Abstract
Telomerase represents an attractive target for a mechanism-based therapeutic approach because its activation has been associated with unlimited proliferation in most cancer cells. Recently, a nonnucleosidic small molecule inhibitor, BIBR1532 (2-[(E)-3-naphtalen-2-yl-but-2-enoylamino]-benzoic acid), has been identified that is highly selective for inhibition of telomerase, resulting in delayed growth arrest of tumor cells. Here we examined the effects of BIBR1532 in different leukemia cell lines as well as in primary cells from patients with acute myeloid leukemia (AML) and chronic lymphocytic leukemia (CLL) in short-term culture assays. We observed a dose-dependent direct cytotoxicity in concentrations ranging from 30 to 80 μM. Interestingly, cell death was not dependent on the catalytic activity of telomerase but was delayed in cells with very long telomeres. We observed time-dependent individual telomere erosion, which was associated with loss of telomeric repeat binding factor 2 (TRF2) and increased phosphorylation of p53. Importantly, the proliferative capacity of normal CD34+ cells from cord blood and leukapheresis samples was not affected by treatment with BIBR1532. We conclude that using this class of telomerase inhibitor at higher concentrations exerts a direct cytotoxic effect on malignant cells of the hematopoietic system, which appears to derive from direct damage of the structure of individual telomeres and must be dissected from telomerase-suppressed overall telomere shortening. (Blood. 2005; 105:1742-1749)
Introduction
Maintenance of telomeres is essential for unlimited cellular proliferation and confers immortality in cancer cells.1,2 Telomeres in human cells consist of repetitive double-stranded repeats of the sequence TTAGGG, which terminate in a single-stranded 3′ extension overhang of the G-rich strand.3 Their major function is to cap the ends of chromosomes and to provide genetic stability. This capping function is mediated by a special architecture in which the 3′ overhang participates with telomere binding proteins in a large loop structure called T-loop.4 In addition, a number of proteins constitute the mammalian telomere and serve to regulate the telomere structure.5 The telomeric repeat binding factors TRF1 and TRF2 bind to double-stranded telomeric DNA and are involved in telomere length regulation and telomere protection.6,7 Since TRF2 assists in formation of the T-loop, it is thought to play a major role in stability of the telomere structure.8 In fact, loss of TRF2 has been associated with induction of apoptosis and senescence in human cells.9
Telomerase is a ribonucleoprotein enzyme that synthesizes telomere repeats de novo.10 In human cells, the telomerase holoenzyme consists of a high-molecular-weight complex with a template-containing RNA subunit,11 hTR, and protein components including the catalytic subunit human telomerase reverse transcriptase,12 hTERT. In most normal somatic cells, telomerase activity is absent and telomere repeats are lost with cell division and with aging. Telomere attrition beyond a certain threshold is assumed to uncap chromosome ends, which subsequently induces DNA damage13 and onset of replicative senescence.14,15 In contrast, about 80% to 90% of cancer cells have detectable telomerase activity, which leads to stabilization of telomeres and unlimited growth potential.16 Likewise, telomerase activity has been detected in the majority of acute and chronic leukemias, particularly with disease progression.17 Interestingly, low levels of activity have also been found in activated lymphocytes18 and hematopoietic stem cells,19 which appear to confer extended proliferative capacity but do not prevent overall telomere shortening.20
Since most cancer cells are reliant on telomerase for their survival, telomerase has become an attractive target for the development of new cancer therapeutics.21 Indeed, during recent years a variety of different classes of telomerase inhibitors, which block the catalytic activity of the enzyme or the access of telomerase to the telomere, have been evaluated.22-24 “Proof of principle” for validation of telomerase inhibition as therapeutic concept was demonstrated in human tumor cells using dominant-negative mutant forms of hTERT.25,26 In these experiments, telomerase activity was abolished that was associated with continuous telomere shortening leading to senescence or apoptosis. It is assumed that indirect inhibition of telomerase induces growth arrest but subsequent loss of telomere function, which derives from uncapping of individual short telomeres.27 A challenge for potential clinical application of pharmaceutically useful telomerase inhibitors is its therapeutic window. Since the antiproliferative effect is dependent on the telomere length of a given tumor cell, clinical response will be delayed until chromosomes run out of their telomeres. This will require a series of cell divisions to become apparent, and treatment may have to be given continuously for weeks to months, potentially in conjunction with other modalities.
Recently a novel structural class of nonpeptidic, nonnucleosidic inhibitors of human telomerase has been described28 that is specific in inhibition of the catalytic activity of telomerase. One example of this class of compounds, designated BIBR1532 (2-[(E)-3-naphtalen-2-yl-but-2-enoylamino]-benzoic acid), inhibits the in vitro processivity of telomerase in a dose-dependent manner, with halfmaximal inhibitory concentrations (IC50) of 93 nM.29 The selectivity of BIBR1532 was assessed in a panel of DNA and RNA polymerases, showing that none of these enzymes was inhibited in concentrations up to 100 μM.
In this work, we investigated the activity of BIBR1532 in normal and malignant hematopoietic cells. In addition to its known role in telomerase inhibition, we provide evidence that high-dose BIBR1532 has a direct cytotoxic effect in leukemia cells but not in normal hematopoietic stem cells. The mechanism that induces immediate rather than delayed growth arrest is most likely due to direct damage to the telomere structure, which possibly reflects a novel approach for cancer therapy.
Patients, materials, and methods
Reagents and antibodies
BIBR1532 was synthesized by Boehringer Ingelheim (Biberach, Germany) and prepared as described previously.28 A stock solution of BIBR1532 at a concentration of 1 mM was prepared by dissolving the compound in sterile dimethyl sulfoxide (DMSO) and stored at -20°C until use. The TRF2, p53, phosphorylated p53 (phospho-p53), and β-actin antibodies were purchased from Biocarta (Hamburg, Germany).
Cells and cell lines
The JVM13 cell line represents an immortalized B-cell line from a patient with prolymphocytic leukemia, HL-60 is an acute myeloid leukemia (AML) cell line, Jurkat is a T-cell lymphoma cell line, and Nalm1 was derived from a patient with chronic myeloid leukemia (CML) in blastic phase. All cell lines were purchased from DFMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany). Cells were cultured in RPMI 1640 medium (Invitrogen, Karlsruhe, Germany) supplemented by 10% heat-inactivated fetal calf serum (Invitrogen), 2 mM l-glutamine, and 1% penicillin/streptomycin (complete medium) in a humidified 5% CO2 atmosphere. Human diploid fibroblast cells (HK1 line) were cultured as described.30 In addition to telomerase-negative wild-type cells, immortalized HK1 cells were used expressing ectopic hTERT.31,32
Patient population and isolation of leukemia cells
Ethylene diamine tetraacetic (EDTA) venous blood samples were collected from consenting CLL and AML patients and separated over Ficoll-hypaque (Biochrome, Berlin, Germany). For the CLL samples, the mononuclear cell fraction was washed twice and adjusted to 1 to 3 × 107/mL. Cells were cultured in 3 mL medium using 6-well plates, and the following concentrations of BIBR1532 were used: 10 μM, 30 μM, 50 μM, and 80 μM. Likewise, corresponding concentrations of DMSO were used as another negative control in addition to the negative control (no inhibitor). Cultures were maintained for a minimum of 14 days, and every 4 days aliquots were taken (volume replaced by medium) for different assays. Cord blood and leukapheresis products following stem cell mobilization were separated over Ficoll-hypaque and washed. CD34+ cells were purified by positive selection using a magnetic-activated cell sorter (MACS) progenitor enrichment kit according to the manufacturer's protocol (Miltenyi Biotec, Bergisch Gladbach, Germany) and adjusted to 0.5 to 1 × 105/mL. Cells were cultured in CellGro serum-free medium (CellGenix, Freiburg, Germany) supplemented with stem cell factor (SCF) 300 ng/mL, Fms-like tyrosine kinase 3 ligand (Flt-3 L) 300 ng/mL, interleukin-3 (IL-3) 100 ng/mL, IL-6 10 ng/mL (each provided by CellGenix), and granulocyte colony-stimulating factor (G-CSF) 10 ng/mL (Amgen, Thousand Oaks, CA). Cells were maintained for 21 days in culture at 37°C and 5% CO2; medium was changed at a 7-day interval, at which the cells were counted and reseeded at 3 to 6 × 106/mL, and the rest was harvested for different assays.
Colony-forming assays
CD34+ cells from cord blood or normal bone marrow were plated at 1 × 103/mL in serum-free methylcellulose medium supplemented with cytokines (MethoCult, GF H4434; Stemcell Technologies, Vancouver, BC) with different concentrations of BIBR1532 as mentioned before. Burstforming unit erythroid (BFU-E) and colony-forming unit granulocyte/monocyte (CFU-GM) were counted by day 14 of culture.
Measurement of cell death
Determination of CLL cell viability in this study is based on the analysis of mitochondrial transmembrane potential by 3,3′ dihexyloxacarbocyanine iodine (DiOC6; Molecular Probes, Eugene, OR) and cell membrane permeability to propidium iodide (PI; Molecular Probes), as previously described.33,34 For comparison, chronic lymphocytic leukemia (CLL) cell viability was also examined using the different relative size and granularity (forward scatter and side scatter) characteristics of vital and dead cells. Trypan blue exclusion was also used to determine the number of viable cells.
Western blot analysis
Protein was extracted with recombinant immunoblotting assay (RIBA) lysis buffer. An equal amount of protein (10-50 μg) of the samples was separated on 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel and transferred onto transfer membrane (Immobilon-P; Millipore, Bedford, MA). After blocking (in 3% nonfat dry milk, 3% bovine serum albumin [BSA] in phosphate-buffered saline [PBS]), the membranes were incubated with primary and secondary antibodies, and the bands were detected by the chemiluminescence method according to the manufacturer's recommendations (Amersham Biosciences, Little Chalfont, United Kingdom).
Telomere length analysis using flow-fluorescence in situ hybridization (Flow-FISH)
To determine mean telomere length in individual cells, cells were hybridized in situ with a fluorescent telomere-specific peptide nucleic acid (PNA) probe as described recently.31,35 Telomere length was expressed in telomere fluorescence units (TFUTRF) based on calibration experiments using telomere restriction fragment (TRF) length analysis by Southern blotting.
Telomerase activity measurements
Telomerase activity of cell populations was determined using the Telo-TaGGG polymerase chain reaction enzyme-linked immunosorbent assay (PCR ELISAPLUS) kit (Roche, Mannheim, Germany) according to the manufacturer's protocol (telomeric repeat amplification protocol [TRAP] assay). Heat-treated cellular lysates (85°C for 10 minutes) were used as negative controls for each sample. Samples are considered as telomerase positive if the difference in absorbance is higher than 2-fold background activity.
WST-1 assays
Cells (0.5 to 5 × 104) were plated as triplicates in complete RPMI 1640 medium with various concentrations of BIBR1532. After 24 to 72 hours, water-soluble tetrazolium (WST-1) (Roche) was added, which is transformed into formazan by mitochondrial reductase systems. The increase in the number of viable cells results in an increase of activity of mitochondrial dehydrogenases, leading to an increase of formazan dye formed, which was quantified by ELISA reader after 2, 3, and 4 hours of incubation.
Fluorescence in situ hybridization and quantitative image analysis
Metaphase chromosomes from JVM13 cells were prepared after 2 hours of treatment with 0.1 μg/mL colcemid (Invitrogen) followed by hypotonic solution in 75 mM KCl (Sigma-Aldrich, Steinheim, Germany) according to standard methods. The number of analyzed metaphases was between 8 and 15. Individual telomere length was analyzed by quantitative fluorescence in situ hybridization (Q-FISH) as described previously using a Cy3-labeled, telomere-specific probe (Applied Biosystems, Foster City, CA).15,28 Digital images of metaphase spreads embedded in Vectashield mounting medium (Vector Laboratories, Burlingame, CA) were recorded with a digital camera (Sensys; Photometric, Tucson, AZ) on an Axioplan II fluorescence microscope (Zeiss, Jena, Germany) using the Vysis workstation QUIPS (Vysis, Downers Grove, IL). Final images were prepared using Adobe Photoshop 6.0 software (Adobe Systems, San Jose, CA).
Statistical analysis
Data analysis was performed using Microsoft Excel (Microsoft, Redmond, WA) with WinSTAT module (R. Fitch Software, Staufen, Germany) and Microcal Origin software (Origin Lab, Northampton, MA). Results are shown as means ± SE of values obtained in independent experiments. Student t test was used to determine statistical significance.
Results
Antiproliferative effect of BIBR1532 in leukemia cell lines
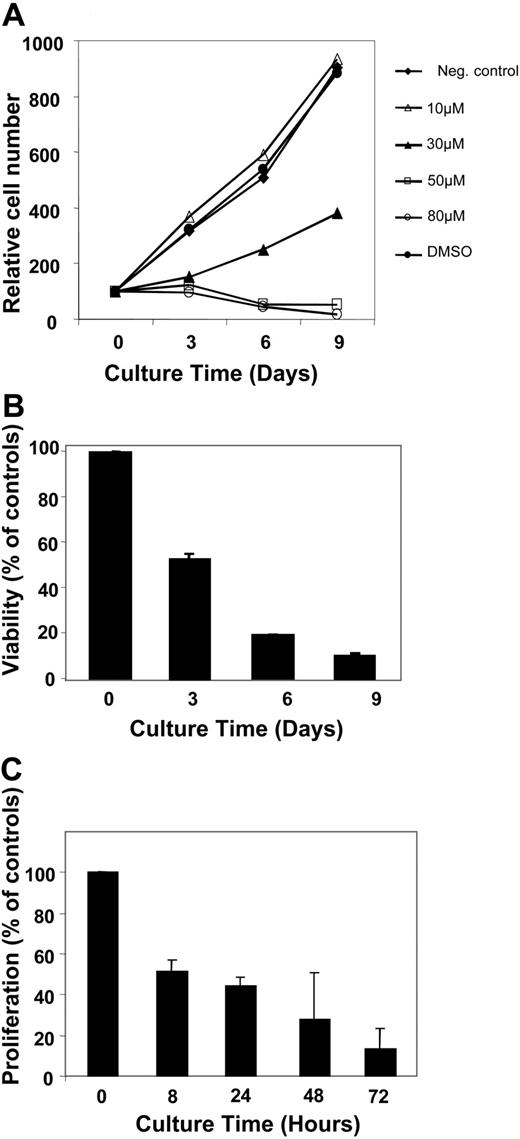
The telomerase inhibitor BIBR1532 has been shown to inhibit telomerase activity in several human cancer cell lines, resulting in progressive shortening and subsequent dysfunction of telomeres when used at concentration of 10 μM.28 Here we analyzed short-term effects of BIBR1532 using concentrations between 10 and 80 μM in the CLL cell line, JVM13. Unlike a delayed effect that was dependent on substantial telomere erosion, a direct antiproliferative effect was observed in a dose-dependent range using inhibitor concentrations higher than 20 μM (Figure 1A). With treatment of 80 μM BIBR1532, we found more than 80% decrease in viability of JVM13 cells within 9 days (Figure 1B-C). In addition, increasing concentrations of BIBR1532 showed a dose-dependent inhibition of proliferation using the WST-1 proliferation assay, which translated into an IC50 value of 52 μM. Similar results were obtained for other leukemia cell lines such as Nalm-1, HL-60, and Jurkat (data not shown). Thus, increasing the doses of BIBR1532 in cell cultures (to more than the concentration that has been reported to induce telomere shortening) does result in a direct antiproliferative effect in contrast to delayed growth arrest.
Antiproliferative effect of BIBR1532 in JVM13 leukemia cell line. (A) JVM13 cells were cultured for 9 days in RPMI medium supplemented with increasing concentrations of BIBR1532. Cells were counted at indicated time points using trypan blue exclusion assay. (B) The viability of JVM13 cells treated with 80 μM was determined at indicated time points using forward scatter (FSC)/side scatter (SSC) pattern of FACS analysis. A gradual decrease in viability was observed with cells cultured in the presence of 80 μM BIBR1532. Values represent means and SEM from triplicates. (C) Proliferation of JVM13 cells was determined using WST-1 assay.36 Bars represent the proliferation of cells cultured in the presence of 80 μM BIBR1532 as percentage of controls at indicated time points. Values are means and SEM from triplicates.
Antiproliferative effect of BIBR1532 in JVM13 leukemia cell line. (A) JVM13 cells were cultured for 9 days in RPMI medium supplemented with increasing concentrations of BIBR1532. Cells were counted at indicated time points using trypan blue exclusion assay. (B) The viability of JVM13 cells treated with 80 μM was determined at indicated time points using forward scatter (FSC)/side scatter (SSC) pattern of FACS analysis. A gradual decrease in viability was observed with cells cultured in the presence of 80 μM BIBR1532. Values represent means and SEM from triplicates. (C) Proliferation of JVM13 cells was determined using WST-1 assay.36 Bars represent the proliferation of cells cultured in the presence of 80 μM BIBR1532 as percentage of controls at indicated time points. Values are means and SEM from triplicates.
Antiproliferative effect of BIBR1532 on primary leukemia cells
In order to explore the antiproliferative effect of BIBR1532 on primary leukemia cells in vitro, we treated peripheral blood cells from patients with CLL in short-term cultures as previously described.34 Samples from 20 different CLL patients were used in these experiments (Table 1). The mean telomere length for patients with early-stage CLL, RAI 0 to II (n = 12), was found to be 7.0 ± 0.6 kilobase (kb), whereas telomeres were relatively shorter in those with late-stage CLL, RAI III to IV (n = 7) (6.3 ± 1.2 kb) although the difference was not significant (P = .1). Interestingly, only 33% of patient samples with early-stage CLL exhibited telomerase activity, while in those with late-stage CLL, 57% were telomerase positive.
Summary of CLL samples
Patient code . | TL . | TA . | RAI . | Viability index . |
|---|---|---|---|---|
| 001TA | 7.4 | − | NA | 25 |
| 002GB | 7.1 | + | IV | 97 |
| 003ZW | 5.4 | − | III | 1 |
| 004LW | 5.9 | + | II | 9 |
| 005GP | 6.9 | − | 0 | 2 |
| 006GK | 7.2 | + | II | 1 |
| 007TK | 6.6 | + | IV | 2 |
| 008RH | 6.9 | − | II | 34 |
| 009SH | 6.9 | + | II | 1 |
| 010IK | 6.9 | − | II | 4 |
| 011HH | 6.5 | − | II | 31 |
| 012CG | 5.7 | + | IV | 80 |
| 013SE | 8.5 | − | IV | 89 |
| 014TP | 7.4 | − | II | 1 |
| 015BJ | 7.8 | − | I | 1 |
| 016VG | 7.7 | − | I | 98 |
| 017FL | 7.3 | − | II | 2 |
| 018DM | 5.4 | + | IV | 8 |
| 019WA | 5.2 | − | IV | 5 |
| 020MH | 6.2 | + | 0 | 1 |
Patient code . | TL . | TA . | RAI . | Viability index . |
|---|---|---|---|---|
| 001TA | 7.4 | − | NA | 25 |
| 002GB | 7.1 | + | IV | 97 |
| 003ZW | 5.4 | − | III | 1 |
| 004LW | 5.9 | + | II | 9 |
| 005GP | 6.9 | − | 0 | 2 |
| 006GK | 7.2 | + | II | 1 |
| 007TK | 6.6 | + | IV | 2 |
| 008RH | 6.9 | − | II | 34 |
| 009SH | 6.9 | + | II | 1 |
| 010IK | 6.9 | − | II | 4 |
| 011HH | 6.5 | − | II | 31 |
| 012CG | 5.7 | + | IV | 80 |
| 013SE | 8.5 | − | IV | 89 |
| 014TP | 7.4 | − | II | 1 |
| 015BJ | 7.8 | − | I | 1 |
| 016VG | 7.7 | − | I | 98 |
| 017FL | 7.3 | − | II | 2 |
| 018DM | 5.4 | + | IV | 8 |
| 019WA | 5.2 | − | IV | 5 |
| 020MH | 6.2 | + | 0 | 1 |
Indicated is the classification for the patients according to RAI staging system. Telomerase activity (TA) was determined using the TRAP (PCR ELISAPLUS) assay, the mean telomere length (TL) was measured using flow-FISH. The response to 80 μM BIBR1532 is expressed as viability index representing the viable cells as percentage of controls after 2 weeks of cell culture. All experimental conditions were performed in duplicates.
— indicates negative; NA, not available; and +, positive.
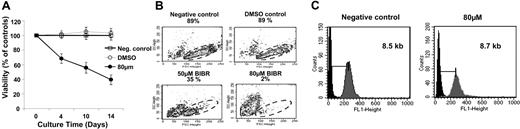
The cytotoxic effect of BIBR1532 on the CLL samples was examined using the DiOC6/PI assay and fluorescence-activated cell sorter (FACS) analysis of forward/side scatter (Figure 2A-B). Of the 20 samples tested, 16 showed more than 50% loss of viability with 80 μM BIBR1532 within 14 days of culture, whereas 14 samples showed more than 80% loss of viability during that time period compared with DMSO-treated and negative controls (Table 1). Similar to the analysis on leukemic cell lines, the IC50 value was found to be in the range of 50 μM (53 ± 3.8 μM). Notably, 4 samples responded poorly to the treatment with BIBR1532, demonstrating less than 50% loss of viability. Of the 4 nonresponders, 3 had progressive disease, including 1 (002GB) who had a long history of different treatment modalities and was refractory to chemotherapy upon time point of analysis.
Antiproliferative effect of BIBR1532 in primary CLL cells. (A) Primary CLL cells were cultured in complete RPMI for 14 days supplemented with 80 μM BIBR1532. As previously reported,34 viability was determined using the DiOC6/PI assay at indicated time points and is expressed as percentage of the controls. Values are means ± SEM (n = 20). (B) FSC/SSC pattern is shown for a representative CLL patient sample measured by day 14 using FACS analysis. Gated cells represent the viable cells at the end of culture. (C) The mean telomere length was measured using flow-FISH technique.35 Representative telomere fluorescence histograms of one CLL patient sample are demonstrated following treatment of BIBR1532 and of DMSO (control cells) for 14 days. Telomere fluorescence intensity was calculated by subtracting the mean background fluorescence (gray) from the corresponding mean fluorescence provided by the telomere-specific probe (black).
Antiproliferative effect of BIBR1532 in primary CLL cells. (A) Primary CLL cells were cultured in complete RPMI for 14 days supplemented with 80 μM BIBR1532. As previously reported,34 viability was determined using the DiOC6/PI assay at indicated time points and is expressed as percentage of the controls. Values are means ± SEM (n = 20). (B) FSC/SSC pattern is shown for a representative CLL patient sample measured by day 14 using FACS analysis. Gated cells represent the viable cells at the end of culture. (C) The mean telomere length was measured using flow-FISH technique.35 Representative telomere fluorescence histograms of one CLL patient sample are demonstrated following treatment of BIBR1532 and of DMSO (control cells) for 14 days. Telomere fluorescence intensity was calculated by subtracting the mean background fluorescence (gray) from the corresponding mean fluorescence provided by the telomere-specific probe (black).
The cytotoxic effect was not dependent on telomerase activity as measured by TRAP assay, because telomerase-negative samples responded as well. Furthermore, the response did not appear to be influenced by differences in overall telomere length, since samples with relatively long telomeres such as 014TP (7.4 kb) or 015BJ (7.8 kb) showed marked reduction in viability as did samples with short telomeres such as 003ZW (5.4 kb) and 019WA (5.2 kb). The mean telomere length did not change significantly within the culture period of 14 days as measured by flow-FISH (Figure 2C).
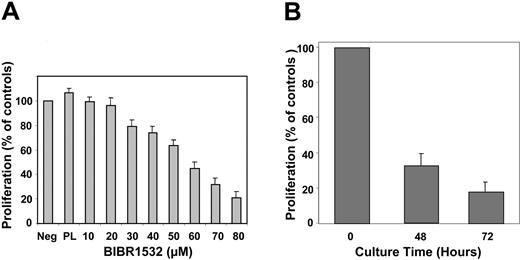
We also investigated the antiproliferative effect of BIBR1532 for acute myeloid leukemia (AML). Mononuclear bone marrow cells from 9 different patients (Table 2) were cultured with increasing concentrations of BIBR1532 for 48 and 72 hours. Again, concentrations higher than 20 μM resulted in a direct antiproliferative effect (Figure 3). Of 9 AML samples, 8 showed more than 70% inhibition of proliferation with 80 μM BIBR1532. One sample from a patient with secondary AML responded poorly to the treatment, showing less than 50% inhibition of proliferation. The IC50 value was 56 ± 5 μM (Figure 3A). As described for CLL cells, the cytotoxic effect of high-dose BIBR1532 was not dependent on the presence of telomerase activity. This supports the hypothesis of an additional cytotoxic mechanism.
Summary of samples from patients with AML
Patient code . | TA . | TL . | CD34 % . | Blast % in BM . | FAB . | Karyotype . | Inhibition of proliferation, % . |
|---|---|---|---|---|---|---|---|
| 101 | + | 6.1 | + | 90 | M2 | ND | > 70 |
| 102 | + | ND | + | 75 | M4 | Normal | > 70 |
| 103 | + | 7.3 | − | 75 | M5 | Monosomy 12, 16, 17 | > 70 |
| 104 | − | 7.6 | − | 50 | ND | Normal | > 70 |
| 105 | − | ND | ND | 94 | M2 | Complex | > 70 |
| 106 | + | 6.3 | − | 85 | M4/2ryAML | Complex | < 50 |
| 107 | ND | ND | + | 80 | M4/M5 | Normal | > 70 |
| 108 | ND | ND | + | 80 | M2/MDS | Normal | > 70 |
| 109 | ND | ND | + | 75 | M5 | Normal | > 70 |
Patient code . | TA . | TL . | CD34 % . | Blast % in BM . | FAB . | Karyotype . | Inhibition of proliferation, % . |
|---|---|---|---|---|---|---|---|
| 101 | + | 6.1 | + | 90 | M2 | ND | > 70 |
| 102 | + | ND | + | 75 | M4 | Normal | > 70 |
| 103 | + | 7.3 | − | 75 | M5 | Monosomy 12, 16, 17 | > 70 |
| 104 | − | 7.6 | − | 50 | ND | Normal | > 70 |
| 105 | − | ND | ND | 94 | M2 | Complex | > 70 |
| 106 | + | 6.3 | − | 85 | M4/2ryAML | Complex | < 50 |
| 107 | ND | ND | + | 80 | M4/M5 | Normal | > 70 |
| 108 | ND | ND | + | 80 | M2/MDS | Normal | > 70 |
| 109 | ND | ND | + | 75 | M5 | Normal | > 70 |
Telomerase activity (TA) was determined using TRAP assay; telomere length (TL) was analyzed using flow-FISH. The response to 80 μM BIBR1532 is expressed as percentage of proliferation inhibition of the controls within 72 hours of culture.
FAB indicates French-American-British; +, positive; −, negative; ND, not detected.
Antiproliferative effects of BIBR1532 in primary AML blasts. (A) Mononuclear bone marrow cells from AML patients (n = 9; Table 3) were plated in 96-well plates with indicated concentrations of BIBR1532. WST-1 reagent was added after 72 hours for the determination of proliferation, which is expressed as a percentage of the controls ± standard deviation of the mean (SDM). Experiments were performed in triplicate. (B) BIBR1532 (80 μM) induced a gradual decrease of proliferation within 72 hours (> 80%). Bars represent means of percentage of proliferation measured by WST-1 assay at the indicated time points, ± SDM (n = 9). PL indicates placebo (DMSO).
Antiproliferative effects of BIBR1532 in primary AML blasts. (A) Mononuclear bone marrow cells from AML patients (n = 9; Table 3) were plated in 96-well plates with indicated concentrations of BIBR1532. WST-1 reagent was added after 72 hours for the determination of proliferation, which is expressed as a percentage of the controls ± standard deviation of the mean (SDM). Experiments were performed in triplicate. (B) BIBR1532 (80 μM) induced a gradual decrease of proliferation within 72 hours (> 80%). Bars represent means of percentage of proliferation measured by WST-1 assay at the indicated time points, ± SDM (n = 9). PL indicates placebo (DMSO).
BIBR1532 does not affect the proliferative capacity of normal hematopoietic progenitor cells
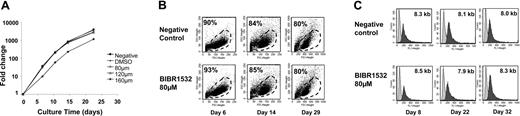
Since some normal cells, including those with stem cell-like properties, retain the ability to activate telomerase physiologically, inhibition of the enzyme may potentially have detrimental effects on their proliferative capacity. Therefore, we investigated the effects of BIBR1532 on hematopoietic, nonmalignant progenitor cells. CD34+ cells from cord blood (n = 7) and leukapheresis products (n = 5) were cultured for 21 days in a serum-free medium supplemented with cytokines and with increasing concentrations of BIBR1532. Cells were harvested periodically for cell count, telomere length measurement, and viability assays. We did not observe any negative effect of BIBR1532 on viability or cell proliferation using concentrations up to 120 μM as determined by trypan blue exclusion assay and the DiOC6/PI assay (Figure 4A-B).
Proliferation and viability of normal hematopoietic progenitor cells. (A) CD34+ cells enriched from cord blood samples (n = 7) were cultured in the presence of BIBR1532 in serum-free medium supplemented with cytokines (SCF, Flt, IL-3, IL-6, and G-CSF). At the indicated time intervals, viable cells were counted using trypan blue exclusion assay. Note that cell expansion was moderately reduced only at drug concentrations of 160 μM. Experiments were performed in duplicates. (B) Viability was measured using FSC/SSC pattern and DiOC6/PI staining at the indicated time points. FACS diagrams represent the nontreated control cells, the DMSO-treated cells (not shown), and the cells cultured with 80 μM BIBR1532. There was no decrease in cell viability of cells cultured under presence of 80 μM BIBR1532 compared with negative and DMSO controls at any time point of culture. (C) The mean telomere length was measured in peripheral blood stem cells (CD34+ leukapheresis sample) following treatment with BIBR1532. Representative telomere fluorescence histograms analyzed at indicated time points are shown, which illustrate no substantial difference in the mean telomere length of treated compared with untreated cells.
Proliferation and viability of normal hematopoietic progenitor cells. (A) CD34+ cells enriched from cord blood samples (n = 7) were cultured in the presence of BIBR1532 in serum-free medium supplemented with cytokines (SCF, Flt, IL-3, IL-6, and G-CSF). At the indicated time intervals, viable cells were counted using trypan blue exclusion assay. Note that cell expansion was moderately reduced only at drug concentrations of 160 μM. Experiments were performed in duplicates. (B) Viability was measured using FSC/SSC pattern and DiOC6/PI staining at the indicated time points. FACS diagrams represent the nontreated control cells, the DMSO-treated cells (not shown), and the cells cultured with 80 μM BIBR1532. There was no decrease in cell viability of cells cultured under presence of 80 μM BIBR1532 compared with negative and DMSO controls at any time point of culture. (C) The mean telomere length was measured in peripheral blood stem cells (CD34+ leukapheresis sample) following treatment with BIBR1532. Representative telomere fluorescence histograms analyzed at indicated time points are shown, which illustrate no substantial difference in the mean telomere length of treated compared with untreated cells.
In addition, no significant differences in the mean telomere length were measured by flow-FISH at different time points of suspension culture compared with control cultures with DMSO and without any treatment (Figure 4C). Finally, we investigated the effects of BIBR1532 in clonogenic assays. These were initiated from suspension culture cells by day 10. Again, no inhibition of colony formation was observed using increasing concentrations of BIBR1532 (Table 3).
Clonogenic assay for cord blood samples in the presence of BIBR1532
. | BFU-E . | CFU-GM . | Total count . |
|---|---|---|---|
| Negative control | 128 ± 111 | 13 ± 8 | 138 ± 117 |
| DMSO | 126 ± 82 | 24 ± 14 | 148 ± 90 |
| 10 μM | 114 ± 98 | 12 ± 8 | 125 ± 105 |
| 30 μM | 133 ± 78 | 24 ± 13 | 155 ± 86 |
| 50 μM | 120 ± 67 | 25 ± 13 | 144 ± 75 |
| 80 μM | 132 ± 82 | 22 ± 11 | 152 ± 89 |
. | BFU-E . | CFU-GM . | Total count . |
|---|---|---|---|
| Negative control | 128 ± 111 | 13 ± 8 | 138 ± 117 |
| DMSO | 126 ± 82 | 24 ± 14 | 148 ± 90 |
| 10 μM | 114 ± 98 | 12 ± 8 | 125 ± 105 |
| 30 μM | 133 ± 78 | 24 ± 13 | 155 ± 86 |
| 50 μM | 120 ± 67 | 25 ± 13 | 144 ± 75 |
| 80 μM | 132 ± 82 | 22 ± 11 | 152 ± 89 |
CD34+ cord blood cells (n = 7) were harvested from suspension culture by day 10, and cultured in serum-free methylcellulose medium supplemented with cytokines (SCF, GM-CSF, G-CSF, IL-3, IL-6, and erythropoietin [EPO]) and with increasing concentrations of BIBR1532 for 14 days. Experiments for each individual specimen were performed in triplicate.
Note: There is no significant difference observed in number or type of colonies at day 14 for those cultured with up to 80 μM BIBR1532 compared with the controls.
BFU-E indicates burst-forming unit erythroid; CFU-GM, colony-forming unit granulocyte/monocyte.
High-dose BIBR1532 induces rapid telomere dysfunction and apoptosis
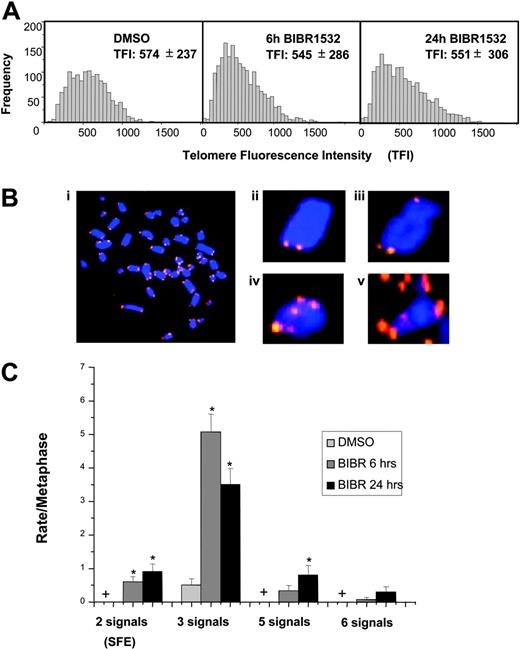
Because higher concentrations of BIBR1532 directly induced growth arrest in AML and CLL cells without detectable changes in overall telomere length as measured by flow-FISH, we thought to analyze individual telomere length dynamics in JVM13 cells using the Q-FISH technique, which is more sensitive to identify subtle changes in telomere dynamics. Although it is a major challenge to generate metaphase spreads from cells entering cell cycle exit, we were able to obtain 8 to 15 metaphases from this cell line following incubation with 80 μM BIBR1532 for different time points. In contrast to the flow-FISH analysis, a small but significant reduction of the mean telomere fluorescence (P < .001) was found between control (telomere fluorescence intensity [TFI], 574 ± 237) and BIBR1532-treated JVM13 cells (6 hours: TFI, 545 ± 286; 24 hours: TFI, 551 ± 306) (Figure 5A). Furthermore, we observed that the distribution of telomere fluorescence values was increasingly skewed as it has been described upon replicative aging of normal diploid fibroblasts.15 Strikingly, the Q-FISH analysis revealed that individual telomeres were progressively lost in a time-dependent manner (Table 4). Whereas telomere spots were present on both chromatid arms in JVM13 cells treated with DMSO, the rate of chromosomes with signal-free ends increased gradually following treatment with high-dose BIBR1532.
High-dose BIBR1532 induces telomere dysfunction in JVM13 cells. Individual telomere length was analyzed by quantitative fluorescence in situ hybridization (Q-FISH) as described previously.28,37 The number of analyzed metaphases was between 8 and 15 (Table 4). (A) The histograms represent telomere fluorescence intensity values (TFI) of JVM13 cells treated with 80 μM BIBR1532 compared with DMSO control at indicated time points. Values represent means ± SD. (B) The presence of telomere spots was counted at individual sister chromatids. (i) Representative metaphase spread from JVM13 cells treated with 80 μM BIBR1532. (ii) Chromosome missing 2 telomere signals. (iii) Chromosome missing one telomere signal from 1 chromatid (= 3 signals). (iv) Chromosome showing 5 telomere signals. (v) Chromosome showing 6 telomere signals (end-to-end fusion). (C) Graphic summary of number of telomere spots/chromosome deviating from the normal distribution of 4 telomeres per chromosome. The values represent mean ± standard error. * indicates a significant difference from cells treated with DMSO (P < .05); + indicates zero events for DMSO-treated cells.
High-dose BIBR1532 induces telomere dysfunction in JVM13 cells. Individual telomere length was analyzed by quantitative fluorescence in situ hybridization (Q-FISH) as described previously.28,37 The number of analyzed metaphases was between 8 and 15 (Table 4). (A) The histograms represent telomere fluorescence intensity values (TFI) of JVM13 cells treated with 80 μM BIBR1532 compared with DMSO control at indicated time points. Values represent means ± SD. (B) The presence of telomere spots was counted at individual sister chromatids. (i) Representative metaphase spread from JVM13 cells treated with 80 μM BIBR1532. (ii) Chromosome missing 2 telomere signals. (iii) Chromosome missing one telomere signal from 1 chromatid (= 3 signals). (iv) Chromosome showing 5 telomere signals. (v) Chromosome showing 6 telomere signals (end-to-end fusion). (C) Graphic summary of number of telomere spots/chromosome deviating from the normal distribution of 4 telomeres per chromosome. The values represent mean ± standard error. * indicates a significant difference from cells treated with DMSO (P < .05); + indicates zero events for DMSO-treated cells.
Analysis of the signal-free ends in JVM13 cells treated with HD-BIBR1532
. | SFE/Metaphase . | No. of Metaphases . |
|---|---|---|
| DMSO | 0 | 8 |
| BIBR 3 hrs | 0.33 | 9 |
| BIBR 6 hrs | 0.46 | 15 |
| BIBR 24 hrs | 0.70 | 10 |
. | SFE/Metaphase . | No. of Metaphases . |
|---|---|---|
| DMSO | 0 | 8 |
| BIBR 3 hrs | 0.33 | 9 |
| BIBR 6 hrs | 0.46 | 15 |
| BIBR 24 hrs | 0.70 | 10 |
Metaphase spreads were prepared from JVM13 cells treated with 80 μM BIBR1552 at different time points, namely 3, 6, and 24 hours. Missing telomere signals at both sister chromatids were counted as signal-free ends (SFE) and expressed as mean SFE per metaphase.
In addition to loss of telomeres of sister chromatids, we also observed a striking variation in telomere signals between 3 and 6 spots per chromosome (Figure 5B-C). Whereas the frequency of chromosomes with 3 signals was higher after 6 hours of treatment compared with 24 hours, we found more chromosomes with 5 and 6 telomere spots after 24 hours relative to 6 hours. This finding suggests that high-dose BIBR1532 induces progressive loss of individual telomeres and formation of end-to-end fusions, which is compatible with telomere dysfunction.
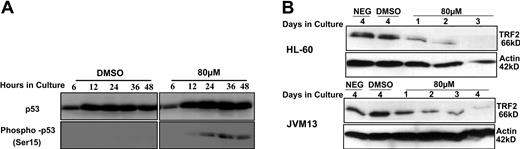
In order to evaluate if individual telomere damage is associated with up-regulation of p53, protein expression was analyzed following treatment with 80 μM BIBR1532. Whereas the levels of total p53 did not change over time, we observed an increase of phosphorylated p53 at Ser15 in treated compared with control cells (Figure 6A), indicating functional activation of p53 in response to DNA damage. Furthermore, we also found a reduction of TRF2 protein expression following exposure to BIBR1532, which is compatible with induction of telomere dysfunction and loss of capping function (Figure 6B). Given the delay in protein reduction, this effect may also be secondary due to loss of individual telomeres and their associated proteins.
Western blot analysis of TRF2 and p53/phospho-p53 in leukemia cells treated with BIBR1532. Protein was extracted using RIBA buffer from HL-60 and JVM13 treated with 80 μM BIBR1532 at indicated time points. Western blot analysis was performed using a monoclonal (A) anti-p53 and anti-phosphorylated p53 (dilution 1:1000) and (B) anti-TRF2 antibody (dilution 1:250). Actin expression levels were used as internal control.
Western blot analysis of TRF2 and p53/phospho-p53 in leukemia cells treated with BIBR1532. Protein was extracted using RIBA buffer from HL-60 and JVM13 treated with 80 μM BIBR1532 at indicated time points. Western blot analysis was performed using a monoclonal (A) anti-p53 and anti-phosphorylated p53 (dilution 1:1000) and (B) anti-TRF2 antibody (dilution 1:250). Actin expression levels were used as internal control.
Delayed cytotoxic effect of high-dose BIBR1532 in normal diploid fibroblasts
Providing evidence that high-dose BIBR1532 induces telomere damage associated with loss of TRF2, we wanted to analyze the cytotoxic effects in normal diploid fibroblasts. Therefore, we used HK1 fibroblasts, which have no detectable telomerase activity and enter replicative senescence after about 55 to 60 population doublings (PD).31 In addition, we also used a subline expressing ectopic human telomerase reverse transcriptase (hTERT), which results in telomere elongation of more than 2 kb and subsequent immortalization.
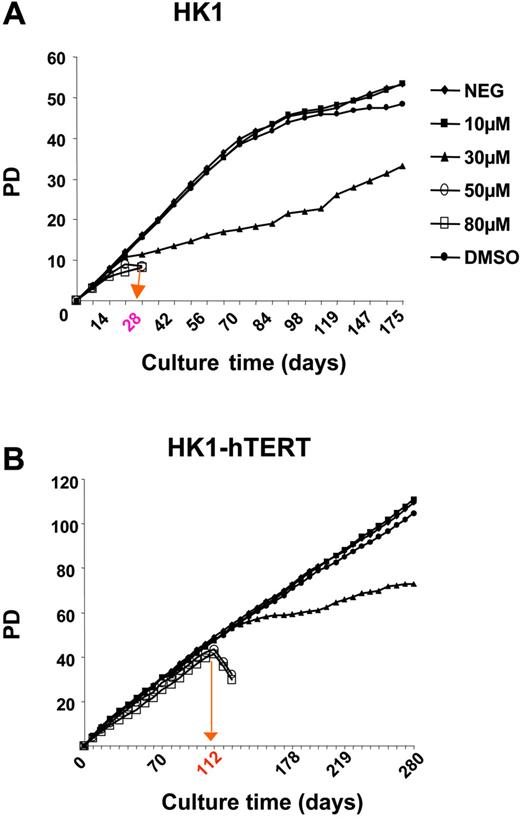
Both, wild-type and hTERT-HK1 cells were exposed to different concentrations of BIBR1532 upon long-term culture (Figure 7A,C). Interestingly, a differential outcome following incubation with various concentrations of BIBR1532 was observed. Whereas 50 μM and 80 μM induced growth arrest in wild-type HK1 cells after approximately 8 population doublings, a diminished proliferation rate was observed for 30 μM of the compound after 12 PDs. Using flow-FISH analysis, a substantial telomere shortening (∼ 2 kb) was found from day 7 to day 56 of cell culture. The mean telomere loss was in the range of 56 to 80 bp per PD for control cells as well as for those treated with 10 μM and 30 μM BIBR1532 (Table 5).
Effects of high-dose BIBR1532 on human fibroblast cells (HK1). Proliferation of HK1 fibroblasts upon exposure of indicated concentrations of BIBR1532. (A) Wild-type HK1 cells. (B) HK1 cells expressing ectopic hTERT. The red arrow and label on the x-axis indicate the time point of cell death. Summary of corresponding telomere length values of wild-type HK1 and HK1-hTERT is presented in Tables 5 and 6, respectively.
Effects of high-dose BIBR1532 on human fibroblast cells (HK1). Proliferation of HK1 fibroblasts upon exposure of indicated concentrations of BIBR1532. (A) Wild-type HK1 cells. (B) HK1 cells expressing ectopic hTERT. The red arrow and label on the x-axis indicate the time point of cell death. Summary of corresponding telomere length values of wild-type HK1 and HK1-hTERT is presented in Tables 5 and 6, respectively.
Loss of telomeric pairs per population doubling, HK1 cells
. | Telomere length . | . | No. population doublings . | . | . | |||
|---|---|---|---|---|---|---|---|---|
. | 7 d in culture . | 56 d in culture . | 7 d in culture . | 56 d in culture . | bp loss per PD . | |||
| Negative control | 8.8 | 6.9 | 3.9 | 32.7 | − 66.6 | |||
| DMSO | 8.4 | 6.8 | 3.5 | 31.5 | − 55.5 | |||
| BIBR1532 | ||||||||
| 10 μM | 9.0 | 6.7 | 3.2 | 31.7 | − 80.1 | |||
| 30 μM | 7.8 | 7.0 | 3.7 | 16.0 | − 68.7 | |||
| 50 μM | 8.3 | — | 3.3 | — | — | |||
. | Telomere length . | . | No. population doublings . | . | . | |||
|---|---|---|---|---|---|---|---|---|
. | 7 d in culture . | 56 d in culture . | 7 d in culture . | 56 d in culture . | bp loss per PD . | |||
| Negative control | 8.8 | 6.9 | 3.9 | 32.7 | − 66.6 | |||
| DMSO | 8.4 | 6.8 | 3.5 | 31.5 | − 55.5 | |||
| BIBR1532 | ||||||||
| 10 μM | 9.0 | 6.7 | 3.2 | 31.7 | − 80.1 | |||
| 30 μM | 7.8 | 7.0 | 3.7 | 16.0 | − 68.7 | |||
| 50 μM | 8.3 | — | 3.3 | — | — | |||
— indicates not done.
Interestingly, the telomerized fibroblasts (hTERT-HK1) with a mean telomere length of approximately 11 kb were not affected in proliferation rate when cultured in the presence of 10 and 30 μM BIBR1532 for more than 100 days (Figure 7C). In contrast, a sudden loss of cell viability was observed for 50 and 80 μM after a culture period of 112 days and 42 PDs. In general, a continuous increase in telomere length was found in control as well as in BIBR1532-treated hTERT-HK1 cells. However, the gain in telomere repeats was substantially reduced with increasing concentrations of the compound (Table 6), which resulted in a difference of the mean telomere length of almost 3 kb after more than 40 population doublings. These data indicate that higher concentrations of BIBR1532 are also cytotoxic to normal somatic cells, although the effect is delayed and appears to be dependent on the initial length of telomeres. Attrition beyond a distinct threshold seems to be necessary to facilitate growth arrest.
Gain of telomeric pairs per population doubling, HK1-hTERT
. | Telomere length . | . | No. population doublings . | . | . | ||
|---|---|---|---|---|---|---|---|
. | 84 d in culture . | 105 d in culture . | 84 d in culture . | 105 d in culture . | bp gain per PD . | ||
| Negative control | 11.1 | 14.0 | 37.1 | 46.0 | + 344 | ||
| DMSO | 11.3 | 13.8 | 36.0 | 45.1 | + 280 | ||
| BIBR1532 | |||||||
| 10 μM | 10.7 | 11.7 | 35.3 | 44.5 | + 110 | ||
| 30 μM | 9.7 | 10.4 | 35.0 | 44.5 | + 76 | ||
| 50 μM | 9.4 | 9.6 | 33.6 | 42.1 | + 32 | ||
. | Telomere length . | . | No. population doublings . | . | . | ||
|---|---|---|---|---|---|---|---|
. | 84 d in culture . | 105 d in culture . | 84 d in culture . | 105 d in culture . | bp gain per PD . | ||
| Negative control | 11.1 | 14.0 | 37.1 | 46.0 | + 344 | ||
| DMSO | 11.3 | 13.8 | 36.0 | 45.1 | + 280 | ||
| BIBR1532 | |||||||
| 10 μM | 10.7 | 11.7 | 35.3 | 44.5 | + 110 | ||
| 30 μM | 9.7 | 10.4 | 35.0 | 44.5 | + 76 | ||
| 50 μM | 9.4 | 9.6 | 33.6 | 42.1 | + 32 | ||
Discussion
The selective activity of telomerase in tumor cells represents a promising target for a rational therapy approach in cancer therapy.21 Inhibition of telomerase activity by pharmacologic or genetic interventions has been demonstrated to result in continuous telomere erosion, which ultimately induces replicative senescence or apoptosis.22 In this study, we have demonstrated that BIBR1532, which represents a potent specific inhibitor of hTERT, exhibits a selective cytotoxicity against primary leukemia cells from AML and CLL patients. Importantly, this effect was observed when BIBR1532 was used at higher concentrations than reported for inhibition of the catalytic activity. This suggests that this compound exerts a direct cytotoxic potential, which must be dissected from telomerase inhibition alone. Interestingly, the malignant cells of the hematopoietic system were 3-fold more sensitive than normal hematopoietic progenitor cells in which the growth potential was severely compromised only when the drug concentration exceeded 160 μM.
In tumor cells, inhibition of telomerase activity does not typically result in immediate growth arrest, but will lead to a delayed effect that is dependent on sufficient telomere decapping upon proliferation. The small molecule inhibitor BIBR1532 has been shown to specifically inhibit the native and recombinant human telomerase comprising the hTERT and hTR components by primarily interfering with the processivity of the enzyme.28,29 The IC50 value for the purified enzyme has been determined to be 93 nM. In contrast, concentrations higher than 100 μM were reported to inhibit RNA polymerases I to III. In the original paper,28 a standard concentration of 10 μM was shown to induce growth arrest in several solid tumor cell lines after a significant lag period (> 100 PDs) that was associated with substantial telomere shortening. Here, the antiproliferative effect occurred within 72 hours when leukemia cells were exposed to the drug at higher concentrations (30-80 μM). The IC50 value for this direct effect was found to be in the range of 50 to 60 μM. Interestingly, this cytotoxicity was independent of measured telomerase activity, because 12 of 20 CLL and 2 of 6 AML samples were negative in the TRAP assay. Thus, what could be the mechanism that mediates the direct cytotoxic effect of BIBR1532 in contrast to delayed induction of senescence? Our data provide evidence that treatment with concentrations from 30 to 80 μM interfered with the capping function of telomeres. Although flow-FISH was unable to detect changes in the mean telomere length during the short-term culture, the more sensitive Q-FISH analysis demonstrated a small but significant loss of the mean telomere length and—more importantly—a time-dependent erosion of individual telomeres. This was associated with a trend toward increased chromosome fusions and loss of TRF2 protein, which suggests that the primary function of telomeres was severely compromised. In agreement with this theory was our observation that high-dose BIBR1532 had also a delayed antiproliferative effect on normal cells without detectable telomerase activity such as diploid fibroblasts. In these cells, growth arrest did not occur within 72 hours but after 28 days. The lag period was even extended in a subline with overlong telomeres and ectopic expression of hTERT (> 100 days). Therefore, a distinct threshold of overall telomere length may exist, which exerts a protective function toward treatment with high-dose BIBR1532.
It is believed that telomeres form a higher-order chromatin structure that physically protects the 3′ end from cellular activities.38 The telomere binding protein TRF2 plays a key role in the protective activity of telomeres and loss of function leads to apoptosis and telomere-induced senescence.7 Increasing evidence is accumulating that suggests that the physical presence of telomerase or its transient activity is part of the integrity of the telomere cap.39,40 In fact, even normal, early passage fibroblasts have been described as showing low expression of hTERT, which is upregulated during their transit through S phase. Such periodic expression of hTERT is supposed to partially maintain the 3′ telomeric overhang and to facilitate cell proliferation.
The molecular effects of telomerase inhibition by different approaches are not completely understood. It has been speculated that ectopic expression of a dominant-negative mutant of hTERT (DN-hTERT) may titrate telomerase or telomere components from the chromosome end, thereby perturbing capping, while the small molecule inhibitor BIBR1532 may simply decrease the catalytic activity of the enzyme without effecting the telomere structure.41 Based on the studies presented here, it is possible that high-dose BIBR1532 not only suppresses the catalytic activity of telomerase but also may compromise binding of protein partners of the enzyme resulting in telomere dysfunction. Loss of TRF2 may be a direct consequence of such induced telomere damage. In fact, Karlseder et al42 reported that a change in the status of the telomere complex rather than telomere shortening will induce DNA damage and induction of growth arrest. Furthermore, Masutomi et al40 described that disruption of telomerase activity in human fibroblast cells restricts cell lifespan by destabilization of the telomeric 3′ overhang without changing the rate of overall telomere shortening. Strikingly, hematopoietic progenitor cells seem to be more resistant toward treatment with high-dose BIBR1532. Possibly, differences in telomere structure and binding of associated proteins may exist between normal and malignant cells of the hematopoietic system that make the latter more vulnerable toward telomerase-associated DNA damage. Such difference could be due to the larger size in telomere repeats that are typical for the stem cell compartment. Compatible with this theory is our observation that fibroblast cells with overlong telomeres (> 11 kb) were primarily resistant toward this treatment.
Finally, we cannot exclude additional mechanisms that cause cell death upon high doses of BIBR1532. Since loss of TRF2 has been also described upon exposure to DNA damage-inducing agents such as ionizing radiation,43 it is possible that high-dose BIBR1532 is inducing DNA damage not only at the telomere locus but also elsewhere in the genome. Nevertheless, telomeredamaging agents might represent an interesting avenue for novel cancer therapies, although they might be more toxic than simple inhibition of the catalytic activity of telomerase. However, from an oncologist's perspective, this form of approach could circumvent the long lag period, which is, in our view, not useful for a successful cancer therapy. Importantly, more knowledge about the structure and function of the telomere complex in normal and malignant cells is essential in order to design such telomeredirected therapy approaches.
Prepublished online as Blood First Edition Paper, October 26, 2004; DOI 10.1182/blood-2003-12-4322.
Supported by grants from the European Union, Sonderforschungsbereich 364 (Deutsche Forschungsgemeinschaft [DFG]), Verein zur Leukämieforschung, Freiburg, and from Boehringer Ingelheim Pharma KG, Biberach, Germany.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We gratefully acknowledge the excellent technical assistance from I. Skatulla and the support from Dr J. Burger providing CLL samples. We are indebted to Prof Mertelsmann for his continuous support. We thank Dr J. Lingner for critical comments on the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal