and Vav proteins. Lysates from unstimulated (–) or anti-IgM–stimulated (+) Daudi cells were mixed with GST or GST Vav2 SH3-SH2-SH3 fusion proteins (B), or with GST or GST 3BP2 SH2 fusion proteins (C) that were immobilized on glutathione beads. Bound proteins were detected by immunoblotting with the indicated antibodies.
Binding of 3BP2 to Vav1 in B cells. (A) Interaction between endogenous 3BP2 and Vav1 in B cells. Daudi cells were activated for 0 or 5 minutes with anti-IgM antibody and lysed in NP-40 lysis buffer supplemented with 0.1% SDS. Immunoprecipitates were prepared with anti-3BP2 or irrelevant (control) antibodies and probed with antiphosphotyrosine antibody. The membranes were then stripped and reprobed with antibodies against 3BP2 and Vav1. (B) Interaction between 3BP2 and Vav1 in lipid raft-dissociating conditions. Unstimulated Daudi cells were lysed in 1% octylglucoside lysis buffer, and immunoprecipitates were prepared with anti-Vav1 or irrelevant (control) antibodies and probed with anti-3BP2 antibody. The membranes were then stripped and reprobed with antibodies to Vav1. (C-D) In vitro interaction between 3BP2 and Vav proteins. Lysates from unstimulated (–) or anti-IgM–stimulated (+) Daudi cells were mixed with GST or GST Vav2 SH3-SH2-SH3 fusion proteins (B), or with GST or GST 3BP2 SH2 fusion proteins (C) that were immobilized on glutathione beads. Bound proteins were detected by immunoblotting with the indicated antibodies.
In vivo mapping of Vav1/2 binding domain on 3BP2
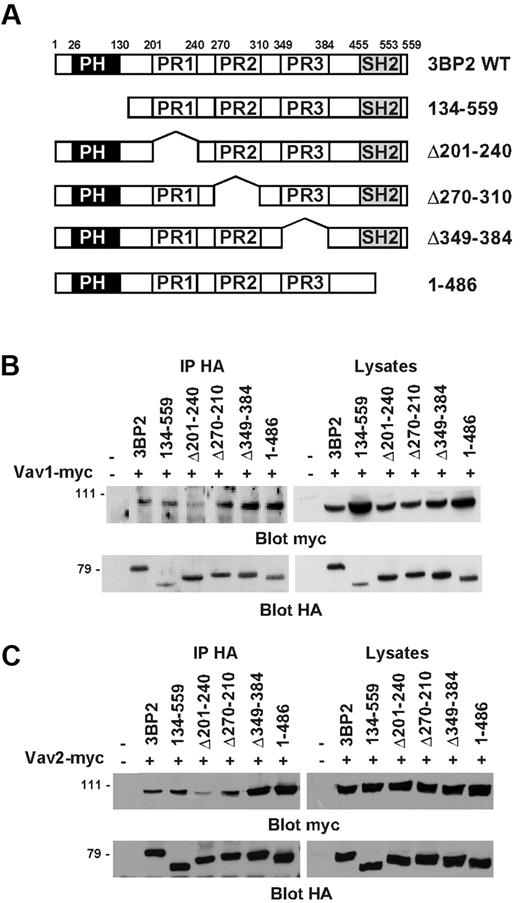
Next, we performed coimmunoprecipitations in COS-1 cells in order to map the region of 3BP2 that binds Vav proteins. 3BP2 is composed of an N-terminal plekstrin homology (PH) domain, a central proline-rich (PR) region with 3 domains (PR1, PR2, PR3) containing SH3 binding consensus sequences,24 and a C-terminal Src homology 2 (SH2) domain. COS-1 cells were cotransfected with plasmids encoding different HA-tagged 3BP2 deletion mutants (Figure 4A), and either myc-tagged Vav1 (Figure 4B) or myc-tagged Vav2 (Figure 4C). HA-immunoprecipitates from unstimulated cells were then probed with anti-myc antibody. As shown in Figure 4B-C, the PH domain, the PR2 and PR3 regions, and the SH2 domain of 3BP2 were not required for interacting with Vav1 or Vav2. In contrast, deletion of PR1 domain (aa 201 to aa 240) significantly decreased the interaction between 3BP2 and Vav proteins. Interestingly, the deletion of either PR3 or SH2 domains apparently reinforced 3BP2/Vav proteins binding, suggesting a conformational regulation of this interaction. Note that identical results were obtained in yeast binding assays (data not shown). These data indicate that, in addition to a phosphotyrosine/SH2 domain–dependent interaction, the first PR domain of 3BP2 (PR1) also binds Vav proteins in intact cells.
Mapping of the constitutive binding domain of Vav1/2 on 3BP2. (A) Schematic structures of the 3BP2 mutants used in these experiments. The PH, proline-rich (PR), and SH2 domains are represented by black, white, and gray boxes, respectively. COS-1 cells were transiently cotransfected with expression plasmids encoding myc-tagged Vav1 (B) or Vav2 (C), and various HA-tagged 3BP2 mutant constructs. Immunoprecipitates were prepared with anti-HA antibody and probed with anti-myc antibody. The membranes were then stripped and reprobed with anti-HA antibody to control 3BP2 protein expression.
Mapping of the constitutive binding domain of Vav1/2 on 3BP2. (A) Schematic structures of the 3BP2 mutants used in these experiments. The PH, proline-rich (PR), and SH2 domains are represented by black, white, and gray boxes, respectively. COS-1 cells were transiently cotransfected with expression plasmids encoding myc-tagged Vav1 (B) or Vav2 (C), and various HA-tagged 3BP2 mutant constructs. Immunoprecipitates were prepared with anti-HA antibody and probed with anti-myc antibody. The membranes were then stripped and reprobed with anti-HA antibody to control 3BP2 protein expression.
3BP2 is involved in BCR-mediated activation of NFATs
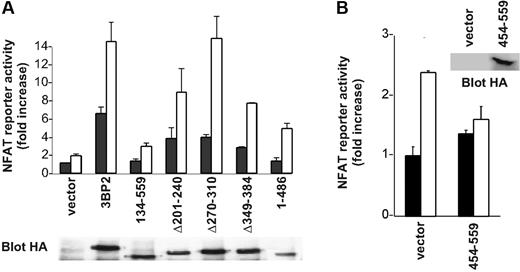
In order to clarify the function of 3BP2 in BCR signaling, we first examined the effect of overexpression of 3BP2 mutants on BCR-mediated NFAT activation. 3BP2 constructs were overexpressed in Daudi cells and NFAT activation was determined using luciferase assays. Figure 5A shows that deleting the SH2 domain of 3BP2 dramatically reduced both basal and anti-IgM–mediated NFAT activation, whereas PR1 deletion led to approximately 50% of diminution of NFAT activities compared with wild-type 3BP2 protein. Other functionally important domains of 3BP2 included PH and PR3 domains, whereas PR2 apparently did not participate in 3BP2-mediated NFAT activation. Overexpression of 3BP2 SH2 domain (3BP2 454-559) in Daudi B cells also decreased the activation of NFATs induced by BCR engagement (Figure 5B).
Involvement of 3BP2 in BCR-mediated activation of NFATs. (A) Daudi cells were transiently transfected with various 3BP2 mutant constructs, NFAT reporter plasmid, and Renilla luciferase plasmid (pRL-TK) used as internal control reporter. At 24 hours after transfection, cells were either left unstimulated, or stimulated with anti-IgM (2 μg/mL) for 6 hours, and harvested for the dual-luciferase assay. The luciferase activity was normalized by the Renilla luciferase activity and expressed as fold increase relative to the basal activity seen in unstimulated mock-transfected cells. Values represent the mean plus or minus the standard deviation from triplicate samples. The lower panel shows the relative expression of each of the 3BP2 constructs. (B) Daudi cells were transfected with an expression plasmid encoding HA-tagged 3BP2 SH2 (aa 454-aa 559), NFAT reporter plasmid, and Renilla luciferase plasmid used as internal control reporter. Cell stimulation and luciferase assay were performed as in panel A.
Involvement of 3BP2 in BCR-mediated activation of NFATs. (A) Daudi cells were transiently transfected with various 3BP2 mutant constructs, NFAT reporter plasmid, and Renilla luciferase plasmid (pRL-TK) used as internal control reporter. At 24 hours after transfection, cells were either left unstimulated, or stimulated with anti-IgM (2 μg/mL) for 6 hours, and harvested for the dual-luciferase assay. The luciferase activity was normalized by the Renilla luciferase activity and expressed as fold increase relative to the basal activity seen in unstimulated mock-transfected cells. Values represent the mean plus or minus the standard deviation from triplicate samples. The lower panel shows the relative expression of each of the 3BP2 constructs. (B) Daudi cells were transfected with an expression plasmid encoding HA-tagged 3BP2 SH2 (aa 454-aa 559), NFAT reporter plasmid, and Renilla luciferase plasmid used as internal control reporter. Cell stimulation and luciferase assay were performed as in panel A.
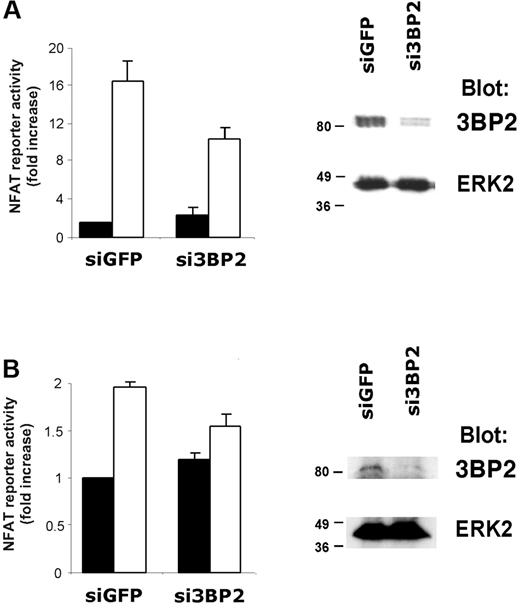
To further confirm the role of 3BP2 in BCR signaling, we used RNA interference (RNAi). Transfection efficiencies are very low in B-cell lines. We therefore first tested siRNA duplexes capable of knocking down 3BP2 expression in transfected Jurkat T cells (Figure 6A). Immunoblot analysis show that siRNA against 3BP2 specifically reduced the expression of 3BP2 protein, compared with control siRNA against GFP (Figure 6A, right panel). Quantification of the immunoblot signals showed that 3BP2 expression was reduced up to 80% (data not shown). NFAT luciferase assays revealed that reducing 3BP2 expression in Jurkat T cells decreased TCR-mediated NFAT activation by 35% to 40% as compared with control (Figure 6A, left panel).
RNA interference of 3BP2 decreases BCR signaling to NFAT. Jurkat (A) or Raji (B) cells were transfected with 3BP2 siRNA (si3BP2) or control siRNA (siGFP) in the presence of fluorescein-conjugated control siRNA. After 24 hours, cells were sorted by FACS, lysed, and protein expression was determined by immunoblotting with antibodies against 3BP2 and ERK2 (right panels). For determination of NFAT activity in 3BP2 knock-down cells, Jurkat (A) or Raji (B) cells were electroporated with 2 μg siRNA, 15 μg NFAT reporter, and 3 μg Renilla luciferase plasmid. After 18 hours, cells were stimulated or not for 6 hours and luciferase activity was measured and normalized (left panels). Values represent the mean plus or minus the standard deviation from triplicate samples.
RNA interference of 3BP2 decreases BCR signaling to NFAT. Jurkat (A) or Raji (B) cells were transfected with 3BP2 siRNA (si3BP2) or control siRNA (siGFP) in the presence of fluorescein-conjugated control siRNA. After 24 hours, cells were sorted by FACS, lysed, and protein expression was determined by immunoblotting with antibodies against 3BP2 and ERK2 (right panels). For determination of NFAT activity in 3BP2 knock-down cells, Jurkat (A) or Raji (B) cells were electroporated with 2 μg siRNA, 15 μg NFAT reporter, and 3 μg Renilla luciferase plasmid. After 18 hours, cells were stimulated or not for 6 hours and luciferase activity was measured and normalized (left panels). Values represent the mean plus or minus the standard deviation from triplicate samples.
We next transiently transfected Raji B cells with si3BP2 or siGFP oligonucleotides together with NFAT-driven luciferase reporter. The cells were stimulated or not with anti-IgM antibodies, and normalized luciferase activities were measured. As shown in Figure 6B (left panel), interfering with 3BP2 expression in B cells resulted in a significant reduction of BCR-mediated NFAT activation (20% to 25% reduction compared with control). Immunoblot analysis on the siRNA-transfected population of Raji cells show that 3BP2 expression was abrogated by si3BP2 (Figure 6B, right panel). Together with overexpression experiments, these data suggest that 3BP2 participates in BCR-mediated transcriptional activation.
Functional cooperation between 3BP2 and Vav family proteins for Rac and NFAT activation
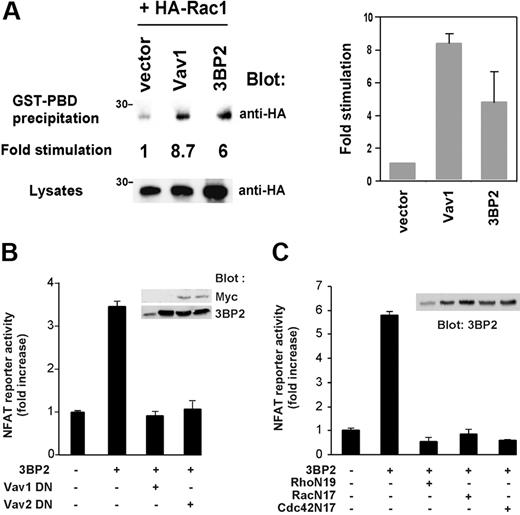
Vav proteins are important regulators of Rho GTPases and NFAT activities in lymphoid cells,7,8 and we previously found that overexpression of 3BP2 leads to activation of NFATs in T cells.18 We thus questioned a role of 3BP2 in Rac activation and whether 3BP2/Vav protein interaction participate in BCR-mediated NFAT activation. For the same technical limitation described in Figure 6, we used Jurkat T cells to test the effect of 3BP2 overexpression on Rac1 activity using a Rac pull-down assay.29 Jurkat T cells were transfected with HA-tagged Rac1 together with 3BP2 or Vav1 cDNAs, then lysed and incubated with GST-PDB fusion protein and glutathione-Sepharose beads. Activated Rac1 was then detected by anti-HA immunoblotting. As shown in Figure 7A, 3BP2 activated Rac1 (6-fold increase compared with basal activity). Under the same conditions, overexpression of Vav1 led to an 8-fold increase of Rac1 activity. We next examined whether 3BP2 required the activity of Vav proteins and Rho GTPases to activate NFATs in B cells. Overexpression of GEF-inactive forms of Vav1 or Vav2 (L213A and L212A, respectively; Figure 7B), and of dominant-negative mutant forms of RhoA, Rac1, and Cdc42 (RhoN19, RacN17, and Cdc42N17, respectively; Figure 7C) blocked 3BP2-induced NFAT activation in Daudi B cells. These data suggest that 3BP2 acts upstream of Vav and Rho proteins in the signaling pathways required for gene activation by BCR.
Functional cooperation between 3BP2, Vav proteins, and Rho GTPases during NFAT activation. (A) 3BP2 activates Rac1 in Jurkat T cells. Cells were transfected with the indicated plasmids. After 24 hours, active forms of HA-Rac1 were captured by incubation of lysates with a GST-PDB fusion protein and glutathione beads. After extensive washings, bound HA-Rac1 proteins were detected by anti-HA immunoblotting. Expression of total HA-Rac1 was assessed on aliquots of cell lysates. Fold stimulation of GTP-bound HA-Rac1 was determined following normalization against total HA-Rac1 (left panel). The average of 2 representative experiments is shown (right panel). Daudi cells were cotransfected with 5 μg NFAT luciferase reporter and HA-tagged 3BP2 in the presence of either (B) 30 μg Vav1 L213A (Vav1 DN) or Vav2 L212A (Vav2 DN) or (C) 30 μg Rho N19, Rac N17, or Cdc42 N17, and luciferase activity was measured. Values represent the mean plus or minus the standard deviation from triplicate samples. Samples from the same lysates were immunoblotted with anti-myc or anti-3BP2 antibodies (insets).
Functional cooperation between 3BP2, Vav proteins, and Rho GTPases during NFAT activation. (A) 3BP2 activates Rac1 in Jurkat T cells. Cells were transfected with the indicated plasmids. After 24 hours, active forms of HA-Rac1 were captured by incubation of lysates with a GST-PDB fusion protein and glutathione beads. After extensive washings, bound HA-Rac1 proteins were detected by anti-HA immunoblotting. Expression of total HA-Rac1 was assessed on aliquots of cell lysates. Fold stimulation of GTP-bound HA-Rac1 was determined following normalization against total HA-Rac1 (left panel). The average of 2 representative experiments is shown (right panel). Daudi cells were cotransfected with 5 μg NFAT luciferase reporter and HA-tagged 3BP2 in the presence of either (B) 30 μg Vav1 L213A (Vav1 DN) or Vav2 L212A (Vav2 DN) or (C) 30 μg Rho N19, Rac N17, or Cdc42 N17, and luciferase activity was measured. Values represent the mean plus or minus the standard deviation from triplicate samples. Samples from the same lysates were immunoblotted with anti-myc or anti-3BP2 antibodies (insets).
Discussion
In this study, we analyzed the role of the cytoplasmic adaptor 3BP2 (also known as SH3BP2) in BCR signaling. We provide biochemical and functional evidence that 3BP2 expression in B cells plays an important role in BCR-mediated activation of NFAT transcriptional activities through its interaction with members of the Vav family of Rho GTPase activators.
Immunoreceptor engagement on leukocytes activates nonreceptor tyrosine kinases of the Src, Syk, and Tec families, which promote the assembly of signaling complexes composed of key enzymes connected by adaptor proteins.15 3BP2 is an ubiquitous adaptor with preferential expression in hematopoietic cells,18,20 whose function remains ill-defined. However, both its original cloning as a binding protein of Abl SH3 domain,19 its subsequent isolation as a Syk-kinase interacting protein in lymphoid cells,18 and recent works in NK and mast cells,21,22 suggested an important role in hematopoietic cells. In further support to this idea, 3BP2 was shown here to play a direct and active role in BCR signaling. 3BP2, which is abundantly expressed in various B-cell lines (Figure 1) and normal B lymphocytes (data not shown), is rapidly and transiently tyrosine phosphorylated after BCR aggregation. The kinase responsible for this phosphorylation was not identified, but we show that 3BP2 is tyrosine phosphorylated following expression of Fyn and Syk, but not Btk, in COS cells. 3BP2 contains several tyrosine phosphorylation sites, one of which (Y183LEP) is predicted to be a Vav1 and PLC-γ binding motif, 2 proteins found to interact with 3BP2 in stimulated B cells. The function of 3BP2 in FcR signaling in NK and mast cells apparently involves its tyrosine phosphorylation by kinases of the Src and Syk families,21,22 and 3BP2 binds the Src PTKs Fyn18 and Lyn.23 Together these observations suggest that 3BP2 is a critical partner and substrate of Src and Syk PTKs in the signaling pathways of multiple immunoreceptors, including BCRs.
To gain understanding of 3BP2's role in BCR signaling, we screened a yeast 2-hybrid B-cell library for 3BP2 interacting proteins. 3BP2 was found to constitutively interact with Vav1 and Vav2, 2 guanine nucleotide exchange factors for Rho family GTPases which play a critical role in the development and growth of T and B lymphocytes.8-10,30 The third member of the Vav family, Vav3, was not found during our screen. However, we found that it coimmunoprecipitates with 3BP2, suggesting that 3BP2 is common partner of Vav proteins in hematopoietic cell signaling. The mapping of the 3BP2-Vav1/2 interaction indicates that it involves a PR domain of 3BP2 (aa 201 to aa 240 or PR1) corresponding to the previously identified Abl SH3 binding site.19 Studies have shown that Vav also associates Bcr-Abl in leukemic cells.31 An interesting issue would be to examine whether the occupation of PR1 domain by Vav proteins could prevent the simultaneous binding of Abl SH3 domain to 3BP2 and how this could affect cell signaling.
The interaction between 3BP2 and Vav proteins appears far more complex since 3BP2 SH2 domain binds tyrosine phosphorylated Vav proteins from BCR-stimulated B lymphocytes (Figure 3). Importantly, 3BP2 and Vav1 also coprecipitate in alkyl-glycosidic detergent conditions under which membrane lipid rafts are dissociated,32 showing that this interaction is not due to indirect associations with rafts. A predicted binding sequence for 3BP2 SH2 domain was determined to be YEN, a motif found in LAT but not within the Vav1 and Vav2 sequences. Further studies are thus required to determine the location of the 3BP2 SH2 binding site on Vav proteins. Recently, Jevremovic et al21 described that Vav1 and PLC-γ SH2 domains associate 3BP2 upon phosphorylation of its tyrosine residue 183 in activated NK cells. Tyr183 lies within the tyrosine phosphorylation site Y183LEP, which constitutes a Vav1 and PLC-γ binding motif, according to previous studies.33 We also detected a Vav2 SH2-dependent interaction with tyrosine phosphorylated 3BP2 in B cells, in addition to the constitutive, SH3-dependent interaction. Interestingly, mutation of 3BP2 Tyr183 indicates that this residue is required for optimal adaptor function of 3BP2 in B cells (I.F. and M.D., unpublished data) and for FcR-mediated NK cell cytotoxicity.21 Thus, there are apparently 3 different mechanisms of interaction between 3BP2 and Vav proteins: first, a basal interaction mediated by a PR domain of 3BP2 and the SH3 domain(s) of Vav proteins; second, an activation-induced interaction between the SH2 domain of 3BP2 and phosphorylated Vav1/2; and third, an interaction between tyrosine phosphorylated 3BP2 and the SH2 domain of Vav1. This multiplicity of interactions could explain why the deletion of the PR domain PR1 only partially reduced 3BP2-induced NFAT activation in B cells (Figure 5) and has almost no effect in T cells.18 Alternatively, this domain of 3BP2 could be implicated in distinct cellular functions through other molecular partners like c-Abl, Fyn, or Lyn.
Finally, what could be the function of the 3BP2/Vav complex in B cells? Several observations point to a role in a PLC-γ–dependent pathway involved in NFAT activation. First, Vav proteins play pivotal roles in PLC-γ, calcium, and NFAT regulation, and the major defect of vav1–/– cells is a reduced PLC-γ activation and calcium movements following immunoreceptor stimulation. Similar to Vav1/2 proteins,8 3BP2 overexpression induced NFAT activation in B cells. Consistent with the reduced NFAT activities seen in 3BP2 knock-down T and B cells (Figure 6), expression of 3BP2 SH2 domain decreased NFAT activation both in T18 and B cells (Figure 5). 3BP2 interacts with several proteins involved in PLC-γ pathways, including Syk and PLC-γ itself. In T, NK, and basophilic cells, 3BP2 SH2 domain also associated phosphorylated LAT, a 36 kDa to 38 kDa raft-enriched adapter protein required for PLC-γ signaling.34 Thus, the function of 3BP2 in BCR signaling shares similarities with those required for PLC-γ activation in other immunoreceptor-stimulated hematopoietic cells. Interestingly, pull-down assays show that 3BP2 interacted with an unidentified 30 kDa to 35 kDa phosphorylated protein in activated B cells. Whether this protein could be LAB/NTAL, the counterpart of LAT in B cells,35,36 remains to be determined. Vav proteins also bind BLNK/SLP-65, a central adaptor of B-cell signaling.16 Importantly, we did not observe the presence of BLNK in the 3BP2/Vav complex (data not shown), suggesting that 3BP2 coordinates the assembly of distinct molecular complexes involved in B-cell signaling. Finally, 3BP2-dependent NFAT activation in B cells is blocked by GEF inactive forms of Vav1 and Vav2, and by dominant-negative mutants of Rho GTPases, including Rac1. Rac1 has been involved in the regulation of NFAT activities in lymphocytes,37,38 and we show here that 3BP2 can increase Rac1 activity in T cells (Figure 7). Because Rho GTPases regulate a broad range of cellular functions, one could speculate that an important function of 3BP2 in hematopoietic cells is to promote the assembly of signaling complexes implicated in gene regulation through Vav proteins and Rho GTPases.
Prepublished online as Blood First Edition Paper, September 2, 2004; DOI 10.1182/blood-2003-08-2965.
Supported by INSERM, the Ministère de l'Enseignement et de la Recherche, and the Association pour la Recherche sur le Cancer (grant 4756 to M.D.). I.F. was a recipient of a fellowship from the Association pour la Recherche sur le Cancer. I.F. and S.L.B. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank T. Trenkle and M. McClelland for the Vav3 cDNA; E. Lemichez and P. Boquet for HA-tagged Rac1 expression plasmids; and M. Ticchioni and S. Tartare-Deckert for discussion.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal