Abstract
CD8+ T-cell responses are an essential antiviral host defense in persistent viral infections, and their sustained effectiveness is thought to be critically dependent on CD4+ T-helper cells. To determine the relationship between HIV-1–induced CD4+ T-cell depletion and hepatitis C virus (HCV)–specific CD8+ T-cell responses during viral persistence, we studied 103 persons positive for HCV, 74 coinfected with HIV-1. CD8+ T-cell responses to the entire HCV polyprotein were determined by using an interferon-γ enzyme-linked immunospot (ELISpot) assay. Although HIV-1 infection by itself was not associated with a diminished HCV-specific response, HIV-1–associated CD4+ depletion was associated with significantly lower HCV-specific CD8+ T cells (R = 0.48, P < .0001). In contrast, declining CD4+ counts over the same range were not associated with diminished Epstein-Barr virus (EBV)– (R = 0.19, P = .31) or HIV-1–specific (R = –0.13, P = .60) CD8+ T-cell responses in persons infected with all viruses. These data indicate that frequencies of circulating HCV-specific CD8+ T-cell responses are sensitive to absolute CD4+ T-cell counts and provide a possible explanation for the accelerated HCV disease course in persons coinfected with HIV-1 and HCV.
Introduction
Hepatitis C virus (HCV) and human immunodeficiency virus type 1 (HIV-1) share routes of transmission, are often harbored within the same host, and establish chronic infections characterized by high plasma viral loads. In humans coinfected with HCV and HIV-1, the disease course for HCV infection is accelerated particularly at lower CD4+ counts, with higher HCV viral loads, and more rapid liver fibrosis, although the mechanisms for this differential pathogenicity are unclear.1,2 HCV has thereby emerged as an important cause of morbidity and mortality in persons infected with HIV-1. Since responses to standard anti-HCV therapy remain suboptimal, particularly for individuals infected with HIV-1, novel approaches such as immunotherapy are needed for this population.3 Given this need for novel therapies, a better understanding of disease pathogenesis in coinfected persons is of paramount importance.
The role of virus-specific immune responses in both HCV infection and HIV-1 infection remains controversial. In both, early containment (HIV-1) or clearance (HCV) of infection is associated with induction of strong CD4+ and CD8+ T-cell responses (reviewed in McMichael and Rowland-Jones4 and Racanelli and Rehermann5 ). However, these same responses have been postulated to play a role in disease pathogenesis, due to accelerated destruction of hepatocytes in the case of HCV and lymphocytes in the case of HIV-1.6-11 Some have proposed that the phenomenon of accelerated fibrosis in individuals coinfected with HCV and HIV-1 may be related to loss or dysregulation of these immuneresponses.12,13
In murine models of persistent viral infection, CD4+ T cells are vital for maintenance of cytotoxic T lymphocyte (CTL) effector functions such as cytokine secretion and secondary expansion upon encounter with antigen, and the development of virus-specific memory responses is impaired in the absence of CD4+ T cells at the time of exposure.14,15 Moreover, recent depletion experiments in chimpanzees highlight the importance of both CD4+ and CD8+ T cells in control of HCV viremia and illustrate the vital role of CD4+ cells in supporting HCV-specific CTLs.16,17 Such observations raise the possibility that CD4+ cell depletion that accompanies progressive HIV-1 infection might lead to loss of critical adaptive immune responses to HCV in coinfected persons, and thereby facilitate HCV disease progression.
In the present study, we quantified CD8+ T-cell responses to HCV, HIV-1, and Epstein-Barr virus (EBV) to test the hypothesis that CD4+ cell depletion would be associated with impaired virus-specific immune responses. We compared breadth, magnitude, and specificity of HCV-specific CD8+ T-cell responses between individuals monoinfected with HCV and individuals coinfected with HCV and HIV-1. Next, we determined the effect of CD4+ T-cell depletion secondary to HIV-1 on the frequency of HCV-specific CD8+ T cells, as measured by ex vivo CD8+ T lymphocyte interferon-γ (IFN-γ) responses to a comprehensive set of potential antigens spanning the whole expressed genome of HCV. Using parallel techniques, we also determined the responses to HIV-1 and EBV. Overall, we find a highly significant correlation between the absolute CD4+ T-cell count and the frequency of CD8+ T cells against HCV but not those against HIV-1 and EBV over the same range of CD4+ T cells.
Patients, materials, and methods
Study subjects
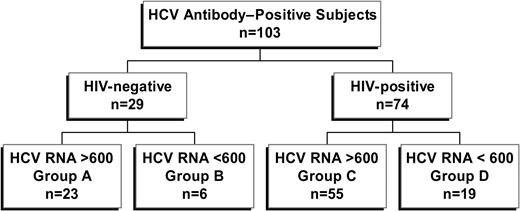
Study subjects were recruited from the Massachusetts General Hospital and the Lemuel Shattuck Hospital. The local institutional review board at each institution approved this study, and all individuals gave written informed consent according to the ethical guidelines of the 1975 Declaration of Helsinki. Peripheral blood samples were obtained from a total of 103 individuals positive for HCV antibodies by third-generation enzyme immunoassay (EIA 3.0; Abbott Diagnostics, Abbott Park, IL) and investigated in this study. Twenty-nine study subjects were negative for antibodies against HIV-1 and thus considered HCV monoinfected. Seventy-four were antibody positive for HIV-1 and HCV and were considered coinfected. Each group was further subdivided into spontaneous controllers of HCV (defined as HCV RNA < 600 IU/mL; Figure 1). Clinical and demographic information is summarized in Table 1. To exclude the possibility that treatment with interferon-α for chronic HCV altered HCV-specific responses, subjects with a history of such treatment were excluded, whereas subjects taking antiretroviral therapy were included. To ensure a relative steady state for CD4+ count and other parameters, inclusion criteria dictated that no changes in antiretroviral therapy occurred during the 3 months prior to analysis.
Patient characteristics of 103 individuals infected with HCV
. | Group A . | Group B . | Group C . | Group D . |
|---|---|---|---|---|
| No. | 23 | 6 | 55 | 19 |
| % IVDU | 68 | 67 | 87 | 67 |
| Prior interferon | None | None | None | None |
| Mean age, y (range) | 44 (19-59) | 45 (39-50) | 45 (28-56) | 44 (33-56) |
| % men | 75 | 67 | 61 | 76 |
| % white | 83 | 100 | 61 | 71 |
| Median HCV VL, IU/mL*,† | 454 000 | < 600 | 1 620 000 | < 600 |
| % genotype 1 | 60 | NA | 72 | NA |
| Mean ALT, U/L (range)† | 75 (17-201) | 19 (4-27) | 75 (6-342) | 37 (10-95) |
| Biopsy, inflammatory score | 4.5¶ | NA | 5.4# | NA |
| Biopsy, fibrosis score | 2.5¶ | NA | 2.6# | NA |
| CD4+/mm3 (range)‡,§ | 940 (227-1677) | 1267 (772-1887) | 526 (64-1340) | 521 (4-1430) |
| CD8+/mm3 (range)‡,§ | 434 (41-784) | 692 (303-1361) | 980 (267-2514) | 1033 (510-2325) |
| CD4+/CD8+ ratio‡,§ | 2.4 | 2.1 | 0.61 | 0.66 |
| ART status (%)∥ | NA | NA | 26/55 (47) | 14/19 (74) |
| Median HIV-1 VL, copies/mL∥ | NA | NA | 3170 | < 50 |
| % HIV-1 VL < 50 copies/mL (%) | NA | NA | 17/55 (31) | 11/21 (52) |
. | Group A . | Group B . | Group C . | Group D . |
|---|---|---|---|---|
| No. | 23 | 6 | 55 | 19 |
| % IVDU | 68 | 67 | 87 | 67 |
| Prior interferon | None | None | None | None |
| Mean age, y (range) | 44 (19-59) | 45 (39-50) | 45 (28-56) | 44 (33-56) |
| % men | 75 | 67 | 61 | 76 |
| % white | 83 | 100 | 61 | 71 |
| Median HCV VL, IU/mL*,† | 454 000 | < 600 | 1 620 000 | < 600 |
| % genotype 1 | 60 | NA | 72 | NA |
| Mean ALT, U/L (range)† | 75 (17-201) | 19 (4-27) | 75 (6-342) | 37 (10-95) |
| Biopsy, inflammatory score | 4.5¶ | NA | 5.4# | NA |
| Biopsy, fibrosis score | 2.5¶ | NA | 2.6# | NA |
| CD4+/mm3 (range)‡,§ | 940 (227-1677) | 1267 (772-1887) | 526 (64-1340) | 521 (4-1430) |
| CD8+/mm3 (range)‡,§ | 434 (41-784) | 692 (303-1361) | 980 (267-2514) | 1033 (510-2325) |
| CD4+/CD8+ ratio‡,§ | 2.4 | 2.1 | 0.61 | 0.66 |
| ART status (%)∥ | NA | NA | 26/55 (47) | 14/19 (74) |
| Median HIV-1 VL, copies/mL∥ | NA | NA | 3170 | < 50 |
| % HIV-1 VL < 50 copies/mL (%) | NA | NA | 17/55 (31) | 11/21 (52) |
Group A was monoinfected, HCV chronic, HIV-1 Ab negative, and HCV Ab positive with HCV RNA greater than 600. Group B was monoinfected, HCV controller, HIV-1 Ab negative, and HCV Ab positive with HCV RNA fewer than 600. Group C was HIV-1+, HCV chronic, HIV-1 Ab positive, and HCV Ab positive with HCV RNA greater than 600. Group D was HIV-1+, HCV controller, HIV-1 Ab positive, and HCV Ab positive with HCV RNA fewer than 600. Cutoff for significance was P < .05, two-tailed. No patient had a prior history of interferon treatment for chronic hepatitis C. Biopsy scores are tabulated on the Ishak staging system.18 The maximum inflammatory score is 18, and the maximum fibrosis score is 6. Genotype data were unavailable for 12 subjects in group C. Ab indicates antibody; VL, viral load; IVDU, intravenous drug use; IU, international units; ALT, alanine aminotransferase; ART, antiretroviral therapy; NA, not applicable
Significant difference between groups A and C.
Significant difference between groups A and C (RNA > 600) and groups B and D (RNA < 600).
Significant difference between groups A and B (all HIV-1 negative) versus groups C and D (all HIV-1 positive).
Data regarding T-cell subset frequencies were collected for 26 of 29 HCV monoinfected individuals.
Significant difference between groups C and D.
n = 18.
n = 21.
Viral load quantification and HCV genotyping
Plasma viral loads were measured using either the Roche Amplicor Monitor assay (detection limit of 400 HIV-1 RNA copies/mL, 600 HCV RNA IU/mL; Roche Molecular Systems, Branchburg, NJ) or the Roche Ultradirect assay (detection limit of 50 HIV-1 RNA copies/mL plasma), according to the manufacturer's specifications. HCV genotypes were determined by INNO-LiPA HCV II (Bayer, Tarrytown, NY).
HLA typing
HLA genotyping was performed by Dynal Biotech (Bromborough, United Kingdom) and the Massachusetts General Hospital Tissue Typing Laboratory (Boston, MA), using standard serologic and molecular techniques.
Synthetic peptides
Peptides corresponding to the HCV strain H77, spanning the entire HCV polyprotein, were synthesized as free acids using the 9-fluorenylmethoxy-carbonyl method (Research Genetics, Huntsville, AL). A total of 407 peptides, ranging in length from 15 to 20 amino acids (aa) and overlapping by 10 aa, were adapted at the C-terminal end to exclude certain residues rarely found at this anchor position of class I–restricted epitopes, since these peptides may be marginally better at detecting CD8+ responses.19 Additionally a set of 87 class I–restricted optimal HCV epitopes ranging in size from 8 to 11 aa were tested, to correspond with the overlapping peptide data.20 For HIV-1 responses, an overlapping peptide set of 410 synthetic HIV-1 peptides, ranging in length from 16 to 19 aa and overlapping by 10 aa, spanning all expressed HIV-1 proteins was used (Applied Biosystems, Foster City, CA). Full genome approaches are not feasible for EBV due to its large size; therefore a comprehensive set of 80 EBV previously defined CD8+ epitopes divided into pools was used (Tonia Woodberry, Leah Henry, Todd Suscovich, Fred Wang, B.D.W., David T. Scadden, and Christian Brander, manuscript submitted, November 2004).
Single cell ELISpot assay for IFN-γ–secreting cells
ELISpot (enzyme-linked immunospot) assays were performed exactly as previously described.21 Briefly, fresh or previously frozen peripheral blood mononuclear cells (PBMCs) were added at a concentration of 200 000 cells/well in polyvinylidene 96-well plates (Millipore, Billerica, MA). Peptides were added directly to the wells at a final concentration of 14 μg/mL. The plates were incubated for 14 to 16 hours at 37°C, 5% CO2. Plates were washed, labeled with 0.25 μg/mL biotin-labeled anti–IFN-γ (Endogen, Rockford, IL), developed by incubation with streptavidin-alkaline phosphatase (Bio-Rad, Hercules, CA), followed by incubation with 5-bromo-4-chloro-indolylphopsphate (BCIP)–nitroblue tetrazolium (NBT; Bio-Rad) in Tris (tris(hydroxymethyl)aminomethane) buffer (pH 9.5). The reaction was stopped by washing with tap water, plates were dried overnight, prior to counting on an automated ELISpot reader (AID, Strassberg, Germany). Responses were considered positive if the number of spots per well minus the background was at least 50 spot-forming cells (SFC)/106 PBMCs with a background of fewer than 15 SFC/106 PBMCs.22,23 Phytohemagglutinin (PHA) served as a positive control. All HCV-specific ex vivo responses were evaluated at least in duplicate. HIV-1–specific responses were determined comprehensively in analogous manner as previously described, using interferon-γ secretion as a readout, with input cells of 50 000 to 100 000 PBMCs/well.22-24 The set of EBV class I optimal epitopes was also divided into pools and analyzed via ELISpot as a surrogate for the EBV-specific CD8+ T-cell response.20
Intracellular cytokine assays
Intracellular cytokine staining (ICS) for IFN-γ was performed as described previously.21,25 Briefly, 106 PBMCs or cells from peptide-stimulated lines were incubated with 4 μM peptide and anti-CD28 and anti-CD49d monoclonal antibodies (1 μg/mL each; Becton Dickinson, San Jose, CA) at 37°C and 5% CO2 for 1 hour before the addition of brefeldin A (1 μL/mL; Sigma-Aldrich, St Louis, MO). Cells were incubated for an additional 5 hours at 37°C and 5% CO2. Cells were then washed and stained with surface antibodies: allophycocyanin (APC)–conjugated anti-CD8 and phycoerythrin (PE)–conjugated anti-CD3 (Becton Dickinson) at 4°C for 30 minutes. After washing, cells were fixed and permeabilized (Caltag, Burlingame, CA), and fluorescein isothiocyanate (FITC)–conjugated anti–IFN-γ monoclonal antibody (Becton Dickinson) was added. Cells were then washed, collected using a FACSCalibur flow cytometer, and analyzed using CellQuest software (BD Biosciences, San Jose, CA).
Class I tetramer assays
Analysis and quantification of CD8+ T-cell responses were determined by using HLA class I peptide tetrameric complexes (“tetramers”) incorporating CD8+ T-cell epitopes. Tetramers were synthesized as previously described26 and included 3 epitopes restricted by HLA-A2 (NS3 peptide 1073-1081 CINGVWCTV7 ; NS4 peptide 1406-1415, KLVALGINAV27 ; NS5B peptide 2594-2603, ALYDVVTKL26 ) and 4 additional epitopes restricted by other class I alleles: HLA A1 (NS3 peptide 1435-1443 ATDALMTGY21 ), HLA B7 (core 41-49 GPRLGVRAT6 ), HLA B8 (NS3 1395-1403 HSKKKDEL6 ), and HLA B35 (NS3 1359-1367 HPNIEEVAL28 ). PBMCs (0.5-1 million) were stained as described.26 Briefly, tetramer staining was performed for 20 minutes at 37°C. After washing for 5 minutes with phosphate-buffered saline (PBS) containing 1% fetal calf serum (FCS) at room temperature, cells were pelleted and directly stained with combinations of the following antibodies: CD8–peridinin chlorophyll A protein (PerCP), CD3-FITC (Becton Dickinson). Flow cytometric analysis was performed with a BD fluorescence-activated cell sorter (FACS Calibur), and data analysis was performed using CellQuest software. Staining was considered positive if tetramer-positive cells formed a cluster distinct from the tetramer-negative CD8+ T-cell population, and the frequency of tetramer positive cells was more than 0.02% of the total CD8+ population.
Bulk stimulation of peripheral blood mononuclear cells
Polyclonal CD8+ T-cell lines were established from cryopreserved or fresh PBMCs (4-10 × 106) stimulated with 1 μg/mL synthetic HCV peptide and 0.5 μg/mL costimulatory antibodies anti-CD28 and anti-CD49d (Becton Dickinson) in R10 medium. Irradiated feeder cells (20 × 106 allogeneic PBMCs) were added to the culture in a 25-cm2 flask (Costar, Cambridge, MA). The culture was kept at 37°C, and recombinant interleukin-2 (IL-2; 25 IU/mL) was added on day 2 and twice a week thereafter. Lines were harvested for further analysis after day 9.
Statistical analysis
Nonparametric Mann-Whitney or Kruskal-Wallis tests, Spearman correlations, and linear regression analyses were performed using GraphPad Prism 3.0 software (GraphPad Software, San Diego, CA). To avoid overestimation of the total breadth and magnitude of antiviral responses, responses to adjacent overlapping peptides were counted as responses to 1 epitopic region, with only the stronger response used for calculating cumulative magnitude.22,23
Results
Study population
A total of 103 subjects positive for HCV were divided into 4 different groups for these studies, stratified by HIV-1 infection status and HCV viral load (Figure 1). These included HCV monoinfected subjects with persistent (group A, n = 23) or controlled (group B, n = 6) HCV viremia, as well as persons coinfected with HCV and HIV-1 with persistent (group C, n = 55) or controlled (group D, n = 19) HCV viremia. Clinical and demographic data of these 4 subject groups are listed in Table 1. Significantly higher CD4+ cell counts were observed when comparing HCV monoinfected persons (groups A and B) to persons coinfected with HCV and HIV-1 (groups C and D, median, 986 versus 475, respectively; P < .0001, Mann-Whitney), whereas CD8+ T-cell counts were lower in persons with HCV monoinfection compared with HCV and HIV-1 coinfection (median, 444 versus 911, respectively; P < .0001). There was substantial overlap in these 2 parameters observed between these 2 groups. In persons with chronic HCV viremia (groups A and C), HIV-1 infection was associated with an overall higher median HCV viral load (P = .03, Mann-Whitney), while viral loads were similar in coinfected subjects taking or not taking antiretroviral therapy. Age, sex, race, and HCV genotype did not differ among groups, nor was there a statistically significant difference between groups regarding HLA class I (data not shown).
Comprehensive ELISpot frequently detects circulating HCV- and HIV-1–specific CD8+ T lymphocytes that secrete interferon-γ in coinfected patients
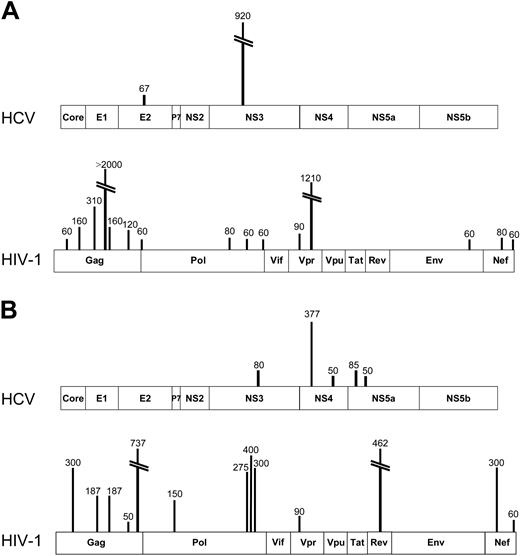
To determine the relative magnitude of immune responses to HCV and HIV-1 in coinfected persons, we assessed antigen-specific CD8+ T-cell responses using a matrix ELISpot assay21-23 and peptides spanning all expressed HCV and HIV-1 proteins. For HCV, we used peptides representing the HCV 1a strain, and, for HIV-1, we synthesized peptides corresponding to the consensus clade B virus strain. Representative data showing breadth, magnitude, and specificity of responses are shown for 2 coinfected subjects in Figure 2A-B.
Sample comprehensive maps of responses to HCV and HIV-1 in 2 subjects coinfected with HCV and HIV-1. (A) From subject D7-02-18 (HCV VL, < 600 IU/mL; HIV-1 VL, < 50 copies/mL on antiretroviral therapy; CD4 T-cell count, 533/mm3), ELISpot detected 2 ex vivo specificities against HCV, 1 each in E2 and NS3, both previously described epitopes. Therefore, the cumulative magnitude of CD8+ IFN-γ–secreting cells was 987 SFC/106 PBMCs or 0.099% of circulating PBMCs. In contrast, 15 specificities were detected against HIV-1, totaling 4610 SFC/106 PBMCs or 0.46% of circulating PBMCs. (B) In subject C27-02M (HCV VL, 405 000 IU/mL; HIV-1 VL, 15 000 copies/mL; CD4 T-cell count, 528/mm3), ELISpot revealed 5 targeted HCV epitopes, all representing novel specificities, while HIV-1 responses also exceeded those against HCV both in breadth and magnitude.
Sample comprehensive maps of responses to HCV and HIV-1 in 2 subjects coinfected with HCV and HIV-1. (A) From subject D7-02-18 (HCV VL, < 600 IU/mL; HIV-1 VL, < 50 copies/mL on antiretroviral therapy; CD4 T-cell count, 533/mm3), ELISpot detected 2 ex vivo specificities against HCV, 1 each in E2 and NS3, both previously described epitopes. Therefore, the cumulative magnitude of CD8+ IFN-γ–secreting cells was 987 SFC/106 PBMCs or 0.099% of circulating PBMCs. In contrast, 15 specificities were detected against HIV-1, totaling 4610 SFC/106 PBMCs or 0.46% of circulating PBMCs. (B) In subject C27-02M (HCV VL, 405 000 IU/mL; HIV-1 VL, 15 000 copies/mL; CD4 T-cell count, 528/mm3), ELISpot revealed 5 targeted HCV epitopes, all representing novel specificities, while HIV-1 responses also exceeded those against HCV both in breadth and magnitude.
Of 23 subjects coinfected with HCV and HIV-1 tested, 100% had detectable HIV-1–specific CD8+ responses. Not surprisingly, the peptides targeted were largely those that have already been shown to be targeted in HIV-1 monoinfection (data not shown).22-24,29 In contrast, analysis of HCV-specific responses in these coinfected persons revealed positive responses in 37 (50%) of 74 subjects tested. This was a significantly higher rate of detection than the 17% rate in previous studies using vaccinia ELISpot assays (P = .023, chi-square test).30 Of the 112 HCV-specific responses detected among the 103 subjects, 41 (36%) were responses to peptides that had not been previously characterized to contain HCV CTL epitopes.20 For persons with novel HCV-specific epitopes, the number of responses not corresponding to previously described epitopes ranged between 17% and 100% (median, 50%) of the responses within each individual. This ELISpot assay does not distinguish between CD8+ or CD4+ T-cell responses; therefore, for each novel HCV-specific CD8+ T-cell response detected, generation of antigen-specific cell lines verified their classification as CD8+ (example, Figure 3). CD4+ ELISpot responses were detected in only a small proportion of individuals (15 of 103, or 14.6%).
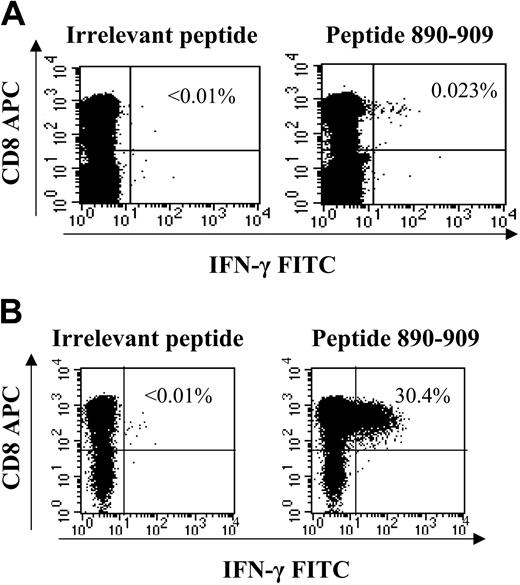
Ex vivo ICS of novel specificity and generation of short-term cell lines directed against HCV antigens. All novel responses (detected within peptide matrix but not corresponding to previously known optimal epitopes) were determined to be CD8+ T cells by generation of T-cell lines. The response ex vivo, which represented 355 SFC/106 PBMCs via ELISpot (not shown), was detectable by ex vivo ICS staining (A) and was expanded via peptide stimulation in presence of interleukin-2 to 30% of CD8+ cells within 14 days in coinfected subject C23-02C (B). All novel responses were verified in the latter manner as the majority fell below the magnitude threshold to be detected by ex vivo ICS.
Ex vivo ICS of novel specificity and generation of short-term cell lines directed against HCV antigens. All novel responses (detected within peptide matrix but not corresponding to previously known optimal epitopes) were determined to be CD8+ T cells by generation of T-cell lines. The response ex vivo, which represented 355 SFC/106 PBMCs via ELISpot (not shown), was detectable by ex vivo ICS staining (A) and was expanded via peptide stimulation in presence of interleukin-2 to 30% of CD8+ cells within 14 days in coinfected subject C23-02C (B). All novel responses were verified in the latter manner as the majority fell below the magnitude threshold to be detected by ex vivo ICS.
The use of individual peptides rather than expressed proteins allowed us to also determine the number of targeted epitopes in this stage of infection. The breadth of HCV-specific CD8+ responses was 0 to 6 epitopes per individual, with cumulative magnitudes ranging as high as 4535 SFC/106 PBMCs. The breadth (range, 2-55 epitopes per individual) and magnitude (range, 322-29 390 SFC/106 PBMCs) of HIV-1–specific responses were comparable to previous reports in persons monoinfected with HIV-1 using these techniques22,23,29 and were both higher than the HCV-specific responses (P < .001). For only 2 of 23 individuals (subjects D3-01-49 and C-23-02C) did the breadth and magnitude of the HCV-specific response exceed that of the HIV-1–specific response; however both subjects had suppressed HIV-1 viral loads on antiretroviral therapy for more than 6 years, a situation known to be associated with a decline in HIV-1–specific CD8+ T-cell frequencies.23,31-34 These data confirm other studies,30,35 indicating that the magnitude and breadth of HCV-specific responses are lower than those against HIV-1 in persons with coinfection when tested within the same individuals.
HIV-1 serostatus does not alter the breadth, magnitude, and specificity of HCV-specific CD8+ T lymphocytes
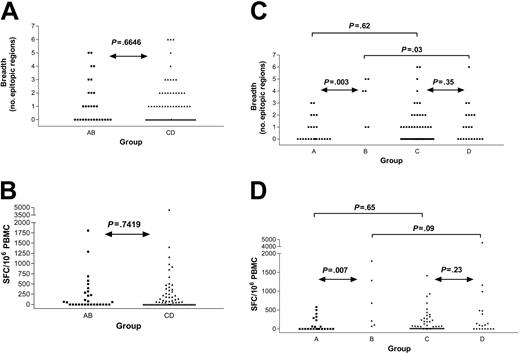
Next, we tested the hypothesis that the presence of HIV-1 would diminish the frequencies of CD8+ T-cell responses against HCV. We therefore compared HIV-1–negative (groups A and B) versus HIV-1–positive (groups C and D) individuals who were all infected with HCV. The overall rate of detection of HCV-specific responses did not vary with HIV-1 status (52% in HIV-1 negative versus 50% in HIV-1 positive, P = .87), and neither the breadth of responses (P = .66, Mann-Whitney) nor the cumulative magnitude (P = .74) differed (Figure 4A-B). Subjects coinfected with HCV and HIV-1 (groups C and D) were more likely to have higher absolute CD8+ cell counts when compared with individuals negative for HIV-1 and positive for HCV (groups A and B). Analyses were repeated normalizing the magnitude for the absolute CD8+ count and had no significant effect; therefore, we used SFC/106 PBMCs as the commonly used standard to express magnitude of responses. The results indicate that HIV-1 serostatus by itself was not associated with any decrement in CD8+ T-cell frequencies against HCV, although it was associated with higher HCV viral loads.
Breadth and magnitude of HCV-specific immunity by group reveal no differences by HIV-1 serostatus. Comparison of monoinfected (HCV Ab positive, HIV-1 Ab negative, group AB) and coinfected (HCV Ab positive, HIV-1 Ab positive, group CD) subjects reveals no significant difference in number of HCV epitopic regions targeted (A) nor the intensity of the response (B). The breadth (C) and magnitude (D) of HCV-specific CD8+ T-cell responses are shown for the same subjects further subdivided by HCV RNA status (group A: monoinfected, HCV viremic; group B: monoinfected, HCV controller; group C: HIV-1 coinfected, HCV viremic; group D: HIV-1 coinfected, HCV controller), demonstrating higher breadth and magnitude in group B compared with group A.
Breadth and magnitude of HCV-specific immunity by group reveal no differences by HIV-1 serostatus. Comparison of monoinfected (HCV Ab positive, HIV-1 Ab negative, group AB) and coinfected (HCV Ab positive, HIV-1 Ab positive, group CD) subjects reveals no significant difference in number of HCV epitopic regions targeted (A) nor the intensity of the response (B). The breadth (C) and magnitude (D) of HCV-specific CD8+ T-cell responses are shown for the same subjects further subdivided by HCV RNA status (group A: monoinfected, HCV viremic; group B: monoinfected, HCV controller; group C: HIV-1 coinfected, HCV viremic; group D: HIV-1 coinfected, HCV controller), demonstrating higher breadth and magnitude in group B compared with group A.
In the subgroup of persons negative for HIV-1 who cleared HCV infection (group B), we defined a significantly higher breadth (P = .003; Figure 4C) and magnitude (P = .007; Figure 4D) of HCV-specific responses when compared with the persons negative for HIV-1 who developed persistent HCV infection (group A), as reported in previous studies.26,36-38 Although a trend was observed, this distinction did not reach significance when comparing the breadth (P = .35; Figure 4C) and magnitude (P = .23; Figure 4D) of HCV-specific immunity in persons positive for HIV-1 who have spontaneously controlled HCV (group D) with those who have persistent HCV viremia (group C).
We also compared the specificities between monoinfected and coinfected persons by percentage of subjects in the cohort recognizing each HCV protein. Overall nonstructural proteins, particularly NS3, were most frequently recognized, consistent with prior studies establishing NS3 as a frequently targeted protein.21,37 However, no differences in specificity were detected between the monoinfected and coinfected subjects (Table 2).
Percentage of individuals with CTL responses by HCV protein
. | All proteins . | Core . | E1 . | E2 . | P7 . | NS2 . | NS3 . | NS4 . | NS5 . |
|---|---|---|---|---|---|---|---|---|---|
| HCV monoinfected, % | 52 | 7 | 0 | 10 | 0 | 7 | 31 | 14 | 14 |
| HCV/HIV-1 coinfected, % | 50 | 7 | 5 | 5 | 4 | 11 | 28 | 11 | 18 |
| P | .87 | .98 | .20 | .37 | .27 | .54 | .78 | .67 | .64 |
. | All proteins . | Core . | E1 . | E2 . | P7 . | NS2 . | NS3 . | NS4 . | NS5 . |
|---|---|---|---|---|---|---|---|---|---|
| HCV monoinfected, % | 52 | 7 | 0 | 10 | 0 | 7 | 31 | 14 | 14 |
| HCV/HIV-1 coinfected, % | 50 | 7 | 5 | 5 | 4 | 11 | 28 | 11 | 18 |
| P | .87 | .98 | .20 | .37 | .27 | .54 | .78 | .67 | .64 |
For HCV monoinfected, n = 29; for HCV/HIV-1 coinfected, n = 74.
For subjects with persistent HCV viremia (groups A and C combined), there were no correlations between magnitude of HCV-specific T cells detected and ALT or plasma HCV viral load. In 27 patients with biopsies performed within 3 months of analysis, no correlation between the breadth or magnitude of cellular immune responses and histologic parameters was detected (data not shown). It should be noted that only 3 of 14 subjects with CD4+ counts fewer than 200 received liver biopsies; therefore, our ability to make correlations of liver histology with immune parameters is limited for this subgroup.
Breadth and magnitude of functional HCV-specific CD8+ T lymphocytes depend on absolute CD4+ T-cell counts in persons coinfected with HCV and HIV-1
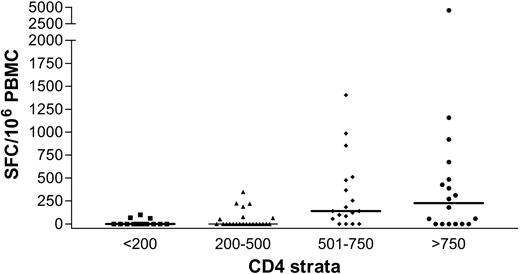
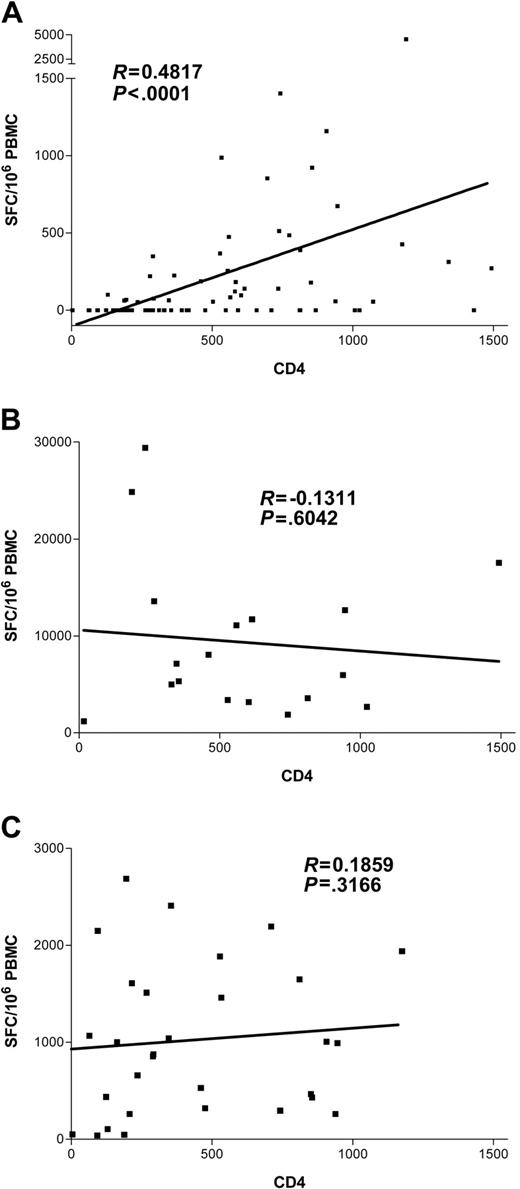
We next evaluated the effect of HIV-1–related CD4+ T-cell depletion on the HCV-specific CD8+ T-cell response. Results showing the magnitude of HCV-specific responses for all coinfected individuals are pictured in Figure 5, stratified by CD4+ count. These results indicate a strong relationship between the absolute frequencies of CD4+ T cells and circulating HCV-specific CD8+ immunity (Kruskal-Wallis, P = .0002). HCV-specific CD8+ T-cell responses were found infrequently in subjects with CD4+ counts fewer than 500 (7 of 23 or 30% of subjects) and were even less frequent when CD4+ counts were fewer than 200 (3 of 14 or 21% of subjects). In contrast, multispecific and sometimes vigorous HCV-specific responses were relatively frequently detected in subjects with CD4+ counts greater than 500 (Figure 5). However, even in the group with the highest CD4+ counts (> 750), only 12 of 18 or 66% of individuals tested had detectable ex vivo CD8+ T-cell responses to HCV. The relationship between cumulative magnitude of IFN-γ–secreting HCV-specific CD8+ cells and CD4+ count was likewise highly significant (Figure 6A; R = 0.48, P < .0001) and remained significant when analyzing each coinfected group separately (group C, R = 0.49, P = .0002; group D, R = 0.49, P = .03). For the subgroup that had been taking antiretroviral therapy for at least 3 months, this correlation also held (n = 40, R = 0.38, P = .01). Interestingly, when analysis was restricted to subjects with known genotype 1 infection, which would be closest to the reference strain of virus used as the basis for synthesis of overlapping peptide sets, the correlation coefficient between absolute CD4+ count and magnitude of HCV-specific CD8+ T-cell responses was strongest (n = 34, R = 0.54, P = .0009). Furthermore, the breadth of HCV responses also strongly correlated with CD4+ counts (R = 0.44, P < .0001).
Relationship of HCV-specific immunity to CD4+ T-cell count in individuals infected with HIV-1. Stratification of magnitude of HCV-specific immunity by CD4+ T-cell count reveals a strong positive relationship.
Relationship of HCV-specific immunity to CD4+ T-cell count in individuals infected with HIV-1. Stratification of magnitude of HCV-specific immunity by CD4+ T-cell count reveals a strong positive relationship.
CD8+ T-cell dependence on CD4+ cell counts differs by viral specificity in individuals coinfected with HCV and HIV-1. Magnitude of virus-specific immunity (SFC/106 PBMCs) plotted against CD4+ count. A significant positive correlation was discovered for HCV-specific CD8+ functional immunity (A) but not for the magnitude of CD8+ cells specific for HIV-1 (B) or EBV (C). Of note, this remains highly statistically significant (P < .0001) after removal of the outlier with the highest level of immunity (subject D3-01-49).
CD8+ T-cell dependence on CD4+ cell counts differs by viral specificity in individuals coinfected with HCV and HIV-1. Magnitude of virus-specific immunity (SFC/106 PBMCs) plotted against CD4+ count. A significant positive correlation was discovered for HCV-specific CD8+ functional immunity (A) but not for the magnitude of CD8+ cells specific for HIV-1 (B) or EBV (C). Of note, this remains highly statistically significant (P < .0001) after removal of the outlier with the highest level of immunity (subject D3-01-49).
Other patient characteristics were stratified and tested for relationships to HCV-specific CD8+ immunity, including age, sex, HLA type, and race, with negative results. HIV viremia, although inversely correlating with CD4 T-cell count (R = –0.49, P < .0001), did not independently correlate with HCV-specific T-cell responses (R =–0.08, P = .5). These data therefore demonstrate that the strongest predictor of HCV-specific CD8+ T-cell magnitude and breadth is the CD4+ T-cell count.
HIV-1 and EBV T-cell responses are preserved even at low CD4+ T-cell counts in coinfected subjects
We next assessed whether decreases in absolute CD4+ T-cell counts would have a similar effect on other viral antigen-specific immune responses. For these studies we examined both HIV-1–specific responses, which are thought to be functionally impaired in the acute stage of HIV-1 infection,39,40 and EBV-specific responses, which would be expected to be activated intermittently during chronic HIV-1 infection.41 Since removal of antigen due to viral suppression on highly active antiretroviral therapy (HAART) consistently results in a decline of HIV-1 responses,23,32 we restricted analysis to 18 patients off anti–HIV-1 therapy to prevent skewing our results. No correlation of HIV-1–specific responses to CD4+ count (R = –0.13, P = .60; Figure 6B) or HIV-1 viral load (R = 0.23, P = .35) was noted. Moreover, when we tested 31 subjects coinfected with HCV and HIV-1 from this cohort with pools containing optimal EBV CD8+ epitopes by ELISpot, immunity was readily detected in all patients, and, again, no significant relationship emerged with CD4+ count (R = 0.19, P = .31; Figure 6C).
In sum, after determining virus-specific CD8+ T-cell responses against HCV, HIV-1, and EBV in this cohort, only the magnitude and breadth of HCV-specific responses were significantly correlated with CD4+ T-cell count (Figure 6A).
Persistence of expandable memory HCV-specific responses in subjects with depressed CD4+ counts
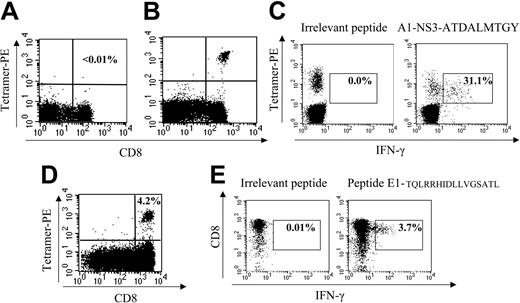
To rule out the possibility that HCV-specific CD8+ cells are present at low CD4+ counts but impaired in their ability to secrete interferon-γ,42,43 tetramer staining was performed in a subset of individuals, including several who demonstrated a lack of ex vivo ELISpot response. This analysis was performed in subjects expressing HLA alleles for which matched HCV tetramers were available (A1, 6 subjects and 1 epitope tested; A2, 7 subjects and 3 epitopes tested; B7, 2 subjects and 1 epitope tested; B8, 3 subjects and 1 epitope tested; B35, 3 subjects and 1 epitope tested; total 35 stainings). Two example stainings are shown in Figure 7A-B. In those persons with detectable ELISpot responses and the appropriate HLA allele, tetramer binding was readily demonstrated (Figure 7B). In contrast, in persons with the appropriate allele but an absence of ELISpot response, tetramer analysis was universally negative (Figure 7A).
CD8+ T cells are detected in subjects with low CD4+ counts after in vitro expansion. (A) Sample negative tetramer stain in subject D19-03-29 with HLA A*0101 allele; tetramer binds A1-NS3-ATDALMTGY. This subject did not have an ELISpot response to this epitope. (B) Immunodominant ex vivo HCV-specific response in subject D3-01-49, tetramer binds A1-NS3-ATDALMTGY, representing 3.6% of CD8+ cells, corresponding with an ELISpot response of 3000 SFC/106 PBMCs. (C) This panel demonstrates that tetramer binding is specific to this antigen because after a 6-hour incubation more than 30% of tetramerhi cells secrete IFN-γ in the presence of appropriate peptide (gated on CD8hi). (D) Tetramer stain after 14 days of in vitro stimulation in subject C9-02-05 (CD4 = 207 cells/mm3) bearing A*0201 allele with peptide 1406-1415, KLVALGINAV. No ex vivo response by ELISpot or tetramer was detected. (E) Expansion of cells specific to novel epitope contained within E1-TQLRRHIDDLVGSATL in subject C41-03T (CD4 = 92 cells/mm3) after 14 days, demonstrated by IFN-γ ICS on cell line. Ex vivo ELISpot response equaled 25 SFC/106 PBMCs, representing an extremely low level response initially below the cutoff of significance for earlier ex vivo analyses but capable of proliferation in vitro to 3.7% of CD8hi cells.
CD8+ T cells are detected in subjects with low CD4+ counts after in vitro expansion. (A) Sample negative tetramer stain in subject D19-03-29 with HLA A*0101 allele; tetramer binds A1-NS3-ATDALMTGY. This subject did not have an ELISpot response to this epitope. (B) Immunodominant ex vivo HCV-specific response in subject D3-01-49, tetramer binds A1-NS3-ATDALMTGY, representing 3.6% of CD8+ cells, corresponding with an ELISpot response of 3000 SFC/106 PBMCs. (C) This panel demonstrates that tetramer binding is specific to this antigen because after a 6-hour incubation more than 30% of tetramerhi cells secrete IFN-γ in the presence of appropriate peptide (gated on CD8hi). (D) Tetramer stain after 14 days of in vitro stimulation in subject C9-02-05 (CD4 = 207 cells/mm3) bearing A*0201 allele with peptide 1406-1415, KLVALGINAV. No ex vivo response by ELISpot or tetramer was detected. (E) Expansion of cells specific to novel epitope contained within E1-TQLRRHIDDLVGSATL in subject C41-03T (CD4 = 92 cells/mm3) after 14 days, demonstrated by IFN-γ ICS on cell line. Ex vivo ELISpot response equaled 25 SFC/106 PBMCs, representing an extremely low level response initially below the cutoff of significance for earlier ex vivo analyses but capable of proliferation in vitro to 3.7% of CD8hi cells.
Finally, we examined whether persons with undetectable ex vivo responses had HCV-specific CD8+ cells capable of proliferation following peptide stimulation with HLA-matched peptides.21 Representative ICS and tetramer stains indicate a significant expansion of specific cells (Figure 7D-E) from 2 subjects. In 7 of 13 subjects with CD4+ counts fewer than 350 tested, we were able to expand these low-level responses that were not detectable either by IFN-γ ELISpot or tetramer assays ex vivo. Therefore, in subjects with low CD4+ counts, HCV-specific CD8+ responses are not entirely absent but rather are present at a frequency below the level of ex vivo detection44 and capable of in vitro expansion.
Discussion
In this cross-sectional study of 103 persons infected with HCV, we demonstrate that HIV-1 serostatus alone does not alter the breadth, magnitude, or specificity of the HCV-specific CD8+ T-cell response per se. However, frequencies of these cells depend on the immune status of the individual as defined by absolute CD4+ T-cell count, independent of whether or not HCV viremia is present or controlled. The finding that HCV-specific CD8+ T-cell responses decline with diminishing absolute CD4+ counts provides a possible explanation for the more rapid HCV disease progression in the setting of HIV-1 coinfection. Moreover, this relationship was unique, as we did not detect a similar effect on HIV-1 or EBV-specific immunity over a similar range of CD4+ T-cell counts. Taken together, our data suggest that the magnitude and breadth of HCV-specific CD8+ cells are sensitive to the T helper lymphocyte environment.
The comprehensive approaches used to investigate HCV-specific immunity in this study defined a higher magnitude in subjects coinfected with HIV-1 than previous reports, including our own using a vaccinia-based assay.30 Two smaller previous studies that used a limited set of optimal peptides in an ELISpot assay found HCV-specific CD8+ responses only in HIV-1 long-term nonprogressors who maintained their CD4+ T-cell counts and not in progressors, including those with CD4+ counts restored via HAART therapy.35,45 In those studies, only a limited number of optimal epitopes were tested as potential antigens; therefore, the full breadth and magnitude of antiviral immunity would be underestimated.21,46-49 Indeed, even our more comprehensive approach is likely to underestimate the full magnitude and breadth of responses, since only those responses crossreactive with the reference strain of HCV used here would be detected. Our utilization of these more sensitive techniques nevertheless shows that HCV-specific CD8+ T cells can be detected in the peripheral blood of a significant number of coinfected persons, at a rate more than twice that of previous studies.
The observed dependence of HCV-specific CD8+ T-cell responses on absolute CD4+ cell number is consistent with recent studies in a chimpanzee model of HCV infection. In those experiments, CD4+ depletion via a monoclonal antibody in 2 animals was associated with a decrement of the CD8+ response, prolonged viremia, and viral escape.17 The clear effect of absolute CD4+ cell count on HCV-specific responses observed in our study is in stark contrast to CD8+ T-cell responses against HIV-1 and EBV, which were detected at high frequencies in many subjects despite low CD4+ counts. Other studies have shown a lack of association between absolute CD4+ count and HIV-1–specific immune responses.22,23,29,50-52 One study found a positive correlation between CD4+ count and EBV-specific responses in individuals infected with HIV-1, but it used a far more limited set of potential epitopes than those used in our study.53 Whether the lack of correlation with EBV-specific responses relates to less frequent reactivation of EBV-specific CD4+ T-cell responses, and therefore their maintenance at lower absolute CD4+ cell numbers relative to those against HCV, is a testable hypothesis for future study. Likewise, the lack of correlation of absolute CD4+ cell count with HIV-1–specific immune responses may relate to the dramatic early effect on HIV-1–specific CD4+ cells in the acute phase of infection,54 leading to early functional impairment and/or deletion of these cells, even when absolute CD4+ cell numbers are relatively preserved. Indeed, high frequencies of HIV-1–specific CD8+ T cells capable of IFN-γ secretion are often found in progressive infection in conditions lacking CD4+ help.23,55 The phenotype of human antiviral CD8+ T cells may vary by viral specificity,56 and further comparative studies may reveal explanations for differential dependence on CD4+ T-cell frequencies.
Despite the CD4+-related decline in frequencies of HCV-specific CD8+ T cells in these coinfected subjects, memory responses remained detectable in the majority of subjects studied. By in vitro stimulation, we were further able to expand antigen-specific CD8+ cells even in subjects with low CD4+ counts. Whether HCV-specific CD8+ cell responses will expand in vivo when CD4+ counts are restored by HAART, as well as how they influence HCV disease progression, need to be studied. To examine the full extent of immune reconstitution, these studies might ideally involve testing immune responses against each subject's autologous virus rather than a reference strain.57 After initiation of antiretroviral therapy, studies have demonstrated variable effects on HCV viral load.58 An early rise in liver function tests and HCV viral load may be secondary to immune reconstitution, based on the assumption that specific lymphocytes are recruited to the liver, induce cytolysis, and elevate viral titers. Emerging reports of spontaneous clearance of chronic HCV viremia in a minority of coinfected subjects on HAART support the hypothesis that HCV-specific immunity may be reconstituted.59-62
While this study was able to include a large number of subjects to provide confidence regarding the primary finding, one limitation is the cross-sectional nature of the analysis, especially as we could not assess the effect of relative timing of acquisition of the 2 infections. Future studies should seek to elucidate longitudinal effects of HIV-1–induced CD4 T-cell depletion and replenishment on preexisting HCV immunity. Conversely, longitudinal analyses of individuals with preexisting HIV-1 infection who subsequently contract HCV are of prime importance. The comprehensive techniques described in the present study, which allow for detection of HCV-specific immune responses in the peripheral blood of the majority of infected persons, will facilitate such investigations.
In summary, these data indicate that IFN-γ–secreting HCV-specific CD8+ T cells are readily detectable in the peripheral blood of persons with chronic HCV and HIV-1 coinfection and reveal that their frequencies are dependent on absolute CD4+ cell numbers. Further studies are needed in the coinfection model to examine whether reversal of immunosuppression can augment HCV-specific immunity, and whether this correlates with clinical events. Finally, these studies in individuals coinfected with HCV and HIV-1 will elucidate the potential for immunotherapeutic intervention in this clinically important population.
Prepublished online as Blood First Edition Paper, September 30, 2004; DOI 10.1182/blood-2004-06-2336.
Supported by grants from the National Institutes of Health (A.Y.K. and B.D.W.), the Campbell Foundation (G.M.L.), Deutsche Forschungsgemeinschaft (J.T.), Deutscher Akademischer Austausch Dienst (J.S.z.W.), the Concerned Parents for AIDS Research (M.M.A.), AmfAR (M.M.A.), the Harvard Center for AIDS Research (CFAR), the Wellcome Trust (P.K.), the European Union (M.L. and P.K.), the Doris Duke Charitable Foundation (B.D.W.), and the Howard Hughes Medical Institute (R.D., J.S.z.W., and B.D.W.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank foremost all subjects who generously enrolled into this study and the dedicated clinical research staff who helped to recruit them. We also thank Almas Rathod and Cori Verrill for valuable technical assistance and Todd Allen, Margaret Feeney, Daniel Kaufmann, and Tonia Woodberry for insightful discussions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal