Abstract
Δ9-Tetrahydrocannabinol (THC) is the active metabolite of cannabis. THC causes cell death in vitro through the activation of complex signal transduction pathways. However, the role that the cannabinoid 1 and 2 receptors (CB1-R and CB2-R) play in this process is less clear. We therefore investigated the role of the CB-Rs in mediating apoptosis in 3 leukemic cell lines and performed microarray and immunoblot analyses to establish further the mechanism of cell death. We developed a novel flow cytometric technique of measuring the expression of functional receptors and used combinations of selective CB1-R and CB2-R antagonists and agonists to determine their individual roles in this process. We have shown that THC is a potent inducer of apoptosis, even at 1 × IC50 (inhibitory concentration 50%) concentrations and as early as 6 hours after exposure to the drug. These effects were seen in leukemic cell lines (CEM, HEL-92, and HL60) as well as in peripheral blood mononuclear cells. Additionally, THC did not appear to act synergistically with cytotoxic agents such as cisplatin. One of the most intriguing findings was that THC-induced cell death was preceded by significant changes in the expression of genes involved in the mitogen-activated protein kinase (MAPK) signal transduction pathways. Both apoptosis and gene expression changes were altered independent of p53 and the CB-Rs.
Introduction
Δ9-Tetrahydrocannabinol (THC) is the active metabolite of cannabis. Although its analgesic and antiemetic effects in cancer patients has been known for some time, its application has been restricted for social reasons.1 Interest in this field has intensified with the findings that THC may selectively induce tumor regression both in vitro and in vivo.2-4 Recently, clinical trials using this drug have commenced in Spain. The mechanism underlying its cytotoxic activity has not been elucidated; however, a number of studies have suggested that cannabinoid receptors are involved in this process.5,6 Two different receptors have been cloned, each with distinct function and distribution. Most is known about the cannabinoid-1 receptor (CB1-R), which is predominantly found in the brain, with highest densities in the hippocampus and cerebellum. It is involved with the psychotropic effects of cannabis; however, CB1-R has also been described at lower levels, in other sites such as the eyes, testis, and lung.7 The second cannabinoid receptor (CB2-R) is expressed in the immune system8 and thought to regulate cellular development and immune function. Both receptor subtypes have been identified in tumors at varying concentrations and have been implicated in the cytotoxic process, although alternative receptor-independent mechanisms have also been suggested.2,4,9,10 There is no clear correlation between these studies, which may be because these studies measured receptor level (by mRNA and with non–functional-specific antibodies) rather than receptor functionality. Indeed, measuring and correlating functional receptors with THC efficacy may be more appropriate than assessing mRNA levels. Nevertheless, the uses of small molecules that specifically agonize and antagonize CB1 and CB2 receptors have indicated that both receptors may be implicated in the cytotoxic process.3,4,10 Consequently, selective CB2-R agonists that cause cell death, but do not induce a psychotropic effect, have been developed as putative cytotoxic agents.1,10
The mechanism of underlying THC-mediated cell death remains unclear; studies have shown THCs have cytostatic, apoptotic, and necrotic effects.2,11,12 Consequently, a number of different mechanisms have been implicated in the activation of this cell death. Both the up- and down-modulation of signal transduction pathways, predominantly those involving mitogen-activated protein kinase (MAPK), has been common to many of these reports.2,3
Overall the mechanism of THC-associated cell kill appears to be more complicated than initially thought. There appears to be both long- and short-term effects of THC, both of which may be important in inducing cell death. As a consequence, this study has investigated the effects of THC on a panel of leukemic cell lines. CB1-R and CB2-R levels and their role in cytotoxicity have also been studied, as have the effects of THC on the cell cycle, induction of apoptosis, gene expression, and DNA binding. As the action of THC appears to be novel, we went on to investigate the potential interaction with established chemotherapeutic agents.
Materials and methods
Cell culture
CEM (acute lymphoblastic), HEL-92 (erythroblastic), HL60 (acute promyelocytic), and MOLT-4 (acute lymphoblastic) leukemic cell lines were obtained from the Cancer Research UK Laboratories (London, United Kingdom) and were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum. No antibiotics were used in our experiments, and all cell lines were incubated in a humidified atmosphere with 5% CO2 in air at 37°C.
To study the effect of THC on cell growth, cells (2 × 105/mL) growing exponentially were reset in fresh culture medium supplemented with THC (0-100 μM; Sigma, Dorset, United Kingdom). Aliquots were removed at 6, 24, and 48 hours for assessment of cell number, cell viability, and cell cycle distribution.
The concentration required to reduce cell viability by 50% (IC50) was determined using the sigmoid Emax model as follows: EP = EC - [(Emax ×Cn)/(IC50n + Cn)], where EP is predicted effect, EC is control effect, Emax is maximum effect, C is concentration of drug, and n is the sigmoid-fit factor.13
As THC binds to both the CB1 and CB2 receptors, we assessed the effects that THC had through individual receptors by culturing THC-treated cells with the respective antagonist. Specifically, cells were cultured with THC at the Bmax concentration (concentration just achieving maximum binding) with the CB1-R antagonist AM28114 (Tocris Cookson, Bristol, United Kingdom) and/or the CB2-R antagonist AM63015 (Tocris) (both 0-100 μM). Cell parameters were then assessed on day 2. The effect on cell viability of culturing cells with the specific CB2-R agonist palmitoylethanolamide (PEA)16 (0-100 μM; Tocris) was also studied in a similar fashion. Additionally, the effect of PEA on THC-induced cytotoxicity was investigated by culturing CEM, HEL-92, and HL60 cells with THC in the presence or absence of PEA (both at IC50 concentrations) prior to assessment of cell viability on day 2.
DNA analysis
The distinct phases of the cell cycle were distinguished by flow cytometry, according to methods described previously.17 Acquisition of data was performed within 1 hour using a FACSCalibur (BD Biosciences, Oxford, United Kingdom). Five thousand cells were analyzed for each data point, and the percentages of cells in sub-G1 (apoptotic fraction, cells with a reduced DNA content but similar morphology), G1, S, and G2/M phases were determined using the cell cycle analysis program WinMDI v2.4 (http://facs.scripps.edu/software.html).
Biotinylation of THC and determination of CB1 and CB2 cannabinoid receptor levels
THC was irreversibly biotinylated with biotin-XX-NHS (CN Biosciences, Nottingham, United Kingdom) using methods reported previously.18 Briefly, a stock concentration of THC (10 mM) was incubated with biotin-XX-NHS (2 mg/mL) at a ratio of 1:5 for 6 hours at room temperature. Phosphate-buffered saline containing 0.1% bovine serum albumin was added and incubated for 1 hour at 4°C. The mixture was then centrifuged in a Microcon YM-100 centrifugal filter unit (Millipore, Watford, United Kingdom). The residue (biotinylated-THC) was then collected from the filter head by reverse centrifugation and made up to a volume of 100 μL with 0.1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) (Solution A).
To assess total receptor binding, cells (1 × 106) were resuspended in 100 μL PBS and incubated with 10 μL biotinylated THC for 1 hour. Following 3 washing steps in Solution A, cells were incubated with streptavidin–fluorescein isothiocyanate (FITC) conjugate (1:100; CN Biosciences, Nottingham, United Kingdom) for 45 minutes at 4°C. Cells were washed thrice and resuspended in 500 μL PBS for flow cytometric analysis using the FACSCalibur. Five thousand events were recorded, and the mean FITC-fluorescence in each sample was measured and assessed using CellQuest v2.8 (BD Biosciences). As there were no good selective CB1-R and CB2-R agonists that could be biotinylated like THC and used to detect the individual cannabinoid receptors, we assessed the amounts of individual cannabinoid receptors by culturing cells with biotinylated THC, in the presence or absence of selective CB1-R and CB2-R antagonists (10 μM AM281 and AM630). The extent of THC binding was then established like before.
Assessment of gene expression
Samples for analyses of gene expression were prepared from CEM cells cultured with an IC50 THC, in a manner previously described.19 Briefly, aliquots (10 × 106 cells) were removed at 3 hours and homogenized in TRIzol reagent (Invitrogen, Paisley, United Kingdom). Total RNA was then chloroform extracted and precipitated using iso-propanol and ethanol. RNA quality was examined using the Agilent 2100 Bioanalyzer (Agilent Technologies UK, Cheshire, United Kingdom). Biotinylated copy RNA transcripts were produced from total RNA and hybridized to the human genome U133A oligonucleotide array comprising around 23 000 genes (Affymetrix UK, High Wycombe, United Kingdom). Samples were run in triplicate, and data analyses were performed using GeneSpring v6.1 (Silicon Genetics, Redwood City, CA). Only consistent changes in gene expression, relative to the untreated control samples that were detected in all 3 replicates, were reported.
Immunoblotting analysis for p53, cleaved PARP, total extracellular signal-related kinase protein (pERK), and phosphorylated pERK
Total cellular protein was solubilized and resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) using 12% acrylamide with a 5% stacking gel as described previously.17 Primary antibody probing was performed with anti-p53 (clone DO-7; 1:500; Dako, Cambridge, United Kingdom), anti-cleaved poly (adenosine diphosphateribose) polymerase (PARP; 1:1000; Cell Signaling Technology, Hitchin, United Kingdom), anti–phospho-p44/42 MAPK (1:1000; Cell Signaling), or anti–p44/42 MAPK (1:100; Cell Signaling). Anti–β-actin was used as a loading control (1:200; Oncogene Research Products, Boston, MA). Following a washing step in 0.1% Tween in tris-buffered saline (100 mM Tris (tris(hydroxymethyl)aminomethane), 150 mM NaCl, pH 7.6) horseradish peroxidase–conjugated anti-species immunoglobulin G1 (IgG1) was used as the secondary antibody (Dako). Bands were visualized by the electrochemiluminescence (ECL) detection system (Amersham Biosciences, Little Chalfont, United Kingdom).
Statistical analysis
All statistical analyses were performed using Minitab version 10 (State College, PA). Data were normally distributed as established by Shapiro-Wilk testing, and parametric analyses were used throughout. Differences between variables and control cultures, as determined by analysis of variance, were further characterized by paired Student t tests.
Results
THC is cytotoxic to leukemic cell lines
Concentration-dependent decreases in cell viability were seen in all cell lines cultured with THC for 2 days (Figure 1A). The concentrations required to reduce cell viability by 50% (IC50) were calculated and were similar in CEM and HL60 cells (26.5 μM and 21.1 μM, respectively), but higher in HEL-92 cells (61.4 μM). There were concomitant increases in apoptotic cells and reductions in the number of cells in the other phases of the cell cycle (Figure 1B-D). Apoptosis was confirmed in CEM cells by the presence of the cleaved PARP in cells cultured with THC as early as 3 hours (Figure 5C). When the subdiploid (sub-G1) portions of the histograms were excluded from the analyses, there was still no evidence of arrests at any phases of the cell cycles (Table 1).
Effect of THC on cell growth parameters. CEM, HEL-92, and HL60 cells were cultured continuously with THC (0-100 μM) for 2 days. (A) Cell viability was assessed by trypan blue dye analyses after 6 hours and 2 days, and the concentration required to cause a 50% reduction in cell viability (IC50) was calculated and is shown in the inset box. The effect of 2-day cultures with THC on cell cycle distribution was also assessed in the cell lines (B-D), and representative cell cycle histograms of cells cultured with 60 μM THC are shown (E). Each data point represents the means of at least 3 separate experiments, and SDs have been omitted for clarity.
Effect of THC on cell growth parameters. CEM, HEL-92, and HL60 cells were cultured continuously with THC (0-100 μM) for 2 days. (A) Cell viability was assessed by trypan blue dye analyses after 6 hours and 2 days, and the concentration required to cause a 50% reduction in cell viability (IC50) was calculated and is shown in the inset box. The effect of 2-day cultures with THC on cell cycle distribution was also assessed in the cell lines (B-D), and representative cell cycle histograms of cells cultured with 60 μM THC are shown (E). Each data point represents the means of at least 3 separate experiments, and SDs have been omitted for clarity.
Effect of THC on p53 protein expression. CEM and HL60 cells were cultured with an IC50 concentration (day 2), and p53 expression was assessed by immunoblot analysis at 6, 24, and 48 hours (A). Similarly, culturing MOLT-4 cells with THC for 2 days also showed no increase in p53 levels, while culturing with cisplatin (cddp) did (B). The culture of CEM cells with THC at 1 × and 5 × IC50 resulted in duration-independent increases in cleaved PARP and decreases in phosphorylated-pERK (C).
Effect of THC on p53 protein expression. CEM and HL60 cells were cultured with an IC50 concentration (day 2), and p53 expression was assessed by immunoblot analysis at 6, 24, and 48 hours (A). Similarly, culturing MOLT-4 cells with THC for 2 days also showed no increase in p53 levels, while culturing with cisplatin (cddp) did (B). The culture of CEM cells with THC at 1 × and 5 × IC50 resulted in duration-independent increases in cleaved PARP and decreases in phosphorylated-pERK (C).
The distribution of cells within G1, S, and G2 phases of the cell cycle in CEM, HEL-92, and HL60 cells
. | CEM, mean % ± SD . | . | . | HEL-92, mean % ± SD . | . | . | HL60, mean % ± SD . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | G1 . | S . | G2 . | G1 . | S . | G2 . | G1 . | S . | G2 . | ||||||
| Control | 37 ± 2 | 28 ± 1 | 35 ± 3 | 58 ± 4 | 13 ± 1 | 28 ± 3 | 49 ± 5 | 12 ± 3 | 39 ± 2 | ||||||
| 6 h | 38 ± 3 | 26 ± 1 | 36 ± 2 | 56 ± 5 | 14 ± 2 | 30 ± 3 | 47 ± 4 | 13 ± 1 | 40 ± 4 | ||||||
| 2 d | 34 ± 1 | 28 ± 2 | 38 ± 3 | 55 ± 3 | 14 ± 2 | 31 ± 4 | 50 ± 6 | 12 ± 2 | 38 ± 5 | ||||||
. | CEM, mean % ± SD . | . | . | HEL-92, mean % ± SD . | . | . | HL60, mean % ± SD . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | G1 . | S . | G2 . | G1 . | S . | G2 . | G1 . | S . | G2 . | ||||||
| Control | 37 ± 2 | 28 ± 1 | 35 ± 3 | 58 ± 4 | 13 ± 1 | 28 ± 3 | 49 ± 5 | 12 ± 3 | 39 ± 2 | ||||||
| 6 h | 38 ± 3 | 26 ± 1 | 36 ± 2 | 56 ± 5 | 14 ± 2 | 30 ± 3 | 47 ± 4 | 13 ± 1 | 40 ± 4 | ||||||
| 2 d | 34 ± 1 | 28 ± 2 | 38 ± 3 | 55 ± 3 | 14 ± 2 | 31 ± 4 | 50 ± 6 | 12 ± 2 | 38 ± 5 | ||||||
Cells were cultured with 60 μM THC (cytotoxic concentration), and cell cycle distribution was assessed by flow cytometry. The sub-G1 population of cells (apoptotic) was excluded from these analyses. Data points are the mean percentages and SDs of 3 separate experiments.
THC was also cytotoxic in the cell lines after only 6 hours of culture (percentage of viable cells [%V] IC50, 48.2, 65.8, 73.2 μM in CEM, HL60, and HEL-92, respectively), and this was associated with varying degrees of apoptosis in the cell lines (percentage of apoptotic cells [%A] at 6 hours, 1.2% ± 0.6% in CEM cells to 38.0% ± 1.1% in HL60 cells; Figure 1E). To establish whether THC cytotoxicity was exclusive to cancer cells, the efficacy of THC was investigated in peripheral blood mononuclear cells. Results showed THC was as potent in these normal cells (day 2 IC50, 48.1 μM).
Leukemic cell lines express varying levels of CB1-R and CB2-R
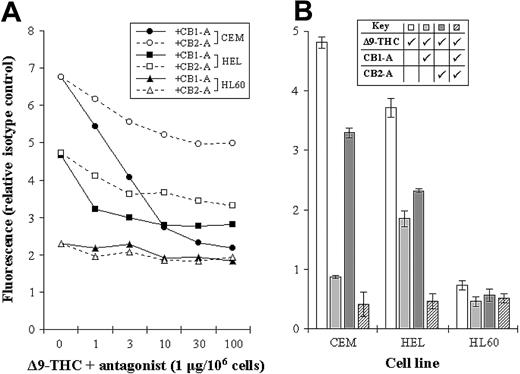
The levels of CB1-R and CB2-R were assessed by a novel nonradioactive method, and results indicated clear differences in their levels in the cell lines tested. Initially, we measured total THC binding to each of the cell lines, without distinguishing the 2 subsets of cannabinoid receptors (total binding) (Figure 2). We next blocked each receptor subset in turn by using specific antagonists, as a way of assessing the level of individual receptor type (Figure 3A-B). As an example, total THC binding to CEM cells was 4.82 ± 0.09 units. Blocking CB1-R with a specific CB1-R antagonist reduced the degree of THC binding to 0.87 ± 0.03 units, while blocking the CB2-R reduced THC binding to 3.29 ± 0.08 units. Blockade of both receptor subtypes, resulted in THC binding of just 0.41 ± 0.21 units (Figure 3B).
Binding profiles for THC. CEM (•), HEL-92 (HEL; ▪), and HL60 (▴) cell lines were cultured with biotinylated THC. Cells were then incubated with FITC-conjugated streptavidin, and the extent of fluorescence was assessed by flow cytometry. The maximum specific binding (Bmax) and the dissociation constant (Kd) values in the cell lines are shown in the inset box. The IgG1 isotype control (control) is also shown, and each data point represents the means and SDs of at least 3 separate experiments.
Binding profiles for THC. CEM (•), HEL-92 (HEL; ▪), and HL60 (▴) cell lines were cultured with biotinylated THC. Cells were then incubated with FITC-conjugated streptavidin, and the extent of fluorescence was assessed by flow cytometry. The maximum specific binding (Bmax) and the dissociation constant (Kd) values in the cell lines are shown in the inset box. The IgG1 isotype control (control) is also shown, and each data point represents the means and SDs of at least 3 separate experiments.
Assessing the levels of CB1-R and CB2-R. (A) CEM, HEL-92 (HEL), and HL60 cell lines were cocultured with a biotinylated THC (at Bmax concentration) and increasing concentrations of either the CB1-R or CB2-R antagonist (CB1-A or CB2-A). Mean fluorescence, as a measure of biotinylated THC binding, was then assessed by flow cytometry. (B) Similarly, the total cannabinoid-receptor level in each cell line was assessed by coculturing cells with biotinylated THC (Δ9-THC) and each antagonist (CB1-A and CB2-A); all were used at a maximum-blocking concentration. Each data point represents the mean of at least 3 separate experiments, and SDs have only been shown in panel B.
Assessing the levels of CB1-R and CB2-R. (A) CEM, HEL-92 (HEL), and HL60 cell lines were cocultured with a biotinylated THC (at Bmax concentration) and increasing concentrations of either the CB1-R or CB2-R antagonist (CB1-A or CB2-A). Mean fluorescence, as a measure of biotinylated THC binding, was then assessed by flow cytometry. (B) Similarly, the total cannabinoid-receptor level in each cell line was assessed by coculturing cells with biotinylated THC (Δ9-THC) and each antagonist (CB1-A and CB2-A); all were used at a maximum-blocking concentration. Each data point represents the mean of at least 3 separate experiments, and SDs have only been shown in panel B.
Cytotoxic effect of THC is not mediated via the CB1-R and CB2-R
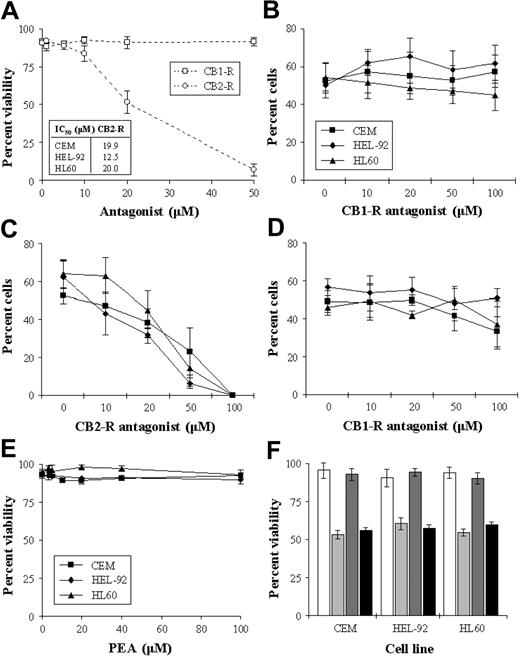
To investigate the role of the cannabinoid receptors in the cytotoxic response, cells were cultured with an IC50 concentration of THC in the presence of the receptor antagonists. Initially, we assessed the cytotoxic effect of the antagonists alone on the cell lines, and results showed the CB1-R antagonist to have no significant effect on cell viability. However, the CB2-R antagonist caused concentration-dependent decreases in cell viability (Figure 4A). Coculturing cells with THC (IC50) and increasing concentrations of the CB1-R antagonist resulted in no loss of cytotoxicity in the cells, indicating that the CB1-R was not responsible for this effect (Figure 4B). The effect of coculturing cells with THC and the CB2-R antagonist was more difficult to interpret as the CB2-R antagonist had a cytotoxic effect in its own right (Figure 4C). However, there was no reversal of THC-induced cytotoxicity at any concentration. The cytotoxic effects of THC were not alleviated by the addition of both antagonists concurrently (%V on day 2 in CEM, HEL-92, and HL60 66.7% ± 6.1%, 72.7% ± 6.4%, and 75.3% ± 3.1%, respectively, in THC-treated cells versus 69.0% ± 8.5%, 68.0% ± 2.0%, and 74.7% ± 3.1%, respectively, in cells cultured with THC and both antagonists). We addressed the possibility that the CB2-R antagonist may have been acting as a putative reverse CB1-R agonist by culturing cells with both the CB2-R and increasing concentrations of CB1-R antagonists. Results showed that the CB1-R antagonist did not alleviate the cytotoxic effect of the CB2-R antagonist (Figure 4D). As the CB2-R antagonist caused cell death on its own, we further explored the role of the CB2-R in mediating cell kill with the use of a specific CB2-R agonist (PEA). Results showed that PEA had no significant cytotoxic effect in the cell lines (Figure 4E). Similarly, PEA did not significantly antagonize/interfere with THC-induced cytotoxicity (Figure 4F).
Effect CB-R antagonists of cell viability. CEM, HEL-92, and HL60 cells were cultured continuously with either the CB1-R or the CB2-R antagonist (0-50 μM) for 2 days. As the trend in the effects of the individual antagonists were similar in that the CB1-R alone had no effect on viability while the CB2-R was cytotoxic, only the results of the effects on CEM are shown graphically, with the IC50s seen in the cell lines shown in the inset box (A). Cells were also cultured for 2 days with a combination of THC (IC50) and increasing concentrations of either the CB1-R antagonist (B) or the CB2-R antagonist (C). The effect of the CB1-R antagonist on the cytotoxic effect of the CB2-R antagonist (IC50) (D), and the cytotoxic effect of the putative CB2-R agonist PEA (E), were also studied. The effect of PEA in CEM, HEL-92, and HL60 cells at IC50 concentrations on THC-induced cytotoxicity were assessed on day 2 (F). White bars show untreated samples; light gray bars, THC at IC50; dark gray bars, PEA alone; and black bars, the effect of both agents used concomitantly. Each data point represents the means and SDs of at least 3 separate experiments.
Effect CB-R antagonists of cell viability. CEM, HEL-92, and HL60 cells were cultured continuously with either the CB1-R or the CB2-R antagonist (0-50 μM) for 2 days. As the trend in the effects of the individual antagonists were similar in that the CB1-R alone had no effect on viability while the CB2-R was cytotoxic, only the results of the effects on CEM are shown graphically, with the IC50s seen in the cell lines shown in the inset box (A). Cells were also cultured for 2 days with a combination of THC (IC50) and increasing concentrations of either the CB1-R antagonist (B) or the CB2-R antagonist (C). The effect of the CB1-R antagonist on the cytotoxic effect of the CB2-R antagonist (IC50) (D), and the cytotoxic effect of the putative CB2-R agonist PEA (E), were also studied. The effect of PEA in CEM, HEL-92, and HL60 cells at IC50 concentrations on THC-induced cytotoxicity were assessed on day 2 (F). White bars show untreated samples; light gray bars, THC at IC50; dark gray bars, PEA alone; and black bars, the effect of both agents used concomitantly. Each data point represents the means and SDs of at least 3 separate experiments.
THC does not increase p53 expression
To investigate whether p53 protein levels were affected by THC treatment, whole-cell lysates from CEM and HL60 cells were analyzed by immunoblotting. Results showed no significant changes to p53 (Figure 5). As these cell lines possess mutated p53 responses, similar immunoblotting experiments were performed using the acute myeloid leukemic cell line MOLT-4, which expressed wild-type p53. These cells were cultured with IC50 concentrations of either THC or cisplatin, and results recapitulated the previous data, showing no increases in p53 levels following culture with THC. In contrast, significant increases in p53 levels were seen following cisplatin treatment (Figure 5).
THC does not interact with common cytotoxic agents
To investigate interactions between THC and established chemotherapeutic agents (cisplatin), cells were cultured with a combination of the 2 at IC25 concentrations. Results from these simple pilot experiments showed additivity in the magnitude of cell kill when using the 2 agents simultaneously. Specifically, the estimated cytotoxicity (numerically the sum of the effects of the drugs used individually) was comparable to the measured cytotoxicity (the effect when the 2 drugs were used concurrently)17 (Figure 6A). Due to the absence of clear hyperadditivity in these early experiments involving IC25 concentrations of both drugs, we next investigated whether or not lower concentration of THC (IC5 and IC10) could enhance cisplatin-induced cytotoxicity. Results showed that there were no significant increases in cell kill when culturing both drugs together at lower doses (Figure 6B).
Effect of combining THC with cisplatin. The viability in the cell lines treated concurrently with IC25 concentrations of THC and cisplatin (▦) were compared with the calculated viability (□) (A). This estimated value was determined as the sum of the viabilities in cells treated with either THC or cisplatin alone. Cells were also cultured with small concentrations of THC (IC5, ⋄; and IC10, ▴) or left untreated (▪) to investigate whether these suboptimal doses could enhance the cytotoxic effects of cisplatin. Each data point represents the means and SDs of at least 4 separate experiments.
Effect of combining THC with cisplatin. The viability in the cell lines treated concurrently with IC25 concentrations of THC and cisplatin (▦) were compared with the calculated viability (□) (A). This estimated value was determined as the sum of the viabilities in cells treated with either THC or cisplatin alone. Cells were also cultured with small concentrations of THC (IC5, ⋄; and IC10, ▴) or left untreated (▪) to investigate whether these suboptimal doses could enhance the cytotoxic effects of cisplatin. Each data point represents the means and SDs of at least 4 separate experiments.
THC induces changes in the gene expression profile
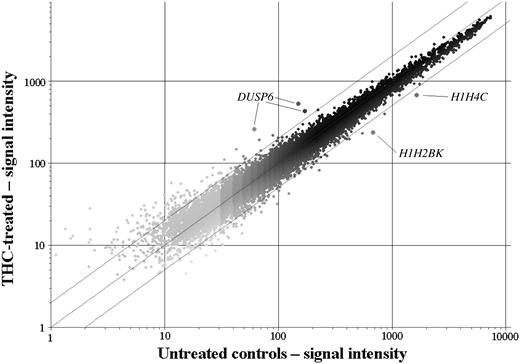
Microarray analyses were performed on CEM cells cultured with THC. We used a low concentration (1 × IC50 at day 2) and a short exposure time, which resulted in a data set with few gene expression changes. However, because of the nature of the treatment, the changes observed were likely to be drug specific and as a direct result of drug exposure, rather than nonspecific secondary effects like cell death or cell cycle arrest. First of all, we looked specifically at the expression of genes that had previously been identified as having a role in THC-induced cell death, such as genes in the ras/MAPK pathway, and those involved in ceramide metabolism. Genes in the MAPK signaling cascade that showed altered expression were DUSP6 (encoding dual specificity phosphatase 6/MAPK phosphatase 3 [MKP3]) and MAP2K2 (mitogen-activated protein kinase kinase 2/MEK2). Interestingly, MKP3 and MAP2K2 have the same intracellular target, the extracellular signal-regulated kinase 2 protein (ERK2/mitogen-activated protein kinase 1). They have, however, opposing functions: MEK2 phosphorylates, and thereby activates ERK2, whereas MKP3 dephosphorylates ERK2, taking it to an inactive state. Accordingly, the expression changes in response to THC observed were an increase in MKP3 expression in all 3 samples and a decrease in MAP2K2 expression (Table 2). Both changes are consistent with decreased MAPK signaling. There were no significant changes seen in genes involved in ceramide metabolism or in any other mechanism associated with THC-induced cell death. Additionally, there were no changes in the cytokine genes associated with THC-induced immune suppression. Many of these genes were in fact absent. Second, we examined the genes exhibiting the greatest degree of change in expression. The top 10 up- and down-regulated genes are listed in Table 2. Of particular note, both MKP3 and MAP2K2 were among the most significantly changed genes. Although 2 genes encoding histone H1 isoforms were the 2 most significantly down-regulated genes, there were no other gene patterns that were immediately apparent. We used a t test and fold-change filtering program (www.cgal.icnet.uk) as a plug-in for the GeneSpring v6.1 software to find the most significantly changed genes. Using a 2-fold cut-off and a delta-value of less than 0.1, 3 genes were found to be significantly changed in the THC-treated samples; these were the DUSP6 gene, encoding MKP3, and the histone H1 isoforms H2BK and H4C. The scatter plot of 1 of the treated samples normalized against the controls (Figure 7) revealed additional genes to these 3 that were outside the 2-fold cut-off lines; however, these outliers were either closer to the 2-fold cut-off or were lower in intensity, which decreased the confidence of the measurements. Although these changes may have been biologically relevant, they were not deemed significant.
The 10 most significantly down- and up-regulated genes following culture with THC in CEM cells
Symbol . | Location . | Function . | Δ, fold . | Key notes . |
|---|---|---|---|---|
| H1H2BK | 6p21.33 | Nucleosome assembly | 0.39 | Histone 1, H2bk; DNA binding capability |
| H1H4C | 6p21.3 | Nucleosome assembly | 0.41 | Histone 1, H4c; DNA binding capability |
| NUBP2 | 16p13.3 | Signaling | 0.55 | Nucleotide binding protein 2 |
| COLA4A3 | 2q36-37 | Matrix structure | 0.59 | Collagen type IV; involved in pathogenesis |
| TFIP11 | 22q12.1 | Secretion | 0.59 | Tuftelin interacting protein 11; involved in protein binding |
| PRDX2 | 13q12 | Oxidation | 0.61 | Peroxiredoxin 2; responder to oxidative stress, and has antioxidant activity |
| LSM7 | 19p13.3 | Unknown | 0.61 | Pre-mRNA splicing factor activity |
| MAP2K2 | 7q32 | Signaling | 0.61 | Mitogen-activated protein kinase kinase 2 |
| C14orf1 | 14q24.3 | Carcinogenesis | 0.61 | Chromosome 14 open reading frame 1; integral to membrane |
| RPS9 | 19q13.4 | Protein synthesis | 0.61 | Ribosomal protein S9; RNA binding capability |
| DUSP6 | 12q22-23 | Apoptosis | 3.36 | Dual specificity phosphatase 6 |
| TUBA2 | 13q11 | Cell motility | 2.36 | Tubulin α2; involved with microtubule-based movement |
| CD69 | 12p13-12 | Signaling | 2.26 | Cell surface receptor linked signal transduction |
| XTP2 | 1q23.3 | Unknown | 2.04 | HbxAg transactivated protein 2 |
| DDX15 | 4p15.3 | RNA processing | 1.87 | DEAD/H box polypeptide 15; has helicase activity |
| FACL4 | Xq22.3-23 | Metabolism | 1.87 | Fatty-acid coenzyme A ligase, long chain 4; involved with fatty acid metabolism |
| TRIM38 | 6p21.3 | Intracellular binding | 1.86 | Tri-partite motif-containing 38; zinc ion binding |
| NTRK2 | 9q22.1 | Signaling | 1.82 | Neutrophic tyrosine kinase receptor, type 2 |
| ALCAM | 3q13.1 | Adhesion | 1.81 | Activated leukocyte cell adhesion molecule |
| TAP2 | 6p21.3 | Transport | 1.77 | Transporter 2, ATP-binding cassette, subfamily B; involved with cellular defense; ABC transporter activity |
Symbol . | Location . | Function . | Δ, fold . | Key notes . |
|---|---|---|---|---|
| H1H2BK | 6p21.33 | Nucleosome assembly | 0.39 | Histone 1, H2bk; DNA binding capability |
| H1H4C | 6p21.3 | Nucleosome assembly | 0.41 | Histone 1, H4c; DNA binding capability |
| NUBP2 | 16p13.3 | Signaling | 0.55 | Nucleotide binding protein 2 |
| COLA4A3 | 2q36-37 | Matrix structure | 0.59 | Collagen type IV; involved in pathogenesis |
| TFIP11 | 22q12.1 | Secretion | 0.59 | Tuftelin interacting protein 11; involved in protein binding |
| PRDX2 | 13q12 | Oxidation | 0.61 | Peroxiredoxin 2; responder to oxidative stress, and has antioxidant activity |
| LSM7 | 19p13.3 | Unknown | 0.61 | Pre-mRNA splicing factor activity |
| MAP2K2 | 7q32 | Signaling | 0.61 | Mitogen-activated protein kinase kinase 2 |
| C14orf1 | 14q24.3 | Carcinogenesis | 0.61 | Chromosome 14 open reading frame 1; integral to membrane |
| RPS9 | 19q13.4 | Protein synthesis | 0.61 | Ribosomal protein S9; RNA binding capability |
| DUSP6 | 12q22-23 | Apoptosis | 3.36 | Dual specificity phosphatase 6 |
| TUBA2 | 13q11 | Cell motility | 2.36 | Tubulin α2; involved with microtubule-based movement |
| CD69 | 12p13-12 | Signaling | 2.26 | Cell surface receptor linked signal transduction |
| XTP2 | 1q23.3 | Unknown | 2.04 | HbxAg transactivated protein 2 |
| DDX15 | 4p15.3 | RNA processing | 1.87 | DEAD/H box polypeptide 15; has helicase activity |
| FACL4 | Xq22.3-23 | Metabolism | 1.87 | Fatty-acid coenzyme A ligase, long chain 4; involved with fatty acid metabolism |
| TRIM38 | 6p21.3 | Intracellular binding | 1.86 | Tri-partite motif-containing 38; zinc ion binding |
| NTRK2 | 9q22.1 | Signaling | 1.82 | Neutrophic tyrosine kinase receptor, type 2 |
| ALCAM | 3q13.1 | Adhesion | 1.81 | Activated leukocyte cell adhesion molecule |
| TAP2 | 6p21.3 | Transport | 1.77 | Transporter 2, ATP-binding cassette, subfamily B; involved with cellular defense; ABC transporter activity |
Each data point represents the mean fold-change of 3 independent experiments. Only genes with consistent detection calls were analyzed. HbxAg indicates hepatitis B virus-encoded X antigen; DEAD, diethyl azodicarboxylate; ATP, adenosine triphosphate; ABC, ATP-binding cassette.
Scatterplot of CEM cells treated with THC. Data are normalized against the controls, showing 2-fold cut-off lines. Only the genes for DUSP6 (increased), H1H2BK, and H1H4C (both decreased) were significantly altered in the THC-treated cells (P < .1).
Scatterplot of CEM cells treated with THC. Data are normalized against the controls, showing 2-fold cut-off lines. Only the genes for DUSP6 (increased), H1H2BK, and H1H4C (both decreased) were significantly altered in the THC-treated cells (P < .1).
THC decreases phosphorylated pERK protein expression
To investigate whether the changes in the gene expression of MAPK-associated species, whole cell lysates from CEM cells cultured with 1 × and 5 × IC50 THC were analyzed by immunoblotting. Results indicated clear decreases in the expression of pERK as early as 3 hours of incubation with 1 × IC50 THC (Figure 5C).
Discussion
This study was undertaken to investigate the cytotoxic effect of THC, the active metabolite of cannabis, in a panel of cell lines, with specific interest in examining the relationship between cannabinoid-receptor expression and activation of cell kill. We confirmed that in these cell lines THC was cytotoxic in a concentration-dependent manner. The most significant finding of this study was that the levels of the cannabinoid receptors were varied in each cell line, and that cytotoxicity was independent of their expression.
In the first part of our investigation, we established that THC induced cell death in the 3 cell lines studied. Previous studies have shown that these concentrations are clinically achievable and well tolerated. More specifically, it has been extrapolated in animal models that THC can be tolerated to several grams per kilogram body weight.20 The cytotoxic effects in this study were seen after only 6 hours of exposure to the drug. In contrast to previous reports that have shown THC-mediated cell death is specific to tumors,1 our data demonstrated significant cell death in nonmalignant cells at equivalent concentrations (peripheral blood mononuclear cells). This may be a tissue-specific effect, as the selectivity was observed in neuronal cells. Flow cytometric analysis showed that a portion of the cell kill was by apoptosis occurring from all phases of the cell cycle as early as 6 hours. No cell cycle arrests were seen. Overall, our findings support the work by Guzman's group, who has also shown the cytotoxic effect of THC in cancer cells.11,21,22
Although the mechanism of THC-induced cell kill has not been fully clarified, there is speculation that the cytotoxicity is mediated by activation of cannabinoid receptors. Consequently, we next assessed the levels of CB-R levels by using a novel nonradioactive method involving the use of common biotin/avidin associations.23 However, our method varied from these standard methods by using extended ester linkages, which reduced the steric hindrance conferred by the bulky FITC-fluorochrome24 and should result in a high degree of specificity. Additionally, using biotinylated whole ligands, rather than monoclonal antibodies, ensured that only structurally competent receptors were measured. Methodologically, receptor levels were expressed relative to the autofluorescence controls (the ratio of fluorescence in tested samples to the nude sample). Our results demonstrated the ease and sensitivity of this novel 2-step method and revealed clear differences in CB-receptor levels in the cell lines tested. In particular, CEM and HEL-92 cells displayed the highest levels of CB-R, while HL60 showed very low receptor levels. The CB-R levels did not correlate well with cytotoxic response.
There are currently 2 subsets of cannabinoid receptors, which have a have distinct structure and function. The importance of the individual receptor in the activation of cell kill is unclear, and previous reports have been contradictory.6,10,21,25 However, those studies assessed receptor quantity rather than function. For this reason, we attempted to separate out the roles of the individual CB-R. We initially cultured cells with anandamide, the endogenous ligand at the CB1-R.26 Unfortunately, like other CB1-R agonists that are currently available, its actions are not exclusively related to binding to the receptor. Indeed, anandamide has recently been shown to induce cell kill independently of the CB1-R.27,28 Thus, in the absence of a definitive CB1-R agonist, we studied the roles of the individual CB-R subtypes by blocking the effect of THC using selective synthetic CB-R antagonists. Results showed that THC bound to both CB1-R and CB2-R, and that the CB1-R binding was the more abundant of the 2. The addition of both antagonists concurrently, abolished virtually all of the THC binding to the cells. However, the addition of CB1-R and CB2-R antagonists together did not negate THC-induced cytotoxicity. Moreover, the HL60 cell line, which expressed the lowest levels of both CB-R, was most sensitive to THC. Together, our results suggested that cytotoxicity was independent of the CB-R. Parenthetically, as the CB2-R has been shown to play a possible role in mediating cell death caused by THC,10 we studied the specific role of the CB2-R by culturing cells with the putative CB2-R agonist PEA.16 Our results showed that PEA had no cytotoxic effect in any of the cell lines, which suggested that activation of the CB2-R alone was not enough to elicit a cell kill event. Additionally, PEA binding to CB2-R did not appear to have any effect on the ability of THC to cause cell death. Together, these results suggested CB2-R to have a minor, if any, role in THC-induced cytotoxicity.
The exact mechanism of THC-induced cell kill is far from understood. In our study, apoptosis appeared to be involved, which has been shown in previous studies.3,10,29 THC-induced cell death did not appear to elicit a p53 response, suggesting the absence of drug/DNA interactions. This was shown first by gene expression profiling, which revealed no changes to p53-responsive genes, and confirmed subsequently by Western blot analysis.
It is important to note that the changes to gene expression were only investigated at one time point, 3 hours after exposure to THC. This time point was selected knowing that THC induces significant cell death after only 6 hours; therefore, we selected this time point to investigate early changes caused by THC rather than nonspecific changes associated with apoptosis. These investigations revealed significant changes to 3 genes, 2 of which were histone H1 isoforms. THC consistently down-regulated these genes. The role of these proteins in cell survival pathways is unclear, although it has recently been shown that another histone H1 isoform, H1.2, plays an important role in positively modulating apoptotic signaling.30 The other significant change observed was a marked up-regulation of the DUSP gene, encoding MKP3. MKP3 is a member of the dual-specificity phosphatase subfamily that negatively regulates members of the MAPK/ERK, stress-activated protein kinase/c-jun N-terminal kinase (SAPK/JNK), and p38 signaling pathways. MKP3 specifically binds and inactivates ERK2.31 Interestingly the MAPKK that positively regulates ERK2, MEK2, was one of the most significantly down-regulated genes in response to THC treatment (Table 1). Consistently, both these changes in gene expression would result in deactivation of the ERK2 protein and downstream MAPK signaling, which was confirmed by immunoblotting analyses. This has an important role in mediating cell survival.32,33 Indeed, inactivation of ERK2, through either MEK2 or MAKP3, has been shown to induce apoptosis.34-36 Interestingly, it was recently shown that MKP3 is a tumor suppressor in pancreatic cancer, over-expression of which resulted in apoptosis.35 Our data suggest the involvement of the MAPK pathway in mediating the cellular effects of THC, possibly through inactivation of ERK2. Together, these data intimate a mechanism of action that may involve modulation of signal transduction pathways, possibly through receptor tyrosine kinases and/or G-protein–coupled receptors.
Although THC has been previously shown to interact with γ-irradiation, enhancing cytotoxicity in leukemic cell lines,37 its effects in combination with common cytotoxic chemotherapy agents has not been assessed. Our results showed no clear hyperadditive interaction between these 2 classes of drugs, and the effects were purely additive.17 Specifically, the degree of cell kill induced by concurrent culture of the agents was not significantly different to the calculated/expected amount. Additionally, our data also showed that smaller, sublethal concentrations of THC did not enhance the cytotoxic effects of the chemotherapy. Although this simplistic approach to investigating synergy/hyperadditivity is limited and less powerful than the usual median-dose effect and isobologram methodologies, strong synergistic interactions would have been detected.17 It would be interesting to see whether there would be any benefit of combining THC with common antileukemic agents, such as the anthracyclines and etoposide.
Overall, these data reaffirm the complexity of the data that are currently available. A cytotoxic response to THC has been studied previously; however, these studies have only correlated cannabinoid receptor mRNA levels with this response. In contrast, we believe that this is the first report to directly correlate ligand-to-receptor binding with cell death. Our data strongly suggest that the cytotoxic effects of THC are independent of both the cannabinoid receptors and of p53. It is important to emphasize that THC was exceptionally efficacious, inducing cell kill as early as 6 hours, and although microarray analysis did not fully elucidate the precise mechanism of cell death, it reinforced the importance of the MAPK pathway in this process. Finally, it is important to stress that THC did not act exclusively on cancer cells and did not appear to interact synergistically with or enhanced platinum chemotherapy. For this reason, additional studies investigating the effect of THC in other cancer cell types, as well as further studies to clarify a mechanism of action, are required.
Prepublished online as Blood First Edition Paper, September 28, 2004; DOI 10.1182/blood-2004-03-1182.
Supported by the Orchid Cancer Appeal (T.B.P. and T.O.).
This work was developed by T.P. and W.M.L. and was independently undertaken by the New Drug Study Group led by W.M.L. and D.P. This work was performed in the Barry Reed Oncology Laboratory, headed by S.J.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Jackie Perry (Barry Reed Oncology Laboratory, London) for technical assistance and Anita Sammugathasan (Cancer Research UK, London) for help in analyzing the microarray data.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal