Abstract
We use lentiviral-delivered RNA interference (RNAi) to inhibit the growth of a model of primary effusion lymphoma (PEL) in vitro and in vivo. RNAi is a phenomenon allowing the sequence-specific targeting and silencing of exogenous and endogenous gene expression and is being applied to inhibit viral replication both in vitro and in vivo. We show that silencing of genes believed to be essential for the Kaposi sarcoma-associated herpesvirus (KSHV) latent life cycle (the oncogenic cluster) has a varied effect in PEL cell lines cultured in vitro, however, concomitant silencing of the viral cyclin (vcyclin) and viral FLICE (Fas-associating protein with death domain-like interleukin-1β-converting enzyme) inhibitory protein (vFLIP) caused efficient apoptosis in all PEL lines tested. We demonstrate that in a murine model of PEL, lentiviral-mediated RNA interference both inhibits development of ascites and can act as a treatment for established ascites. We also show that the administered lentiviral vectors are essentially limited to the peritoneal cavity, which has advantages for safety and dosage in a therapeutic setting. This shows the use of lentiviral-mediated RNA interference in vivo as a potential therapeutic against a virally driven human cancer.
Introduction
RNA interference (RNAi) is being exploited to treat or prevent infection and as a therapeutic against cancer.1,2 Attractive targets include foreign (eg, viral) and mutated, fused, or overexpressed genes (ie, cancer).3-5 In vitro, RNAi has been shown to block infection and replication of various pathogens including human immunodeficiency virus (HIV-1),6,7 influenza,8 and viruses implicated in oncogenesis such as Epstein Barr virus (EBV),9,10 hepatitis B,11,12 hepatitis C,13 and human papilloma virus (HPV).14,15 In experimental in vivo models, RNAi has been shown to prevent chemical- and viral-induced hepatitis.16-18
The efficient delivery of therapies that knock-down specific RNA remains one obstacle to translate RNAi into a realistic treatment option for human disease.19 The direct delivery of antisense RNA to treat or prevent CMV retinitis is one of the few successful clinical applications thus far of RNA-targeted treatment.20,21 Although antisense provides significantly less robust inhibition of gene expression compared to RNAi, the major problem with both therapies remains effective delivery to the site of disease. One use of RNAi is based around injecting large quantities of synthetic double-stranded RNA (dsRNA) or DNA encoding short hairpin RNA (shRNA) intravenously or using hydrodynamic transfection.16,18 This approach is not realistic for treating human disease, except for diseases involving sites where delivery of synthetic dsRNA is more straightforward. The development of shRNA to knock-down gene expression22,23 and the incorporation of shRNA into lentiviral-24,25 or adenoviral-based26,27 vectors offers the opportunity to use these vectors to target RNA efficiently in vivo. Vesicular stomatitis virus-g envelope (VSV-g)-pseudotyped lentiviral vectors have been shown to be able to infect a wide range of cells in vitro and in vivo28 and have potential to deliver therapeutic shRNA.
Viral-associated malignancies could be a particularly attractive target for RNAi, because these cancers are driven by foreign oncogenes.29 Kaposi sarcoma herpesvirus (KSHV or HHV8) is associated with Kaposi sarcoma, primary effusion lymphoma (PEL), and multicentric Castleman disease (MCD).30 PEL is the KSHV-related malignancy with the poorest survival and with very few therapeutic options.31 These lymphomas often present and persist as effusions in the peritoneal (ascites) or pleural cavities.32 A murine model of PEL faithfully replicates the phenotype of this disease.33,34
Like other gammaherpesviruses, the life cycle of KSHV is divided into a lytic and a latent phase.35 Genes expressed during latency are thought to be directly involved in driving cell proliferation, allowing immune escape, and preventing apoptosis. Only a fraction of KSHV genes are expressed during latency; this includes the oncogenic cluster encoded by open reading frames (ORFs) 71-73 (vFLIP, vcyclin, and latency-associated nuclear antigen [LANA]).36 vcyclin promotes cell progression through G1 to S phase by acting like a D-type cyclin,37 but vcyclin also has additional functions including inactivating the cyclin-dependent kinase inhibitor (cdki) p2738 and activating origin replication complex 1 (orc-1).39 vFLIP activates the NFκB pathway, and silencing vFLIP in PEL cells in vitro results in apoptosis.40 LANA maintains the viral episome,41 tethering the episome to DNA during cell division to ensure delivery of viral episomes to all daughter cells42 and interferes with p53,43 pRb,44 and β-catenin45 pathways to prevent apoptosis and to drive cell proliferation.
We hypothesised that targeting the oncogenic cluster of KSHV could be a therapeutic approach for PEL. We tested the efficiency of lentiviral-delivered shRNA to vcyclin, vFLIP, or LANA to induce apoptosis in vitro and prevent or treat PEL in an experimental model.
Materials and methods
Cell culture
Adherent cells (293 and 293t) were cultured in Dulbecco modified Eagle medium (DMEM) (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum without antibiotics. Adherent cells were split by washing with trypsin-EDTA (ethylenediaminetetraacetic acid) solution until the cells were released from the base of the flask. The trypsin-EDTA was removed by centrifugation at 2000g (1200 rpm) for 5 minutes, and cells were seeded 1 in 4 into new tissue culture flasks. The 293 and 293t cells were split every 2 days.
Suspension cells were grown in RPMI 1640 medium (Gibco) supplemented with 10% (Ramos, BC-3, and JSC-1) or 20% (BCP-1 and HBL-6) fetal bovine serum without antibiotics. Live populations were ensured by use of a Ficoll gradient once a week in accordance with the manufacturer's guidelines. When appropriate, puromycin (Invivogen, San Diego, CA) selection was performed at 3 μg/mL 48 hours after infection.
Microscopy was performed using a Nikon Eclipse E600 inverted microscope with Nikon fluorescent lamp. The objective was 10, 20, 60, or 80 × with total final magnification 100 to 800 ×. Images were taken using a Zeiss Axiocam camera and software.
Immunofluorescence
IFA (immunofluorescence assay) was performed on cells as follows. Cells were fixed for 15 minutes at room temperature in 4% PFA (paraformaldehyde; SERVA, Heidelberg, Germany) then washed 3 times with phosphate-buffered saline (PBS) and 3% fetal calf serum (FCS). Cells were permeabilized in PBS and 0.2% TRITON-X (Sigma, Saint Louis, MO). Cells were spotted on glass slides and allowed to dry before being kept at -20°C until use. IFA for LANA was performed by incubating cells with LANA antibody46 diluted 1:120 or negative control (PBS) for 1 hour at 37°C. Cells were washed in PBS and 3% FCS 3 times. Cells were then incubated with anti-rat fluorescein isothiocyanate (FITC) antibody (Dako, Glostrup, Denmark) for 30 minutes at 37°C and then washed in PBS and 3% FCS 3 times. 50% glycerol was dropped onto the slide and a coverslip added. Cells were then visualized using a Nikon Eclipse E600 inverted microscope with Nikon fluorescent lamp (Nikon, Tokyo, Japan). The objective lens aperture was 10, 20, 60, or 80 × with total final magnification 100 to 800 ×. Images were taken using a Zeiss Axiocam Camera and software (Carl Zeiss, Thornwood, NY).
Cell proliferation assays
Cell doubling time was measured by seeding 1 × 105 cells into 1 mL of appropriate solution in a 24-well plate and analyzing the number of cells after 24 hours. The cell doubling time in days was calculated according to the following formula:
where a = number of cells after 24 hours and b = 1 × 105.
This process was repeated to allow the cell doubling time to be calculated over the course of the experiment. Methyl-thiazol tetrazolium (MTT) assays (Roche, Basel, Switzerland) and BrdU Cell Proliferation enzyme-linked immunosorbent assay (ELISA; Roche) were performed according to the manufacturer's guidelines.
Apoptosis assay
Percentage of cells in apoptosis was determined by annexin V-FITC and propidium iodide (PI) staining using the apoptosis detection kit (Molecular Probes, Eugene, OR) in accordance with the manufacturer's guidelines.
RNA interference
Three targets for RNA interference were selected using the Dharmacon sequence selection tool (Dharmacon, Lafayette, CO). DNA oligos containing the target sequence, a TTCG hairpin, the antisense of the target, a 5 T termination sequence, and a CTAG (XbaI site) were synthesised by MWG Biotech (Ebersburg, Germany), annealed, and inserted into the pGEM-U6M plasmid by digestion with XbaI and SmaI (Promega, Madison, WI) and ligation with T4 DNA ligase (New England Biolabs, Beverley, MA) in accordance with the manufacturer's guidelines. pGEM-U6M was created from pGEM-U6L (a gift from Sam Wilson, Windeyer Institute, University College London) and altering the + 1 base pair of the U6 promoter from G to C using Stratagene quickchange site directed mutagenesis kit (Stratagene, La Jolla, CA). Lentiviral RNAinterference plasmids were then generated by subcloning the U6 promoter-hairpin construct from pGEM-U6M into pCSGW (a gift from Adrian Thrasher, Institute of Child Health, University College London, United Kingdom) or pSIN-PAC (a gift from Greg Towers, University College London) by digestion with EcoRI. Insert orientation was found not to affect silencing. The efficacy of the 3 targets was determined by Western blot and the best chosen for subsequent experiments. The most efficient targets were found to be short hairpin targeting LANA(sh-LANA); GTCCCACAGTGTTCACATCCGGGC; sh-vcyclin: GTTCCTGCCAACGTCATTCCGCAG; sh-vFLIP: GTGCTCTGTCAGGTTCTCCCATCG.
Virus production and concentration
Lentiviral virions were produced by transient cotransfection of 1 μg of p8.91, 1 μg of pMD.G (both gifts from D. Trono, Department of Microbiology and Molecular Medicine, University of Geneva, Switzerland), and 1.5 μg of the pSIN vector into a 10-cm dish of 293t cells as previously described.
The media was changed 24 hours after transfection and then supernatant containing virus was harvested every 8 hours (replacing the media with 8 mL of fresh DMEM and 10% FCS) from 48 hours until the 293t cells detached from the plate. Supernatant was stored at -80°C throughout the experiment. Yields were typically 80 to 120 mL from a single 10-cm dish, and the pooled supernatant contained 0.5 to 1 × 108 293t infectious units (293t IU) as determined by serial dilution of virus and detection of green fluorescence protein (GFP) expression 48 hours or puromycin resistance 7 days after infection of a known number of 293t cells.
Lentivirus was concentrated by ultracentrifugation of 35 mL of supernatant at 50 000g for 2 hours at 4°C in a Beckman SW-28 rotor (Beckman Coulter, Marseille, France) and resuspension overnight in 1 mL PBS. This typically increased the titer of the virus 10-fold. Further concentration for in vivo studies was achieved by pooling the concentrated virus after ultracentrifugation and gel ultrafiltration using a Centricon-80 100-kDa molecular-weight (MW) cut-off column (Millipore, Billerica, MA) in accordance with the manufacturer's guidelines.
Using this protocol, titers in excess of 8 × 1010 293t IU were routinely produced by pooling the harvests from 48 10-cm dishes. This virus was subsequently diluted into 300 μL aliquots of 1 × 1010 or 1 × 109 virions for in vivo injections.
Viral infections
Unless otherwise stated in the figures, viral infections were carried out at an MOI (multiplicity of infection) of 10 293t IU in the presence of 8 μg/mL polybrene. All lentiviral knock-downs were selected by culturing cells in the presence of puromycin (3 μg/mL) 48 hours after infection.
Western blotting (sodium dodecyl sulfate-polyacrylamide gel electrophoresis)
A 6% (for LANA) or 15% (for vFLIP, vcyclin or β-actin) polyacrylamide gel was poured with a 5% stack. Protein content of the lysates was assessed by the Bradford assay and equalized to load 20 μg of protein per well. Gels were run until the protein marker (Biorad Protein Plus ladder) reached the bottom of the gel. Transfer was semidry for 20 minutes per gel for small proteins or 1 hour per gel for LANA in standard transfer buffer with 20% methanol (BioRad, Hercules, CA). The resulting membrane was blocked in 4% milk and then probed for protein-specific antibodies overnight at 4°C. The next day, the membrane was washed for 1 hour in PBS and 0.1% Tween-20 and then incubated for 1 hour with appropriate secondary antibody conjugated to horseradish peroxidase (HRP). After further washing the membrane was developed using enhanced chemiluminescence (ECL) (Amersham, Buckinghamshire, United Kingdom) or ECL and (Amersham) for LANA. LN5365 was used at a dilution of 1:1000 to detect LANA, anti-v-cyclin used 1:1000 (Exalpha Biologicals, Maynard, MA), anti-vFLIP 1:1000 (a kind gift from Mary Collins, University College London), and β-actin (Oncogene, Cambridge, MA) used 1:10 000. Secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) (anti-mouse HRP and anti-sheep HRP) and Immunologicals Direct (anti-rat HRP; Oxfordshire, United Kingdom). All secondary antibodies were used at a concentration of 1:5000.
qPCR and qRT-PCR
Quantitative polymerase chain reaction (qPCR) and reverse transcriptase-polymerase chain reaction (RT-PCR) were performed using an ABI TaqMan Prism 7000 machine. Primers used for detection of lentiviral packaging cDNA and latent gene mRNA and DNA are as follows: Lentiviral packaging cDNA; forward 5′-ACTTGAAAGCGAAAGGGAAACCA-3′; reverse 5′-GTGCGCGCTTCAGCAA-3′; concentration 300 pmoles forward/300 pmoles reverse. vcyclin: forward 5′-CATTGCCCGCCTCTATTATCA-3′; reverse 5′-ATGACGTTGGCAGGAACCA-3′; concentration 300 pmoles forward/300 pmoles reverse. vFLIP: forward 5′-TTTCCCCTGTTAGCGGAATGT-3′; reverse 5′-CTAAGTGAAGCAGGTCGCGC-3′; concentration 300 pmoles forward/300 pmoles reverse. LANA: forward 5′-TTGCCACCCACGCAGTCT-3′; reverse 5′-GGACGCATAGGTGTTGAAGAGTCT-3′; probe 5′-TCTTCTCAAAGGCCACCGCTTTCAAGTC-3′; concentration 500 pmoles forward/500 pmoles reverse for cDNA (qRT-PCR) and 700 pmoles forward/700 pmoles reverse for genomic DNA (qPCR). Human glyceraldehyde phosphate dehydrogenase (GAPDH): forward 5′-GGAGTCAACGGATTTGGTCGTA-3′; reverse 5′-GGCAACAATATCCACTTTACCAGAGT-3′; probe 5′-CGCCTGGTCACCAGGGCTGC-3′; concentration 300 pmoles forward/300 pmoles reverse for cDNA (qRT-PCR), and 700 pmoles forward/700 pmoles reverse for genomic DNA (qPCR). Mouse GAPDH: forward 5′-GGCATGGCCTTCCGTGT-3′; reverse 5′-GGTTTCTCCAGGCGGCA-3′; concentration 300 pmoles forward/900 pmoles reverse.
All reactions were performed using SYBR green (Affymetrix, Santa Clara, CA) at the concentrations indicated unless a probe is specified. These conditions were shown through optimisation to be sensitive, specific, and with no pimer-dimer formation. Standard TaqMan cycling conditions were used throughout.
In vivo model
The in vivo model of PEL was based upon previous studies. The immunodeficient mice used were RAG-/-, C3-/-, and common gamma chain-/-. Mice were culled in accordance with the American Type Culture Collection (ATCC)-defined criteria for humane conditions. A weight gain of more than 6 g above the mean in the weight control group was taken as the cut-off for severe ascites, and mice were culled. A palpable solid tumor mass exceeding 1 cm was taken as solid tumor development, and mice were culled. Outliers in experiments were included in any statistical analysis.
Injections of JSC-1 cells were performed intraperitoneally along the midline. Injection of lentiviral vector was intraperitoneal on the left or right side.
Results
Inhibition of expression of KSHV latent genes
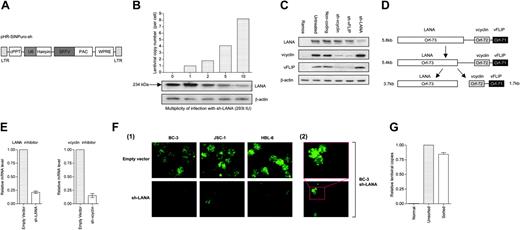
First, we show that lentiviral vectors delivering shRNA efficiently knock down the expression of vcyclin, vFLIP, and LANA. Our lentiviral vectors (Figure 1A) are based upon a self-inactivating HIV-1 backbone47 and coexpress the short hairpin driven by the human U6 promoter and a puromycin selection cassette. All infections were performed with puromycin selection added after 48 hours to select vector-transduced populations. TaqMan quantitative PCR (qPCR) 48 hours after infection shows that the silencing effect of a short hairpin targeting LANA correlates with the number of lentiviral inserts (Figure 1B). The bicistronic nature of vFLIP and vcyclin correlates to simultaneous silencing of both proteins when either is targeted by RNAi (Figure 1C).48 This result has been documented40 and was expected due to the proposed mechanism of action of RNAi.1 The independent silencing of LANA despite its presence on a tricistronic transcript suggests structural separation of the LANA coding region from those of the vcyclin and vFLIP (Figure 1D). This result concurs with current literature.40 We quantified by Taq-Man qRT-PCR the knock-down of expression at the message level after infection with sh-LANA or sh-vcyclin. Expression at the message level for both vcyclin and LANA are reduced to around 20% endogenous levels (Figure 1E). We used IFA to show whether the residual LANA protein and mRNA are attributable to a small number of cells still expressing normal LANA levels or whether all cells show 20% residual levels (Figure 1Fi). The IFA shows that for most cells, nuclear stippling in the knock-down lines is absent, indicating a lack of or very low expression level for LANA. However, in some cells the normal nuclear stippling of LANA can be seen (1Fii). Fluorescence-activated cell sorting (FACS) analysis indicates that in approximately 6.6% of BC-3 cells, LANA expression remains strong after lenti-shRNA infection (data not shown). After cell sorting for LANA-FITC, the LANA-positive cells showed the presence of the lentiviral packaging signal as determined by TaqMan qPCR (Figure 1G), indicating successful lentiviral infection.
Lentiviral short hairpin RNA (shRNA) to knock-down KSHV latent genes. (A) The lentiviral construct used for shRNA production. cPPT, central polypurine tract; SFFV, spleen focus forming virus promoter; PAC, puromycin N-acetyltransferase gene; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element; LTR, long terminal repeat. (B) Graph showing the correlation between MOI measured in 293t IU (293t cell infectious units) per cell, the number of lentiviral inserts per cell determined by TaqMan qPCR for the lentiviral packaging sequence, and the knockdown of LANA expression compared to β-actin control shown by Western blot. (C) Western blot panel for KSHV latent genes (gene name on left side) in BC-3 cells 7 days (sh-vFLIP and sh-vcyclin) or 14 days (sh-LANA) after infection with short hairpin (top label indicate hairpin target). vFLIP and vcyclin expression is simultaneously inhibited when either gene is targeted. (D) Hypothetical representation of the polycistronic nature of the mRNA species of the oncogenic cluster (adapted from Guasparri et al40 and Talbot et al65 and findings from RNAi knock-down results). (E) TaqMan qRT-PCR of BC-3 cells 96 hours after infection with a short hairpin showing decrease in the relative abundance of the mRNA species responsible for LANA and vcyclin production when infected with an appropriate inhibitor. (Fi) IFA for LANA (images at 600 × magnification) in BC-3, JSC-1, and HBL-6 cells shows that 10 days after infection with sh-LANA, the majority of cells lose characteristic nuclear stippling indicative of LANA expression. (Fii) Higher magnification (800 × top, 200 × bottom) of BC-3 cell expressing LANA despite presence of knock-down. (G) TaqMan qPCR for the lentiviral packaging signal shows that BC-3 cells positive for LANA (sorted for LANA-FITC) contain comparable levels of the sh-LANA construct to unsorted infected BC-3 cells. Error bars represent the standard error of the mean.
Lentiviral short hairpin RNA (shRNA) to knock-down KSHV latent genes. (A) The lentiviral construct used for shRNA production. cPPT, central polypurine tract; SFFV, spleen focus forming virus promoter; PAC, puromycin N-acetyltransferase gene; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element; LTR, long terminal repeat. (B) Graph showing the correlation between MOI measured in 293t IU (293t cell infectious units) per cell, the number of lentiviral inserts per cell determined by TaqMan qPCR for the lentiviral packaging sequence, and the knockdown of LANA expression compared to β-actin control shown by Western blot. (C) Western blot panel for KSHV latent genes (gene name on left side) in BC-3 cells 7 days (sh-vFLIP and sh-vcyclin) or 14 days (sh-LANA) after infection with short hairpin (top label indicate hairpin target). vFLIP and vcyclin expression is simultaneously inhibited when either gene is targeted. (D) Hypothetical representation of the polycistronic nature of the mRNA species of the oncogenic cluster (adapted from Guasparri et al40 and Talbot et al65 and findings from RNAi knock-down results). (E) TaqMan qRT-PCR of BC-3 cells 96 hours after infection with a short hairpin showing decrease in the relative abundance of the mRNA species responsible for LANA and vcyclin production when infected with an appropriate inhibitor. (Fi) IFA for LANA (images at 600 × magnification) in BC-3, JSC-1, and HBL-6 cells shows that 10 days after infection with sh-LANA, the majority of cells lose characteristic nuclear stippling indicative of LANA expression. (Fii) Higher magnification (800 × top, 200 × bottom) of BC-3 cell expressing LANA despite presence of knock-down. (G) TaqMan qPCR for the lentiviral packaging signal shows that BC-3 cells positive for LANA (sorted for LANA-FITC) contain comparable levels of the sh-LANA construct to unsorted infected BC-3 cells. Error bars represent the standard error of the mean.
The latent nuclear antigen as a target for inhibiting PEL
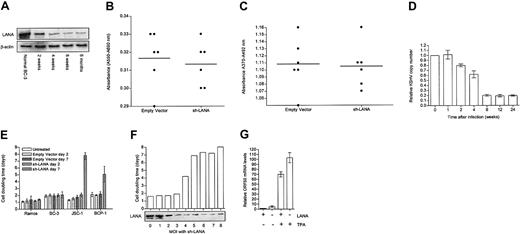
We next showed that stable LANA knock-down can be achieved in PEL lines for up to 6 months in culture under puromycin selection (Figure 2A). These cells continued to divide normally with no change in their proliferation as determined by MTT and BrdU assay (Figure 2B-C). Since LANA tethers the episome to chromatin during cell division,42 we tested whether the KSHV copy number is reduced in the LANA knock-down BC-3 cells. We show by TaqMan qPCR for LANA DNA (Figure 2D) that the KSHV copy number gradually decreases until a stable number of copies remains.
Targeting LANA with shRNA. (A) Western blot shows that after infection with sh-LANA, knock-down remains stable in BC-3 cells in puromycin selection for at least 6 months. (B) MTT assay for cell proliferation showing no significant difference between empty vector and sh-LANA cells 6 months after infection. The horizontal bar indicates the mean value. (C) BrdU assay showing no significant difference in 5-bromo-2′-deoxyuridine uptake between empty vector and sh-LANA cells 6 months after infection. The horizontal bar indicates the mean value. (D) qPCR for LANA DNA as a representation of KSHV DNA copy number shows that KSHV copy number gradually decreases in BC-3 cells until it reaches a plateau. (E) Graph of cell doubling time in KSHV negative B-cell (Ramos) and KSHV-positive PEL cell lines (BC-3, JSC-1, and BCP-1), 2 or 7 days after infection with empty vector (▦, day 2; ▩ , day 7) or with sh-LANA (▨, day 2; ▧, day 7). □ represents untreated cells. LANA knock-down has different effects on different lines. (F) Graph shows effect of increasing MOI (in 293t IU/cell) with sh-LANA on doubling time of JSC-1 cells 10 days after infection. Western blot shows LANA levels in these cells. (G) qRT-PCR for ORF50 (Rta) mRNA as an indicator of activation of lytic replication in JSC-1 cells before and after infection with sh-LANA and with or without the addition of TPA to induce lytic replication. Error bars represent the standard error of the mean.
Targeting LANA with shRNA. (A) Western blot shows that after infection with sh-LANA, knock-down remains stable in BC-3 cells in puromycin selection for at least 6 months. (B) MTT assay for cell proliferation showing no significant difference between empty vector and sh-LANA cells 6 months after infection. The horizontal bar indicates the mean value. (C) BrdU assay showing no significant difference in 5-bromo-2′-deoxyuridine uptake between empty vector and sh-LANA cells 6 months after infection. The horizontal bar indicates the mean value. (D) qPCR for LANA DNA as a representation of KSHV DNA copy number shows that KSHV copy number gradually decreases in BC-3 cells until it reaches a plateau. (E) Graph of cell doubling time in KSHV negative B-cell (Ramos) and KSHV-positive PEL cell lines (BC-3, JSC-1, and BCP-1), 2 or 7 days after infection with empty vector (▦, day 2; ▩ , day 7) or with sh-LANA (▨, day 2; ▧, day 7). □ represents untreated cells. LANA knock-down has different effects on different lines. (F) Graph shows effect of increasing MOI (in 293t IU/cell) with sh-LANA on doubling time of JSC-1 cells 10 days after infection. Western blot shows LANA levels in these cells. (G) qRT-PCR for ORF50 (Rta) mRNA as an indicator of activation of lytic replication in JSC-1 cells before and after infection with sh-LANA and with or without the addition of TPA to induce lytic replication. Error bars represent the standard error of the mean.
We next tested the effects of LANA knockdown in other PEL cells. Compared to BC-3 cells, BCP-1 and JSC-1 cells showed growth inhibition or apoptosis after infection with sh-LANA at an MOI of 10 (Figure 2E). However, titration of the knock-down effect allowed the generation of a JSC-1 line infected at an MOI of 3 with sh-LANA. When the MOI was 4 or more, the cell doubling time increased dramatically, and the JSC-1 cells could not be maintained in long-term culture (Figure 2F).
Since LANA is an inhibitor of Rta and therefore a suppressor of reactivation of lytic replication,49 we theorized that LANA knock-down may result in an increase in spontaneous or tetradecanoyl phorbol acetate (TPA) induced lytic replication. TaqMan qRT-PCR for ORF50 levels in these knockdown cells shows an increase in mRNA levels, suggesting an increase in the number of cells entering lytic cycle. ORF50 levels also were increased after TPA treatment of sh-LANA cells (Figure 2G).
vcyclin and vFLIP as targets for inhibiting PEL
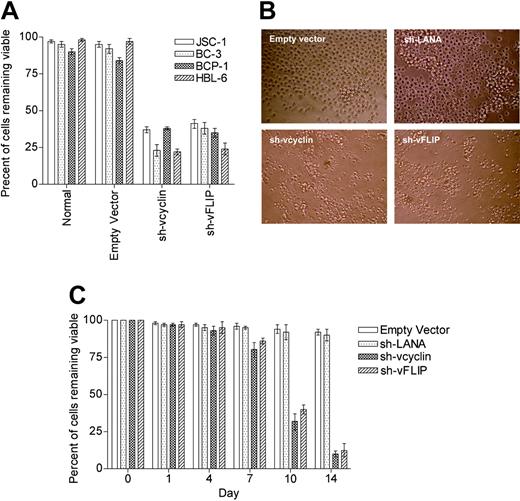
We investigated the effects of vcyclin and vFLIP knock-down on the same PEL lines. vcyclin and vFLIP knock-down induced apoptosis in all PEL lines tested (Figures 3A-B). All hairpins targeting vcyclin and vFLIP silenced both genes, and for further in vivo experiments we selected the most efficient hairpin for silencing as determined by TaqMan qRT-PCR (data not shown). The selected hairpin targets vcyclin.
Knockdown of vcyclin and vFLIP in PEL cells. (A) Graph shows the percent of PEL cells remaining viable as determined by FACS analysis for annexin V-FITC 10 days after infection with sh-vcyclin or sh-vFLIP. □ indicates JSC-1 cells; ▦, BC-3; ▩ , BCP-1; and ▨, HBL-6. (B) Light microscopy (magnification 100 ×) of BC-3 cells 10 days after infection with sh-LANA, sh-vcyclin, or sh-vFLIP shows extensive cell loss with vcyclin or vFLIP knockdown. (C) Graph shows percent of BC-3 cells remaining viable as determined by FACS analysis for annexin V-FITC at timepoints after infection with sh-LANA (▦), sh-vcyclin ( ▩ ), or sh-vFLIP (▨)atanMOI of 10 293t IU/cell. Error bars indicate the standard error of the mean.
Knockdown of vcyclin and vFLIP in PEL cells. (A) Graph shows the percent of PEL cells remaining viable as determined by FACS analysis for annexin V-FITC 10 days after infection with sh-vcyclin or sh-vFLIP. □ indicates JSC-1 cells; ▦, BC-3; ▩ , BCP-1; and ▨, HBL-6. (B) Light microscopy (magnification 100 ×) of BC-3 cells 10 days after infection with sh-LANA, sh-vcyclin, or sh-vFLIP shows extensive cell loss with vcyclin or vFLIP knockdown. (C) Graph shows percent of BC-3 cells remaining viable as determined by FACS analysis for annexin V-FITC at timepoints after infection with sh-LANA (▦), sh-vcyclin ( ▩ ), or sh-vFLIP (▨)atanMOI of 10 293t IU/cell. Error bars indicate the standard error of the mean.
To assess the efficiency of the effect, we analyzed this knock-down on JSC-1 cells at different MOIs. An MOI of 10 293t IU caused significant apoptosis, with further increases in MOI not enhancing the apoptotic effect (data not shown). We next showed that at an MOI of 10 the apoptotic effect starts at day 7 and is optimal at day 14 (Figure 3C). FACS analysis of JSC-1 cells infected with a lentiviral vector expressing eGFP (instead of PAC) and the short hairpin against vcyclin at day 21 indicates fewer than 0.15% of cells were positive for eGFP, compared to more than 95% 48 hours after infection. This indicates that almost no infected cell remained viable (data not shown).
Preventing PEL growth in vivo
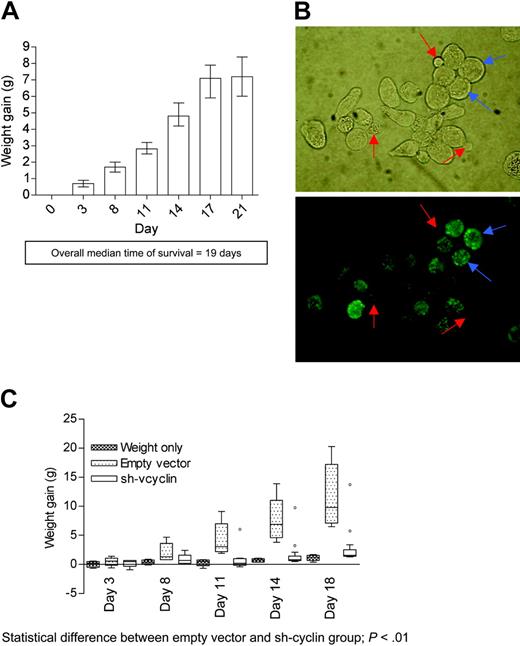
To demonstrate the potential of this effect to inhibit the growth of PEL in an in vivo model, we established a model in immunodeficient mice. We injected 1 × 107 JSC-1 cells intraperitoneally into 10 triple knock-out (RAG-/-,C3-/-, and common gamma chain-/-) mice (Figure 4A). All injected mice developed ascites at a rapid and reliable pace, and samples from the ascitic fluid were positive for all 3 latent genes by Western blot (data not shown) and positive for LANA by IFA (Figure 4B). Since an MOI of 10 was required for efficient inhibition of PEL growth, we assessed the bioavailability of the lentiviral vector by TaqMan qPCR of lentivirions injected into the peritoneum of these mice. The vector infected the cells lining the peritoneal cavity, but systemic distribution remained limited (Table 1). We also determined the cellular density of the ascitic fluid in the mice. These data were used to determine the quantity of lentiviral vectors required to infect all cells in the ascitic fluid. Analysis of 10 mice gave values in the range of 3 × 107 to 1 × 108 cells per gram of ascites.
In vivo model for PEL. (A) Graph of weight gain of a group of 10 mice over the course of 21 days after intraperitoneal inoculation with 1 × 107 JSC-1 cells. (B) LANA IFA of ascitic fluid drained from mice 14 days after inoculation with JSC-1 cells. LANA-positive cells (blue arrow) show typical nuclear stippling and indicate reliable development of appropriate ascites, while the presence of small cells (red arrows), which stain negative for LANA, are murine cells in the ascitic tap. (C) Graph shows the weight gain of 3 groups of 6 mice each: weight-only control group ( ▩ ), IP co-injection with 1 × 107 JSC-1 cells, and 1 × 108 293t IU of empty vector (▦) or sh-vcyclin (□). Outliers marked on the sh-vcyclin group represent 2 treated mice that developed ascites despite administration of sh-vcyclin. Outliers were included in all statistical analyses. Error bars indicate the standard error of the mean.
In vivo model for PEL. (A) Graph of weight gain of a group of 10 mice over the course of 21 days after intraperitoneal inoculation with 1 × 107 JSC-1 cells. (B) LANA IFA of ascitic fluid drained from mice 14 days after inoculation with JSC-1 cells. LANA-positive cells (blue arrow) show typical nuclear stippling and indicate reliable development of appropriate ascites, while the presence of small cells (red arrows), which stain negative for LANA, are murine cells in the ascitic tap. (C) Graph shows the weight gain of 3 groups of 6 mice each: weight-only control group ( ▩ ), IP co-injection with 1 × 107 JSC-1 cells, and 1 × 108 293t IU of empty vector (▦) or sh-vcyclin (□). Outliers marked on the sh-vcyclin group represent 2 treated mice that developed ascites despite administration of sh-vcyclin. Outliers were included in all statistical analyses. Error bars indicate the standard error of the mean.
Analysis of systemic distribution of lentiviral vectors injected intraperitoneally
Organ . | Viral inserts relative to peritoneum . |
|---|---|
| Peritoneum | 1 |
| Liver | 0.01 ± 0.003 |
| Spleen | 0.3 |
| Tail | 0.019 ± 0.002 |
| Heart | 0.0032 ± 0.0017 |
| Brain | Not detected |
| Flank muscle | 0.03 ± 0.004 |
Organ . | Viral inserts relative to peritoneum . |
|---|---|
| Peritoneum | 1 |
| Liver | 0.01 ± 0.003 |
| Spleen | 0.3 |
| Tail | 0.019 ± 0.002 |
| Heart | 0.0032 ± 0.0017 |
| Brain | Not detected |
| Flank muscle | 0.03 ± 0.004 |
To show the inhibition of PEL in this model, we divided mice into 2 groups of 6 mice and co-injected 1 × 108 293t IU of lentiviral vector expressing sh-vcyclin (treated group) or 1 × 108 293t IU of empty vector (untreated group) into the peritoneal cavity with 1 × 107 JSC-1 cells and monitored the weight of the mice over the course of 3 weeks (Figure 4C). All tumor-challenged untreated mice developed ascites and were culled (according to ATCC guidelines) by day 21, however, only 2 mice treated with the sh-vcyclin developed ascites (marked as outliers on Figure 4C). The other mice remained healthy until the end of the experiment at 4 weeks, with similar weights compared to the unchallenged weight control mice. When the treated mice were culled, 3 of 4 had developed small (< 5 mm) solid tumors that stained positive for LANA (data not shown). We theorized these to be due to leakage of JSC-1 cells through the injection site out of the peritoneal cavity where they were unaffected by the lentivirus expressing sh-vcyclin.
Treating established lymphomatous effusions with RNAi
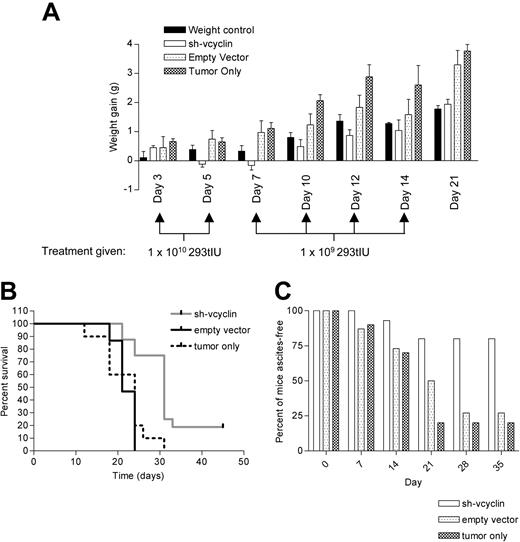
This demonstrates the ability of the vector to inhibit the development of PEL-driven ascites in this model, however, inhibition of developed ascites represented a challenge due to the number of cells present within the ascitic fluid. Since developed ascites contained up to 1 × 108 cells per gram, and some mice gained in excess of 10 g, this would involve the infection of up to 1 × 109 cells. We therefore treated 8 mice that had gained 1 g 3 days after inoculation with JSC-1 cells with 1 × 109 293t IU at day 3 and again at day 7, based on an MOI of 10 infecting up to 1 × 108 cells. This regime failed to prevent development of ascites, although it had a significant effect on the survival curve of the mice (data not shown). We proposed that infection of murine cells in the peritoneum may have reduced the infection rate below that which is sufficient to cause resolution of PEL. We therefore repeated the experiment using an enhanced treatment regime of 1 × 1010 293t infectious units once at day 3, again at day 5, and then treatment with 1 × 109 293t IU 3 times per week for a 2-week period (Figure 5A). The initial weight gain seen at day 3 was almost immediately reversed, and there was a significant difference (P < .001) between the sh-vcyclin group and the empty vector group throughout the experiment. The Kaplan-Meier survival curve for this experiment (Figure 5B) shows a significant difference (P = .004) between the sh-vcyclin and empty vector and sh-cyclin and tumor-only groups. There was no significant survival benefit (P = .43) of treatment with empty vector compared to the tumor-only group. The overall increase in median time of survival was found to be 10 days, with the determined end point 45 days after initial injection. However, 3 of 15 mice in the treated group appeared to be long-term survivors (symptom-free after 8 weeks).
Therapeutic potential of sh-vcyclin. (A) Bar chart shows the weight gain of 8 normal mice (weight control, ▪), 15 mice inoculated with 1 × 107 JSC1 cells (tumor only, ▩ ), 15 mice treated from day 3 after inoculation with 1 × 107 JSC-1 cells with sh-vcyclin (sh-vcyclin, □), or 15 mice treated from day 3 after inoculation with 1 × 107 JSC-1 cells with empty vector (empty vector, ▦). Treatment regime was 1 × 1010 293t IU of lentiviral vector on day 3 and 5 and then 1 × 109 293t IU lentiviral vector 3 times per week until day 14. (B) Kaplan-Meier survival curve shows a significant difference (P =.004) in survival between the empty vector (solid line) and the sh-vcyclin (gray line) groups. There was no significant difference (P = .4) between the tumor only (broken line) and empty vector groups. (C) Bar chart shows percent of mice ascites-free up to 5 weeks after injection of JSC-1 cells. Mice culled for reasons other than ascites are considered ascites-free. Bar shading is as in panel A. Error bars indicate the standard error of the mean.
Therapeutic potential of sh-vcyclin. (A) Bar chart shows the weight gain of 8 normal mice (weight control, ▪), 15 mice inoculated with 1 × 107 JSC1 cells (tumor only, ▩ ), 15 mice treated from day 3 after inoculation with 1 × 107 JSC-1 cells with sh-vcyclin (sh-vcyclin, □), or 15 mice treated from day 3 after inoculation with 1 × 107 JSC-1 cells with empty vector (empty vector, ▦). Treatment regime was 1 × 1010 293t IU of lentiviral vector on day 3 and 5 and then 1 × 109 293t IU lentiviral vector 3 times per week until day 14. (B) Kaplan-Meier survival curve shows a significant difference (P =.004) in survival between the empty vector (solid line) and the sh-vcyclin (gray line) groups. There was no significant difference (P = .4) between the tumor only (broken line) and empty vector groups. (C) Bar chart shows percent of mice ascites-free up to 5 weeks after injection of JSC-1 cells. Mice culled for reasons other than ascites are considered ascites-free. Bar shading is as in panel A. Error bars indicate the standard error of the mean.
We autopsied and analyzed the solid tumors and ascites from all culled mice. Three of four of the solid tumors from the treated mice were midline extraperitoneal and were negative for the lentiviral insert. Three of the mice culled due to solid tumors in the empty vector group also developed ascites (Table 2 and Figure 5B). All ascitic samples also were found to be negative (< 0.01 copies per cell) for the lentiviral insert (data not shown). Since these tumors were therefore uninfected by intraperitoneal (IP) lentiviral injections, this represents a failure of administration, not treatment, and we reanalyzed the survival data in terms of development of ascites (Figure 5C). Mice that were culled due to solid tumors were considered to be ascites-free.
Comparison of development of ascites and solid tumors in infected versus control mice
. | No. mice developing ascites . | No. mice developing solid tumors . | No. alive . | Median survival time, d . |
|---|---|---|---|---|
| sh-vcyclin | 2 | 10 | 3 | 31 |
| Empty vector | 9 | 6 | 0 | 21 |
. | No. mice developing ascites . | No. mice developing solid tumors . | No. alive . | Median survival time, d . |
|---|---|---|---|---|
| sh-vcyclin | 2 | 10 | 3 | 31 |
| Empty vector | 9 | 6 | 0 | 21 |
Discussion
We show here the ability to use RNAi to inhibit the growth of an oncogenic virus-driven malignancy in an experimental model. This murine model faithfully mimics the phenotype of human primary effusion lymphoma.
RNAi is successfully being used to investigate various cellular processes and functions of genes in nonmammalian and mammalian systems.50-52 Although RNAi is the most robust form of gene silencing currently available, its specificity is still debated.53-56 Therefore, the therapeutic potential of RNAi as targeted medicine is being approached with caution, and efficient delivery remains of concern. Targeting one viral or cellular gene also could lead to the development of escape mutants,57,58 therefore the simultaneous targeting of multiple genes may be necessary for effective therapeutics.
We chose here to investigate the potential of RNAi as a therapy against KSHV-driven primary effusion lymphoma (PEL): all PEL are associated with KSHV latent gene expression59-61 ; these lymphomas present, and usually persist, as effusions, meaning gene therapy could be delivered directly and more efficiently than delivery to multiple systemic sites; these lymphomas are resistant to most cytotoxic drugs and their prognosis remains poor; and a murine model resembling human disease can be generated.33,34
We chose lentiviruses to deliver our RNAi, because these vectors have been shown to be effective delivery systems for shRNA,24 and pseudotyped lentiviral vectors infect a broad range of cell types in vitro and in vivo.62 Recent advances improve the safety of these vectors for potential use in HIV-positive individuals, reducing the risk of recombination with wild-type HIV.63
We first investigate the optimum target for RNAi to induce growth arrest of PEL. We focus on the oncogenic cluster in the KSHV genome: ORFs 71-73 encoding LANA, vFLIP, and vcyclin.64 These proteins are encoded on a tricistronic transcript,65 and each protein is implicated in oncogenesis.30 Silencing vFLIP alone induces apoptosis by way of inhibiting the NFκB pathway.40
We are uncertain as to why knock-down of LANA was independent of that of vcyclin and vFLIP (Figure 1C). We propose further RNA processing to turn tricistronic mRNA into LANA monocistronic and vcyclin-vFLIP bicistronic mRNAs (Figure 1D) is responsible for this effect, or the sequence between vcyclin and LANA ORF may form a secondary structure insulating the effect of RNAi.
Targeting either vcyclin or vFLIP leads to decreased expression of both proteins (Figure 1C). The knock-down of both of these oncogenic genes by 1 target makes them attractive for RNAi-based therapeutics. As expected, RNAi silencing was dose-dependent (correlating with number of lentiviral vectors delivered/cell) (Figure 1B) and partial (Figure 1E). In all PEL lines tested, LANA expression was down-regulated in the vast majority of cells, as determined by IFA (Figure 1F), with only a fraction of cells showing residual normal intensity for LANA (Figure 1Fii).
Because LANA maintains the viral episome,41 tethers viral DNA to chromatin during mitosis,42 and is responsible for interfering with crucial cellular pathways including p53,43 pRb/E2F,44 and GSK3B/β-catenin,45 it is proposed as a potential target for therapeutic intervention. However, targeting LANA by lentiviral-delivered RNAi did not affect growth in certain PEL lines tested, and LANA knock-down stable lines were generated for 2 of the lines (Figures 2A,F). As LANA is involved in ensuring the delivery of viral progeny to daughter cells, the KSHV copy number declined after LANA knock-down, but stabilized at approximately 15 copies per cell. We cannot exclude that the KSHV genome, as has been shown for other gammaherpesviruses,66 has integrated into cellular DNA in these lines. This would mean that viral progeny are equally distributed during cell division, without the need of LANA to link viral episomes to chromatin.
In PEL cells there are 50 to 150 copies KSHV per cell, whereas in KS there can be fewer than 10 copies per cell.67,68 Much of LANA protein in PEL cells could be redundant for its critical functions, and without complete silencing, targeting LANA may not be of therapeutic value in this lymphoproliferation.
A further concern of targeting LANA as a therapeutic was our finding that ORF50 expression is increased after LANA knock-down (Figure 2G). ORF50 (immediate-early Rta) is a viral transactivator responsible for the lytic switch in latently infected cells.69-71 Overexpression of Rta induces KSHV lytic replication and therefore the production of progeny virions. Our demonstration that LANA knockdown results in increased spontaneous and TPA-induced ORF50/Rta mRNA levels concurs with findings that LANA directly inhibits viral lytic replication by suppressing ORF50/Rta.49,72 The partial knockdown of LANA that we achieved here with RNAi and that is likely to result from small-molecule interventions may therefore result in increased KSHV virion production, increased viral load, and, subsequently, other KSHV-related pathologies.73
In contrast, targeting vcyclin and vFLIP with one shRNA results in apoptosis in all PEL lines tested (Figure 3A). Apoptosis occurred irrespective of the presence of EBV, confirming that KSHV is the essential oncogenic virus driving these tumors. Non-KSHV cell lines, including Ramos cells, were not affected by these vectors, supporting the specificity of targeting viral cyclin and FLIP. Apoptosis was maximal at day 7 (Figure 3C), consistent with the time for lentiviral integration, expression, and activity of the RNAi and the half-life of vFLIP and vcyclin.
We next investigated these targets in an experimental model of PEL. We used triple-knockout immunodeficient mice, which are negative for RAG, the common gamma chain, and C3, and show that we can faithfully reproduce PEL after intraperitoneal injection of 1 × 107 JSC-1 cells. This experiment also showed that at day 3, mice have already developed ascites, with an average 5% to 10% in weight gain. The vast majority of cells in the ascitic fluid were PEL cells staining positive for LANA by IFA (Figure 4B). Although other animal models for PEL have been established (eg, subcutaneous injection33,34 ), IP injections are the most appropriate model for human PEL. The biodistribution of lentiviruses after IP injection showed that little virus escapes the peritoneum to reach distant sites (Table 1). This is another potential advantage by obtaining efficient local delivery of RNAi and by minimizing any potential distant integration and adverse events.74 Co-inoculation of PEL cells and lentiviral-RNAi prevented ascites formation in 4 of 6 mice and showed an inhibitory effect in the remaining 2 mice (Figure 4C).
We next tested the therapeutic value of RNAi. After optimization experiments (data not shown), we chose a treatment regimen of bi-weekly injections (Figure 5A). We show that most mice remain ascites-free after RNAi treatment, and this translated into a significant survival benefit (Figure 5B). However, many mice developed solid tumors at the site of PEL inoculation (Table 2). This confirms that the lentiviruses injected intraperitoneally do not leak into the abdominal wall at a high enough titer to be of therapeutic value. Unfortunately, the development of solid tumors meant that mice had to be culled without having other evidence of PEL. Most encouragingly, with this regimen, 3 mice in the treatment arm remain completely disease-free at day 56 (8 weeks after initial injection).
Nonviral targets for KSHV-driven lymphoproliferations that also may have therapeutic potential include the NFκB pathway40,75 ; Stat3 signaling76 ; interleukin-2 receptor signaling77 ; the GSK3B/β-catenin pathway45 ; the unfolded protein response, and the vitamin D receptor.77,78
Mice were previously treated with RNAi to prevent hepatitis,17,18 but the high dosage or difficult delivery of dsRNA was necessary for a therapeutic effect. Our data suggest that human PEL should be amenable to intervention with lentiviral-delivered RNAi targeting vcyclin and vFLIP. Our data also indicate that IP inoculation of lentiviral vectors could be an efficient route to obtain therapeutic gene delivery in the peritoneal cavity. Optimizing treatment for PEL may require larger does of lentivirus, prolonged treatment, or combining RNAi with cytotoxic drugs. Targeting specifically PEL cells by altering the lentiviral envelope and therefore reducing infection of the peritoneum also should be tested. Other tools to deliver RNAi such as peptides79 or targeting vcyclin and vFLIP with novel gene knockdown technology80 or small molecule inhibitors also could be further exploited now that suitable targets have been identified within the oncogenic cluster.
Prepublished online as Blood First Edition Paper, November 30, 2004; DOI 10.1182/blood-2004-08-3052.
Funded by the St Stephens AIDS Trust and Cancer Research, United Kingdom.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to acknowledge Adrian Thrasher and Mike Blundell and Ruth Lyons (Institute of Child Health) for producing and caring for the triple knockout mice. We would also like to thank Dimitra Bourmpoulia, Dimitrios Lagos, Hsei-Wei Wang, and Heike Laman from the CRUK Viral Oncology Group for advice and technical assistance, and Yasuhiro Ikeda and Mary Collins (Windeyer Institute) for assistance with plasmids, antibodies, and production of lentiviral vectors.
The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal