Abstract
Erythrocyte invasion by malaria parasites and cytoadherence of Plasmodium falciparum-infected erythrocytes to host capillaries are 2 key pathogenic mechanisms in malaria. The receptor-binding domains of erythrocyte-binding proteins (EBPs) such as Plasmodium falciparum EBA-175, which mediate invasion, and P falciparum erythrocyte membrane protein 1 (PfEMP-1) family members, which are encoded by var genes and mediate cytoadherence, have been mapped to conserved cysteine-rich domains referred to as Duffy-binding–like (DBL) domains. Here, we have mapped regions within DBL domains from EBPs and PfEMP-1 that contain receptor-binding residues. Using biochemical and molecular methods we demonstrate that the receptor-binding residues of parasite ligands that bind sialic acid on glycophorin A for invasion as well as complement receptor-1 and chondroitin sulfate A for cytoadherence map to central regions of DBL domains. In contrast, binding to intercellular adhesion molecule 1 (ICAM-1) requires both the central and terminal regions of DBLβC2 domains. Determination of functional regions within DBL domains is the first step toward understanding the structure-function bases for their interaction with diverse host receptors.
Introduction
The clinical symptoms of malaria are attributed to the blood stage of the malaria parasite's life cycle. The invasion of erythrocytes by malaria parasites is, thus, key to malaria pathogenesis. Of the 4 Plasmodium species that cause human malaria, Plasmodium falciparum is responsible for the vast majority of deaths resulting from malaria. The virulence of cerebral and other forms of severe malaria is mainly due to the sequestration of infected erythrocytes containing P falciparum trophozoites and schizonts in the vasculature of diverse host organs.1 Sequestration of P falciparum in brain capillaries is implicated in the pathologic outcome of cerebral malaria.2-4 In case of infection during pregnancy, P falciparum late stages may sequester in the placenta, resulting in complications that often put the lives of both mother and child at risk.5 Erythrocytes infected with P falciparum can also bind uninfected erythrocytes to form rosettes, a cytoadherence phenotype that is also associated with severe malaria.6 It is important to understand the molecular interactions between malaria parasites and the host that mediate red cell invasion and cytoadherence, 2 important pathogenic mechanisms in malaria.
A family of erythrocyte-binding proteins (EBPs) mediates interactions with erythrocyte receptors during invasion.1,7 The EBP family includes Plasmodium vivax and Plasmodium knowlesi Duffy-binding proteins (PvDBP and PkDBP), P knowlesi β and γ proteins that bind receptors other than Duffy antigen on rhesus erythrocytes, and the 175-kDa erythrocyte-binding antigen from P falciparum (EBA-175), which binds sialic acid residues on glycophorin A.7 The functional receptor-binding domains of EBPs have been mapped to their N-terminal, conserved, cysteine-rich regions that are referred to as region II.8,9 These functional receptor-binding domains are also referred to as Duffy-binding–like (DBL) domains after the first erythrocyte-binding domain identified from PvDBP and PkDBP.8 DBL domains are also found in the P falciparum erythrocyte membrane protein 1 (PfEMP-1) family of variant surface antigens, which are expressed on the surface of P falciparum-infected erythrocytes and bind endothelial receptors to mediate cytoadherence.10-12 Binding domains for endothelial cytoadherence receptors such as intercellular adhesion molecule 1 (ICAM-1), CD31, chondroitin sulfate A (CSA), heparan sulfate, complement receptor 1 (CR1), and immunoglobulin G (IgG) have been mapped to DBL domains of PfEMP-1.13-22
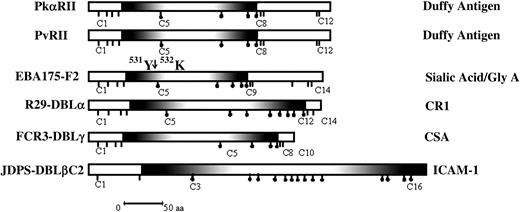
Malaria parasites thus use DBL domains to mediate diverse receptor-ligand interactions that are involved in both red cell invasion and cytoadherence. It is important to understand the structure-function basis for the interaction of DBL domains with the diverse host receptors that they bind to. As a first step in this direction, it is important to map receptor-binding sites within DBL domains. Regions II, the binding domains of PvDBP and PkDBP, which are also referred to as PvRII and PkαRII respectively, contain about 350 amino acids including 12 cysteines.23,24 The binding residues for the Duffy antigen map to the central approximately 170-amino acid stretch of PvRII and PkαRII that includes cysteines C5 to C8 (Figure 1).24 Here, we demonstrate that the binding residues of other DBL domains derived from EBPs and PfEMP-1 that bind diverse host receptors such as sialic acid residues on glycophorin A, CR1, and CSA also map to central regions that are equivalent to the C5 to C8 stretch of PvRII and PkαRII. An exception is a DBLβC2 domain from PfEMP-1 that binds ICAM-1, which requires sequences from both the central region of the DBLβ domain as well as sequences from the adjoining C2 region for binding. Understanding the structure-function bases for the interaction of DBL domains with their receptors may allow the development of novel methods to block red cell invasion and cytoadherence and provide protection against malaria.
DBL domains from EBPs and PfEMP-1. Shown are the DBL domains including the binding domains of PvDBP and PkDBP (PvRII and PkαRII); the binding domain of EBA-175 (F2), which binds sialic acid on glycophorin A; the DBLα domain of R29var1, which binds CR1; the DBLγ domain of FCR3varCSA, which binds CSA; and the DBLβC2 domain of JDP8ICAMvar, which binds ICAM-1. The position of chymotrypsin cleavage of F2 between tyrosine 531 (531Y) and lysine 532 (532K), which results in the 24-kDa F2 fragment that retains binding activity is shown. Shaded regions show the minimal deletion construct for each DBL domain that retains binding activity. Sticks under the domains show positions of cysteines. Sticks with black dots indicate cysteines that are included in the minimal functional deletion construct for each domain. The receptors the domains bind to are shown on the right; aa indicates amino acid.
DBL domains from EBPs and PfEMP-1. Shown are the DBL domains including the binding domains of PvDBP and PkDBP (PvRII and PkαRII); the binding domain of EBA-175 (F2), which binds sialic acid on glycophorin A; the DBLα domain of R29var1, which binds CR1; the DBLγ domain of FCR3varCSA, which binds CSA; and the DBLβC2 domain of JDP8ICAMvar, which binds ICAM-1. The position of chymotrypsin cleavage of F2 between tyrosine 531 (531Y) and lysine 532 (532K), which results in the 24-kDa F2 fragment that retains binding activity is shown. Shaded regions show the minimal deletion construct for each DBL domain that retains binding activity. Sticks under the domains show positions of cysteines. Sticks with black dots indicate cysteines that are included in the minimal functional deletion construct for each domain. The receptors the domains bind to are shown on the right; aa indicates amino acid.
Materials and methods
Production of recombinant F2, the binding domain of P falciparum EBA-175
Plasmid pF2PET1, which contains a DNA fragment that encodes the F2 domain of EBA-175 (amino acids 447-795 of EBA-175) fused with a C-terminal 6-histidine tag cloned downstream of the T7 promoter in the Escherichia coli expression vector pET28a+ (Novagen, Madison, WI), was used for expression of recombinant F2 in E coli BL21(DE3) cells (Novagen) as previously described.25 Recombinant F2 was purified from inclusion bodies under denaturing conditions by metal affinity chromatography, refolded by the method of rapid dilution, and purified to homogeneity by ion-exchange chromatography using SP-Sepharose and gel filtration chromatography using Superdex 75 as previously described.25
Proteolysis of recombinant F2 and pull-down assays to test binding to glycophorin A
Chymotrypsin was used for mild proteolysis of bacterially expressed and refolded recombinant F2 domain of P falciparum EBA-175. Recombinant F2 (5 μg) was digested with 0.4 μg chymotrypsin (Sigma, St Louis, MO) in 10 μL cleavage buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, pH 7.5, 100 mM NaCl, and 0.02% sodium azide) for either 1 hour (incomplete proteolysis) or 2 hours (complete proteolysis) at 21°C. The reaction was stopped by addition of 1 mM phenylmethylsulfonyl fluoride (PMSF). Full-length F2 and proteolytic fragments of F2 were incubated with 5 μg purified glycophorin A (Sigma) at 4°C for 1 hour to allow binding followed by incubation for another 1 hour at 4°C with anti–glycophorin A mouse monoclonal antibodies (Sigma) at 1:100 dilution. The mixture was incubated with 50% slurry of protein A beads (Pharmacia, Uppsala, Sweden) for 1 hour at 4°C. Protein A beads with bound protein-antibody complexes were separated by centrifugation for 10 minutes at 1500g at 4°C. The beads were boiled for 15 minutes in presence of nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and centrifuged at 1500g for 20 minutes at 4°C. Released proteins in the supernatant were separated by SDS-PAGE, transferred to Immuno-Blot polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules, CA), and detected using rabbit sera raised against recombinant F2.25 The identity of F2 fragments transferred to PVDF membranes was determined by protein N-terminal amino acid sequencing at a commercial facility at Michigan State University (East Lansing, MI). In control experiments, recombinant F2 was incubated with 1% bovine serum albumin (BSA) instead of glycophorin A prior to addition of anti–glycophorin A antibodies in pull-down assays. Recombinant PvRII, the binding domain of PvDBP, was produced as described earlier26 and used in pull-down assays with glycophorin A as another negative control. Rabbit serum raised against recombinant PvRII was used to detect PvRII by Western blotting in control pull-down assays.
Constructs for expression of full-length, deletion, and chimeric DBL constructs on the surface of mammalian cells
Plasmids were constructed to express DBL domains derived from EBPs and PfEMP-1 on the surface of mammalian cells as described earlier.8 Briefly, DBL domains were fused to the signal sequence and transmembrane domain of herpes simplex virus glycoprotein D (HSV gD) to allow targeting to the mammalian cell surface.8,27 The plasmid pRE4 (kindly provided by Drs Roselyn Eisenberg and Gary Cohen, University of Pennsylvania, Philadelphia), which contains the gene for HSV gD, was digested with PvuII and ApaI to excise the central region encoding amino acids 33 to 248 of HSV gD.8,27 DNA fragments encoding full-length or truncated DBL domains were amplified by polymerase chain reaction (PCR) using Pyrococcus furiosus DNA polymerase-specific primers (Strategene, La Jolla, CA), based on DBL domain sequences and plasmids containing the DBL domains as templates. PCR products were digested with PvuII and ApaI and cloned in the vector pRE4. Chimeric constructs containing DNA fragments from different DBL domains were made as described previously.23 Table 1 describes the boundaries and primers used to make constructs in the pRE4 vector to express full-length, truncated, and chimeric DBL domains on the surface of mammalian cells.
Constructs used for expression of full-length, truncated, and chimeric DBL domains on surface of mammalian cells
Construct . | Description . | PCR primers* . |
|---|---|---|
| EBA175-F2(C1-C14) | AA 447-795 of EBA-175 | F: 5′ acaagtcagctggaaaagcgtgaacatattgat 3′ |
| R: 5′ tctcgtgggcccatcgtcatcacgttctttttg 3′ | ||
| EBA175-F2(C5-C14) | AA 505-795 of EBA-175 | F: 5′ acaagtcagctgggaaacattgatagaatatac 3′ |
| R: 5′ tctcgtgggcccatcgtcatcacgttctttttg 3′ | ||
| EBA175-F2(C5-C12) | AA 505-736 of EBA-175 | F: 5′ acaagtcagctgggaaacattgatagaatatac 3′ |
| R: 5′ tctcgtgggccctttataatctgaatctaattc 3′ | ||
| EBA175-F2(C5-C9) | AA 505-639 of EBA-175 | F: 5′ acaagtcagctgggaaacattgatagaatatac 3′ |
| R: 5′ tctcgtgggcccatcttcacaaggtttttcttt 3′ | ||
| R29-DBLα(C1-C14) | AA 82-391 of R29var1 | F: 5′ tctcgtcagctggagtacaccgaaggtagaaag 3′ |
| R: 5′ acgagtgggccctttccttttttgcttatcaaattgttttc 3′ | ||
| R29-DBLα(C5-C12) | AA 133-366 of R29var1 | F: 5′ tctcgtcagctggatagaaatttagaatatttgatc 3′ |
| R: 5′ acgagtgggcccacgtggacaatttaaatctataaag 3′ | ||
| FCR3-DBLγ(C1-C10) | AA 1270-1578 of FCR3varCSA | F: 5′ acttgccagctggaaaacgatggaaagaaac 3′ |
| R: 5′ acgagtgggccccctgttcaagtaatctgttg 3′ | ||
| FCR3-DBLγ(C5-C10) | AA 1315-1527 of FCR3varCSA | F: 5′ acttgccagctggtacatttcttggcaaatg 3′ |
| R: 5′ acgagtgggcccttctgtatcacacttcaattg 3′ | ||
| FCR3-DBLγ(C5-C8) | AA 1315-1517 of FCR3varCSA | F: 5′ acttgccagctggtacatttcttggcaaatg 3′ |
| R: 5′ acgagtgggccctttaacgtagtcctcgcatttc 3′ | ||
| JDP8-DBLβC2(C1-C16) | AA 853-1275 of JDP8Icvar | F: 5′ tctcgtcagctgaacaatattccaatgct 3′ |
| R: 5′ acgagtgggcccctcgactttgccgccatc 3′ | ||
| FCR3-DBLβC2(C1-C16) | AA 810-1260 of FCR3varCSA | F: 5′ tctcgtcagctgataaaacattctaatcgtaatctt g 3′ |
| R: 5′ acgagtgggcccttccgtatccttttcttccgc 3′ | ||
| CH1 | AA 853-1141 (DBLβ) of JDP8Icvar fused to AA 1125-1260 (C2) of FCR3varCSA | F1: 5′ tctcgtcagctgaacaatattccaatgct 3′ |
| R1: 5′ gtatttcagttccattttgctcc 3′ | ||
| F2: 5′ aaagaattacatgaacaagcac 3′ | ||
| R2: 5′ acgagtgggcccttccgtatccttttcttccgc 3′ | ||
| CH2 | AA 810-1130 (DBLβ) of FCR3varCSA fused to AA 1128-1275 (C2) of JDP8Icvar | F1: 5′ tctcgtcagctgataaaacattctaatcgtaatcttg 3′ |
| R1: 5′ gtatttatctgatattatattccattg 3′ | ||
| F2: 5′ tgtacttcgaatttggaac 3′ | ||
| R2: 5′ acgagtgggcccctcgactttgccgccatc 3′ | ||
| JDP8-DBLβC2(C3-C16) | AA 905-1275 of JDP8Icvar | F: 5′ tctcgtcagctgacttcgaatttggaacatttac 3′ |
| R: 5′ acgagtgggcccctcgactttgccgccatc 3′ | ||
| JDP8-DBLβ(C3-C7) | AA 905-1098 of JDP8Icvar | F: 5′ tctcgtcagctgacttcgaatttggaacattta 3′ |
| R: 5′ acgagtgggcccaataccacactgcgtctc 3′ |
Construct . | Description . | PCR primers* . |
|---|---|---|
| EBA175-F2(C1-C14) | AA 447-795 of EBA-175 | F: 5′ acaagtcagctggaaaagcgtgaacatattgat 3′ |
| R: 5′ tctcgtgggcccatcgtcatcacgttctttttg 3′ | ||
| EBA175-F2(C5-C14) | AA 505-795 of EBA-175 | F: 5′ acaagtcagctgggaaacattgatagaatatac 3′ |
| R: 5′ tctcgtgggcccatcgtcatcacgttctttttg 3′ | ||
| EBA175-F2(C5-C12) | AA 505-736 of EBA-175 | F: 5′ acaagtcagctgggaaacattgatagaatatac 3′ |
| R: 5′ tctcgtgggccctttataatctgaatctaattc 3′ | ||
| EBA175-F2(C5-C9) | AA 505-639 of EBA-175 | F: 5′ acaagtcagctgggaaacattgatagaatatac 3′ |
| R: 5′ tctcgtgggcccatcttcacaaggtttttcttt 3′ | ||
| R29-DBLα(C1-C14) | AA 82-391 of R29var1 | F: 5′ tctcgtcagctggagtacaccgaaggtagaaag 3′ |
| R: 5′ acgagtgggccctttccttttttgcttatcaaattgttttc 3′ | ||
| R29-DBLα(C5-C12) | AA 133-366 of R29var1 | F: 5′ tctcgtcagctggatagaaatttagaatatttgatc 3′ |
| R: 5′ acgagtgggcccacgtggacaatttaaatctataaag 3′ | ||
| FCR3-DBLγ(C1-C10) | AA 1270-1578 of FCR3varCSA | F: 5′ acttgccagctggaaaacgatggaaagaaac 3′ |
| R: 5′ acgagtgggccccctgttcaagtaatctgttg 3′ | ||
| FCR3-DBLγ(C5-C10) | AA 1315-1527 of FCR3varCSA | F: 5′ acttgccagctggtacatttcttggcaaatg 3′ |
| R: 5′ acgagtgggcccttctgtatcacacttcaattg 3′ | ||
| FCR3-DBLγ(C5-C8) | AA 1315-1517 of FCR3varCSA | F: 5′ acttgccagctggtacatttcttggcaaatg 3′ |
| R: 5′ acgagtgggccctttaacgtagtcctcgcatttc 3′ | ||
| JDP8-DBLβC2(C1-C16) | AA 853-1275 of JDP8Icvar | F: 5′ tctcgtcagctgaacaatattccaatgct 3′ |
| R: 5′ acgagtgggcccctcgactttgccgccatc 3′ | ||
| FCR3-DBLβC2(C1-C16) | AA 810-1260 of FCR3varCSA | F: 5′ tctcgtcagctgataaaacattctaatcgtaatctt g 3′ |
| R: 5′ acgagtgggcccttccgtatccttttcttccgc 3′ | ||
| CH1 | AA 853-1141 (DBLβ) of JDP8Icvar fused to AA 1125-1260 (C2) of FCR3varCSA | F1: 5′ tctcgtcagctgaacaatattccaatgct 3′ |
| R1: 5′ gtatttcagttccattttgctcc 3′ | ||
| F2: 5′ aaagaattacatgaacaagcac 3′ | ||
| R2: 5′ acgagtgggcccttccgtatccttttcttccgc 3′ | ||
| CH2 | AA 810-1130 (DBLβ) of FCR3varCSA fused to AA 1128-1275 (C2) of JDP8Icvar | F1: 5′ tctcgtcagctgataaaacattctaatcgtaatcttg 3′ |
| R1: 5′ gtatttatctgatattatattccattg 3′ | ||
| F2: 5′ tgtacttcgaatttggaac 3′ | ||
| R2: 5′ acgagtgggcccctcgactttgccgccatc 3′ | ||
| JDP8-DBLβC2(C3-C16) | AA 905-1275 of JDP8Icvar | F: 5′ tctcgtcagctgacttcgaatttggaacatttac 3′ |
| R: 5′ acgagtgggcccctcgactttgccgccatc 3′ | ||
| JDP8-DBLβ(C3-C7) | AA 905-1098 of JDP8Icvar | F: 5′ tctcgtcagctgacttcgaatttggaacattta 3′ |
| R: 5′ acgagtgggcccaataccacactgcgtctc 3′ |
AA indicates amino acid.
Two pairs of primers (F1, R1 and F2, R2) were used for PCR amplification of the 2 DNA fragments that were ligated to form the chimeric constructs CH1 and CH2.
Forward (F) and reverse (R) primers used for amplification of DNA encoding DBL domain fragments by PCR are shown
Mammalian cell culture, transfection, and immunofluorescence assays
COS-7 and 293T cells were cultured in Dulbecco modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) with 10% heat-inactivated fetal calf serum (FCS) in a humidified CO2 (5%) incubator at 37°C. Fresh monolayers of 40% to 60% confluent COS-7 and 293T cells growing in 35-mm diameter wells were transfected with 2 to 4 μg plasmid DNA using Lipofectamine Plus reagent (Invitrogen) as described by the manufacturer. Transfected cells were used for immunofluorescence and binding assays 36 to 40 hours after transfection. Immunofluorescence assays using mouse monoclonal antibody DL6 (kindly provided by Drs Roselyn Eisenberg and Gary Cohen), which reacts with amino acids 272 to 279 of HSV gD, were performed as described earlier8,27 to detect expression of the fusion proteins on the surface of transfected COS-7 and 293T cells.
Erythrocyte-binding assays with COS-7 cells expressing DBL domain fragments on the surface
Transfected COS-7 cells expressing various deletion constructs of EBA175-F2 and R29-DBLα were tested for binding to erythrocytes as described previously.8 Transfected COS-7 cells expressing F2 constructs were tested for binding to normal, neuraminidase-treated, and trypsintreated human erythrocytes. Erythrocytes were treated with neuraminidase and trypsin as described previously.9,23 Transfected COS-7 cells expressing R29-DBLα constructs were tested for binding to normal human erythrocytes and CR1-deficient erythrocytes (kindly provided by Dr Karina Yazdanbaksh, New York Blood Centre [NYBC], New York, NY). The copy number of CR1 on CR1-deficient erythrocytes was about 10% of normal levels (personal communication, Dr Karina Yazdanbaksh, January 13, 2004). The number of rosettes of erythrocytes bound to transfected COS-7 cells was scored in 25 fields at × 200 magnification. A cluster of 6 or more erythrocytes bound to a COS-7 cell was scored as a rosette.
Binding of CSA to 293T cells expressing full-length or truncated FCR3-DBLγ domain on the surface
Mammalian 293T cells were grown on coverslips and transfected with constructs designed to express full-length or truncated FCR3-DBLγ domains on the surface. Coverslips with transfected 293T cells were transferred to 12-well culture plates 36 to 40 hours after transfection and used to test binding of CSA as described earlier.21 Briefly, cells were washed once with phosphate-buffered saline (PBS) and fixed with 2% paraformaldehyde. CSA, CSB, and CSC (Calbiochem, San Diego, CA) were biotinylated using a biotinylation kit (Pierce, Rockford, IL) as described by the manufacturer. Transfected 293T cells were incubated with biotinylated CSA (Bio-CSA), Bio-CSB, or Bio-CSC (100 μg/mL) for 1 hour at room temperature, washed 3 times with PBS, and incubated with mouse sera (1:200 dilution) raised against biotin (Sigma). Cells were washed 3 times with PBS and incubated with fluoresceinisothiocyanate (FITC)–conjugated anti–mouse IgG goat antibodies (Sigma) diluted 1:200. Cells were washed 3 times with PBS and mounted with coverslips after addition of Antifade reagent (Sigma). All sera were diluted in PBS containing 0.5% BSA (Sigma) and all incubations with antibodies were for 1 hour at room temperature. Number of fluorescent and total cells was scored in 15 fields at × 1000 magnification to determine the binding efficiency.
Binding of ICAM-1 to COS-7 cells expressing full-length, truncated, and chimeric DBLβC2 domain fragments
ICAM1-Fc was purified from supernatants of transfected COS-7 cells by affinity chromatography on protein G columns and used for binding assays as previously described.19,22 ICAM1-Fc was coated on Dynal magnetic beads and tested for binding to transfected COS-7 cells expressing DBLβC2 constructs on the surface as described earlier.19,22
Results
Binding residues for sialic acids on glycophorin A lie between cysteines 5 and 9 of EBA-175 DBL domain F2
EBA-175 binds sialic acids on glycophorin A to mediate invasion of human erythrocytes by P falciparum merozoites.28 The extracellular domain of EBA-175 contains tandem repeated DBL domains, F1 and F2, within the N-terminal cysteine-rich region, region II.9,29 The receptor-binding domain of EBA-175 maps to DBL domain F2, which contains 14 cysteines9 (Figure 1). Recombinant F2 was expressed in E coli, extracted from inclusion bodies, refolded by the method of rapid dilution, and purified to homogeneity as described previously.25 Refolded F2 migrates on SDS-PAGE as a single band of about 42 kDa and binds normal human erythrocytes but not neuraminidase-treated erythrocytes in erythrocyte-binding assays (data not shown) indicating that it is functional and correctly folded.
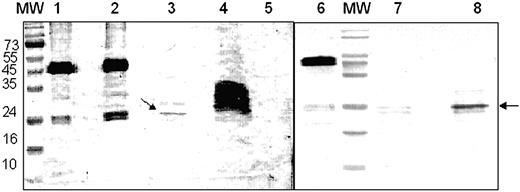
To identify fragments of F2 that retain receptor-binding activity, recombinant F2 was treated with chymotrypsin and both full-length F2 and proteolytic fragments of F2 were tested for binding to glycophorin A in pull-down assays. Both F2 and proteolytic fragments of F2 were incubated with glycophorin A to allow binding. Antibodies to glycophorin A and protein A-coated beads were then used to pull down glycophorin A-F2 fragment complexes. Precipitated complexes were separated by SDS-PAGE, and fragments of F2 that bound glycophorin A were detected by Western blotting using rabbit sera raised against recombinant F2. Full-length F2 as well as a 24-kDa proteolytic fragment of F2 bound glycophorin A in the pull-down assays (Figure 2). Incubation of F2 with BSA instead of glycophorin A did not result in precipitation of F2 in pull-down assays (Figure 2). Moreover, recombinant PvRII, the binding domain of P vivax Duffy-binding protein, did not bind glycophorin A in the pull-down assays (data not shown). The binding of F2 and the 24-kDa F2 fragment to glycophorin A is thus specific. Protein N-terminal sequencing revealed that the 24-kDa band starts with lysine 532 (K532), which lies between cysteines C4 and C5 of EBA-175 (Figure 1). Molecular mass considerations suggest that the 24-kDa fragment is likely to span the region containing cysteines C5 to C12 of F2.
Binding of recombinant F2 and chymotryptic F2 fragments to glycophorin A in pull-down assays. Recombinant F2 was used in pull-down assays with glycophorin A before and after treatment with chymotrypsin. Recombinant F2 and chymotrypsin-treated F2 were incubated with glycophorin A to allow binding. F2 fragments bound to glycophorin A were precipitated using a mouse monoclonal antibody raised against glycophorin A and protein A-Sepharose. Bound F2 fragments were separated by SDS-PAGE and detected by Western blotting using rabbit sera raised against recombinant F2. MW indicates prestained molecular weight markers shown in kDa; lane 1, full-length recombinant F2 used for pull-down assays; lane 2, pull-down assay with recombinant F2 and glycophorin A; lane 3, pull-down assay with chymotrypsin-treated F2 (incomplete proteolysis) and glycophorin A; lane 4, chymotrypsin-treated F2 (incomplete proteolysis) used for pull-down assay; lane 5, control pull-down assay using recombinant F2 and BSA instead of glycophorin A; lane 6, full-length recombinant F2; lane 7, chymotrypsin-treated F2 (complete proteolysis); lane 8, pull-down assay with chymotrypsin-treated F2 (complete proteolysis) and glycophorin A.
Binding of recombinant F2 and chymotryptic F2 fragments to glycophorin A in pull-down assays. Recombinant F2 was used in pull-down assays with glycophorin A before and after treatment with chymotrypsin. Recombinant F2 and chymotrypsin-treated F2 were incubated with glycophorin A to allow binding. F2 fragments bound to glycophorin A were precipitated using a mouse monoclonal antibody raised against glycophorin A and protein A-Sepharose. Bound F2 fragments were separated by SDS-PAGE and detected by Western blotting using rabbit sera raised against recombinant F2. MW indicates prestained molecular weight markers shown in kDa; lane 1, full-length recombinant F2 used for pull-down assays; lane 2, pull-down assay with recombinant F2 and glycophorin A; lane 3, pull-down assay with chymotrypsin-treated F2 (incomplete proteolysis) and glycophorin A; lane 4, chymotrypsin-treated F2 (incomplete proteolysis) used for pull-down assay; lane 5, control pull-down assay using recombinant F2 and BSA instead of glycophorin A; lane 6, full-length recombinant F2; lane 7, chymotrypsin-treated F2 (complete proteolysis); lane 8, pull-down assay with chymotrypsin-treated F2 (complete proteolysis) and glycophorin A.
A series of deletion constructs were designed to express truncated fragments of F2 on the surface of COS cells and test them for binding to erythrocytes. All the deletion constructs include cysteine C5 at the N-terminus and terminate either after cysteines C14, C12, or C9 at the C-terminus. They are referred to as F2(C5-C14), F2(C5-C12), and F2(C5-C9), respectively. Following transfection, erythrocyte-binding assays were used to test the binding of COS cells expressing deletion constructs of F2 to human red cells. In control experiments, binding to neuraminidase-treated human erythrocytes was examined. Constructs designed to express region F2 or region II (F1+F2) were used as positive controls. Regions II (F1+F2) and F2 bind normal human erythrocytes but do not bind neuraminidase-treated human erythrocytes (Table 2). The deletion constructs F2(C5-14), F2(C5-C12), and F2(C5-C9) also bind human erythrocytes but not neuraminidase-treated human erythrocytes (Table 2). COS cells expressing regions F2(C1-C14) and F2(C5-C9) were also tested for binding to trypsin-treated human erythrocytes. Trypsin cleaves glycophorin A but not glycophorin B. Both the full-length domain F2(C1-C14) and F2(C5-C9) did not bind trypsin-treated human erythrocytes that had lost glycophorin A. Critical binding residues for sialic acid residues on glycophorin A thus lie in the central region of F2 between cysteines C5 and C9. The binding efficiency of region II (F1+F2) is significantly higher than the binding efficiency of region F2 alone. Similarly, although F2(C5-C9) binds erythrocytes with correct specificity, it binds poorly compared to regions II (F1+F2), F2, and F2(C5-C12) (Table 2).
Binding of normal and neuraminidase-treated human erythrocytes to deletion constructs of region F2 of EBA-175
. | . | . | No. rosettes* . | . | . | . | Binding efficiency† . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Transfection efficiency, % . | . | Normal . | . | Neu . | . | Normal . | . | Neu . | . | ||||||
| Construct . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | ||||||
| F1 + F2 | 10 | 25 | 101 | 161 | 0 | 0 | 7.0 | 11.0 | 0 | 0 | ||||||
| F2(C1-C14) | 6 | 33 | 62 | 67 | 0 | 0 | 1.5 | 7.4 | 0 | 0 | ||||||
| F2(C5-C14) | 9 | 31 | 51 | 19 | 0 | 0 | 1.7 | 1.9 | 0 | 0 | ||||||
| F2(C5-C12) | 5 | 28 | 48 | 31 | 0 | 0 | 1.0 | 3.0 | 0 | 0 | ||||||
| F2(C5-C9) | 15 | 30 | 8 | 9 | 0 | 0 | 0.5 | 1.0 | 0 | 0 | ||||||
| Untransfected | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
. | . | . | No. rosettes* . | . | . | . | Binding efficiency† . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Transfection efficiency, % . | . | Normal . | . | Neu . | . | Normal . | . | Neu . | . | ||||||
| Construct . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | ||||||
| F1 + F2 | 10 | 25 | 101 | 161 | 0 | 0 | 7.0 | 11.0 | 0 | 0 | ||||||
| F2(C1-C14) | 6 | 33 | 62 | 67 | 0 | 0 | 1.5 | 7.4 | 0 | 0 | ||||||
| F2(C5-C14) | 9 | 31 | 51 | 19 | 0 | 0 | 1.7 | 1.9 | 0 | 0 | ||||||
| F2(C5-C12) | 5 | 28 | 48 | 31 | 0 | 0 | 1.0 | 3.0 | 0 | 0 | ||||||
| F2(C5-C9) | 15 | 30 | 8 | 9 | 0 | 0 | 0.5 | 1.0 | 0 | 0 | ||||||
| Untransfected | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
Neu indicates neuraminidase-treated erythrocytes; NA, not applicable.
The number of COS-7 cells with rosettes of bound erythrocytes was scored in 25 fields at × 200 magnification. Data from 2 independent experiments are reported. The number of rosettes observed was normalized for transfection efficiency of 20% for both experiments. No rosettes were observed in the entire well where number of rosettes is reported as zero
The binding efficiency is calculated as follows: % binding efficiency = no. of COS cells with rosettes × 100/total no. of COS cells
Binding residues for CR1 lie in the central region of the DBLα domain of R29var1, the expressed var gene from rosetting P falciparum strain R29
Red cells infected with P falciparum strain R29 bind CR1 on uninfected erythrocytes to form rosettes.15 R29var1, the var gene expressed in R29, has been identified.15 The functional receptor-binding domain that binds CR1 has been mapped to the N-terminal DBLα domain, R29-DBLα, of R29var1.15 R29-DBLα contains 309 amino acids including 14 cysteines. Transfected COS cells expressing R29-DBLα on the surface bind CR1 on human erythrocytes.15 Here, we have determined if the receptor-binding residues of R29-DBLα map to the central region of R29-DBLa that is equivalent to the central approximately 170-amino acid stretch spanning C5 to C8 of PvRII and PkαRII that contain binding residues for the Duffy antigen.24
A multiple sequence alignment was performed to identify the boundaries of the central region of R29-DBLα that is homologous to the C5-C8 regions of PvRII and PkαRII (data not shown). The multiple sequence alignment demonstrates that cysteines corresponding to C11 and C12 of PvRII and PkαRII are missing in R29-DBLα. In addition, 4 cysteines from the central region of R29-DBLα, C6, C8, C10, and C12, are not found in PvRII or PkαRII. A 233-amino acid stretch of R29-DBLα that includes cysteines C5 to C12 corresponds to the central C5 to C8 stretch of PvRII and PkαRII. A deletion construct, R29-DBLα(C5-C12), was used to express this central fragment of R29-DBLα on the surface of COS cells and test binding to normal human erythrocytes. Binding to CR1-deficient human erythrocytes that express lower levels of CR1 was tested in control experiments. Constructs designed to express full-length R29-DBLα and PvRII were also used as controls. PvRII binds both normal and CR1-deficient cells with similar efficiency. In contrast, both R29-DBLα and R29-DBLα(C5-C12) bind CR1-deficient erythrocytes poorly compared to normal human erythrocytes (Table 3). The deletion construct R29-DBLα(C5-C12) and full length R29-DBLα(C1-C14) thus bind CR1 on human erythrocytes. The binding residues for CR1 lie in the central region of R29-DBLα in the amino acid stretch containing cysteines C5 to C12 (Figure 1).
Binding of normal human erythrocytes and CR1-deficient human erythrocytes to deletion constructs of R29-DBLα
. | . | No. rosettes* . | . | . | . | Binding efficiency, %† . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Normal . | . | CR1-deficient . | . | Normal . | . | CR1-deficient . | . | ||||||
| Construct . | Transfection efficiency, % . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | ||||||
| R29-DBLα(C1-C14) | 33 | 120 | 105 | 40 | 25 | 36 | 39 | 11 | 10 | ||||||
| R29-DBLα(C5-C12) | 33 | 142 | 95 | 40 | 26 | 38 | 36 | 10 | 9 | ||||||
| PvRII | 60 | 217 | 144 | 204 | 153 | 56 | 55 | 53 | 50 | ||||||
| Untransfected | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
. | . | No. rosettes* . | . | . | . | Binding efficiency, %† . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | Normal . | . | CR1-deficient . | . | Normal . | . | CR1-deficient . | . | ||||||
| Construct . | Transfection efficiency, % . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | ||||||
| R29-DBLα(C1-C14) | 33 | 120 | 105 | 40 | 25 | 36 | 39 | 11 | 10 | ||||||
| R29-DBLα(C5-C12) | 33 | 142 | 95 | 40 | 26 | 38 | 36 | 10 | 9 | ||||||
| PvRII | 60 | 217 | 144 | 204 | 153 | 56 | 55 | 53 | 50 | ||||||
| Untransfected | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
The number of COS-7 cells with rosettes of bound erythrocytes was scored in 25 fields at × 200 magnification. Data from 2 independent experiments are reported. Transfection efficiency for experiment 1 is shown. No rosettes were seen in entire well where number of rosettes is reported as zero
The binding efficiency is calculated as follows: % binding efficiency = no. of COS cells with rosettes × 100/total no. of COS cells
Binding residues for CSA lie in the central region of DBLγ from FCR3varCSA
The DBLγ domain, FCR3-DBLγ, derived from the var gene, FCR3varCSA, has been shown to bind CSA.21 FCR3-DBLγ contains 302 amino acids including 10 cysteines. Multiple sequence alignment of FCR3-DBLγ with PvRII and PkαRII revealed that the region spanning cysteines C5 to C8 of FCR3-DBLγ corresponds to the C5 to C8 stretch of PvRII and PkαRII24 (data not shown). Full-length FCR3-DBLγ containing cysteines C1 to C10 (FCR3-DBLγ(C1-C10)) as well as deletion fragments containing cysteines C5 to C10 (FCR3-DBLγ(C5-C10)) and cysteines C5 to C8 (FCR3-DBLγ(C5-C8)) were expressed on the surface of mammalian 293T cells and tested for binding to CSA. In control experiments, binding was tested to CSB and CSC. FCR3-DBLγ(C1-C10), FCR3-DBLγ(C5-C10), and FCR3-DBLγ(C5-C8) bound CSA but not CSB or CSC (Table 4). The binding residues for CSA, thus, lie in the central region of FCR3-DBLγ between cysteines C5 and C8 (Figure 1).
Binding of CSA, CSB, and CSC to deletion constructs of FCR3-DBLγ
. | Transfection efficiency, % . | . | . | Binding efficiency, CSA, %* . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Construct . | Exp 1 . | Exp 2 . | Exp 3 . | Exp 1 . | Exp 2 . | Exp 3 . | ||||
| FCR3-DBLγ(C1-C10) | 64 | 64 | 74 | 60 | 60 | 57 | ||||
| FCR3-DBLγ(C5-C10) | 63 | 60 | 65 | 62 | 62 | 64 | ||||
| FCR3-DBLγ(C5-C8) | 61 | 65 | 67 | 67 | 61 | 67 | ||||
| Untransfected | NA | NA | NA | 0 | 0 | 0 | ||||
. | Transfection efficiency, % . | . | . | Binding efficiency, CSA, %* . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Construct . | Exp 1 . | Exp 2 . | Exp 3 . | Exp 1 . | Exp 2 . | Exp 3 . | ||||
| FCR3-DBLγ(C1-C10) | 64 | 64 | 74 | 60 | 60 | 57 | ||||
| FCR3-DBLγ(C5-C10) | 63 | 60 | 65 | 62 | 62 | 64 | ||||
| FCR3-DBLγ(C5-C8) | 61 | 65 | 67 | 67 | 61 | 67 | ||||
| Untransfected | NA | NA | NA | 0 | 0 | 0 | ||||
Transfected 293T cells were incubated with biotinylated CSA, CSB, and CSC. FITC-conjugated streptavidin was used to detect binding of biotinylated CSA, CSB, and CSC to 293T cells. Between 1500 and 2000 293T cells were scored. The number of 293T cells that are fluorescent was determined. No fluorescent cells were seen in entire well where binding efficiency is reported as zero. The binding efficiency was calculated as follows: % binding efficiency = no. of fluorescent 293T cells × 100/total no. of 293T cells. None of the constructs bound CSB or CSC
Mapping the region that contains binding residues for ICAM-1 within DBLβC2 of JDP8Icvar
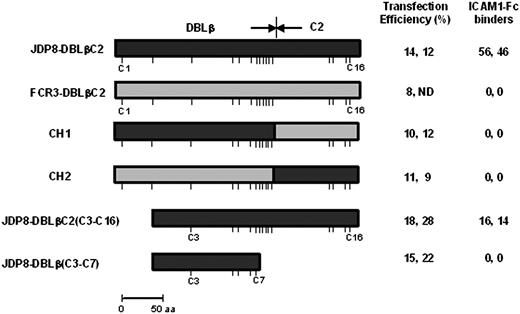
P falciparum field isolate JDP8 binds ICAM-1.22 The ICAM-1–binding domain has been mapped to the DBLβC2 domain of JDP8Icvar, the var gene expressed in P falciparum JDP8.22 The DBLβ and C2 domains of JDP8-DBLβC2 contain 12 and 4 conserved cysteines, respectively. Neither JDP8-DBLβ nor JDP8-C2 bind ICAM-1 when expressed separately.22 Both JDP8-DBLβ and JDP8-C2 are thus necessary for binding to ICAM-1. Here, we have tested the ICAM-1–binding ability of chimeric constructs that contain stretches from JDP8-DBLβC2, which binds ICAM-1, fused to stretches from a homologous DBLβC2 domain, FCR3-DBLβC2, which does not bind ICAM-1 (Figure 3). The chimeric construct, CH1, which contains JDP8-DBLβ fused to FCR3-C2, does not bind ICAM-1 (Figure 3), indicating that the JDP8-C2 domain contains some contact residues that are critical for binding to ICAM-1. Similarly, a chimeric construct, CH2, which contains FCR3-DBLβ fused to JDP8-C2 does not bind ICAM-1 (Figure 3), indicating that JDP8-C2 sequences are necessary but not sufficient for binding to ICAM-1. Both DBLβ and C2 domains thus contain binding residues for ICAM-1. Multiple sequence alignment of JDP8-DBLβC2 with PvRII and PkαRII revealed that cysteines C5 of PvRII and PkαRII correspond to cysteine C3 of JDP8-DBLβC224 (data not shown). Deletion constructs were used to express truncated JDP8-DBLβC2 domains on the surface of COS cells and test binding to ICAM-1. JDP8-DBLβC2(C3-C16) binds ICAM-1 (Figure 3). JDP8-DBLβ(C3-C7), which does not contain the C2 region, fails to bind ICAM-1 (Figure 3). Binding residues for ICAM-1 thus lie in the region spanning cysteines C3-C16 of JDP8-DBLβC2.
Binding of ICAM-1 to DBLβC2 chimeric and deletion constructs. Chimeric and deletion constructs based on the ICAM-1–binding DBLβC2 domain derived from JDP8Icvar (JDP8-DBLβC2) and DBLβC2 domain derived from FCR3varCSA (FCR3-DBLβC2), which does not bind ICAM-1, were expressed on surface of COS-7 cells and tested for binding to ICAM-1. Transfection efficiency for each construct is shown. Number of transfected COS-7 cells that bind ICAM1-Fc–coated magnetic beads scored in 25 fields at × 200 magnification and normalized to transfection efficiency of 20% is shown. No binding was seen in the entire well where zero binding is reported. Positions of cysteines are shown with sticks. ND indicates not done.
Binding of ICAM-1 to DBLβC2 chimeric and deletion constructs. Chimeric and deletion constructs based on the ICAM-1–binding DBLβC2 domain derived from JDP8Icvar (JDP8-DBLβC2) and DBLβC2 domain derived from FCR3varCSA (FCR3-DBLβC2), which does not bind ICAM-1, were expressed on surface of COS-7 cells and tested for binding to ICAM-1. Transfection efficiency for each construct is shown. Number of transfected COS-7 cells that bind ICAM1-Fc–coated magnetic beads scored in 25 fields at × 200 magnification and normalized to transfection efficiency of 20% is shown. No binding was seen in the entire well where zero binding is reported. Positions of cysteines are shown with sticks. ND indicates not done.
Discussion
Malaria parasites use DBL domains to bind diverse host receptors to mediate interactions that are involved in 2 key pathogenic processes, namely, red cell invasion and cytoadherence.1 Receptor-binding domains of both EBPs, which mediate red cell invasion, and PfEMP-1, which mediate cytoadherence, have been mapped to DBL domains.1 It is important to understand how these conserved cysteine-rich domains are able to bind such a diverse range of host receptors. What is the underlying conserved architecture of DBL domains and where do their receptor-binding pockets lie? A clear understanding of the interaction of DBL domains with their receptors may allow the development of novel strategies to inhibit red cell invasion or reverse cytoadherence.
Phylogenetic analysis of DBL domains reveals that DBL domains derived from EBPs group together.30,31 The PfEMP-1 DBL domains have diverged significantly from the EBP DBL domains and cluster into at least 6 distinct sequence subtypes designated α, β, γ, δ, ϵ, and X.30-32 DBL domains of EBPs contain 12 invariant cysteines. The first 10 cysteines from EBP DBL domains can usually be identified in PfEMP-1 DBL domains, although in some cases a subset of cysteines may be missing and novel cysteines may appear. Interspersed between the cysteines are conserved amino acid stretches that form type-specific homology blocks that are flanked by hypervariable amino acid stretches. EBP DBL domains display natural variation in sequence due to point mutations but rarely display variation in length of amino acid stretches between invariant cysteines.30 In contrast, PfEMP-1 DBL domains display great divergence in both sequence and length even among DBL domains of the same subtype.30 Despite the extent of sequence diversity in DBL domains, it is predicted that due to the presence of conserved cysteines and homology blocks between invariant cysteines, DBL domains may have a common fold.30 As a result the receptor-binding pockets may lie in the same region of diverse DBL domains.
We have previously shown that critical binding residues for the Duffy antigen lie in a central stretch of about 170 amino acids spanning C5 to C8 of the prototypical DBL domains, PvRII and PkαRII.23,24 Here, we have tested whether the binding residues of other DBL domains from EBPs and PfEMP-1 also lie in central regions equivalent to the C5 to C8 stretch of PvRII and PkαRII. We have analyzed the interaction of DBL domain F2 from P falciparum EBA-175, which binds sialic acid on glycophorin A to mediate invasion. Proteolytic cleavage of recombinant F2 using chymotrypsin followed by pull-down assays using glycophorin A identified a 24-kDa fragment starting with K532 of EBA-175 at the N-terminus that retained binding activity. The position of the cleavage site resulting in the functionally active 24-kDa fragment indicates that, as previously seen for PvRII and PkαRII,24 the amino acid stretch spanning cysteines C1 to C4 is not required for binding. The use of deletion constructs demonstrated that F2(C5-C9), which contains cysteines C5 to C9, binds red cells with specificity. Binding residues of DBL domain F2 thus lie in the central region as previously seen for PvRII and PkαRII. The deletion construct F2(C5-C9) binds erythrocytes poorly compared to region II (F1+F2), region F2, and region F2(C5-C12). It is likely that sequences outside the central region (C5-C9) play a role in the binding affinity of region F2 either by influencing the folding of F2 or by contributing additional contact residues.
We have also mapped regions containing binding residues in DBL domains derived from PfEMP-1 that mediate cytoadherence. Binding sites were mapped in R29-DBLα, which binds CR1, FCR3-DBLγ, which binds CSA, and JDP8-DBLβC2, which binds ICAM-1. Multiple sequence alignments were performed to identify central regions of these DBL domains that are equivalent to the C5 to C8 stretch of PvRII and PkαRII24 (data not shown). Deletion constructs were used to express the central regions of R29-DBLα, FCR3-DBLγ, and JDP8-DBLβC2 on the surface of mammalian cells and test them for binding to relevant receptors.
R29-DBLα binds CR1 to mediate rosetting of P falciparum-infected erythrocytes with uninfected erythrocytes.15 A deletion construct, R29-DBLα(C5-C12), which corresponds to the C5 to C8 stretch of PvRII and PkαRII,24 binds erythrocytes with the same specificity as full-length R29-DBLα(C1-C14). Binding residues for CR1 thus lie in the central region of R29-DBLα as in case of PvRII, PkαRII, and F2 (Figure 1).
FCR3-DBLγ, which binds CSA, contains 14 cysteines.21 A deletion construct, FCR3-DBLγ(C5-C8), which spans the central region corresponding to the C5 to C8 stretch of PvRII and PkαRII,24 binds CSA with specificity. The binding residues for CSA, thus, also lie in the central region of FCR3-DBLγ (Figure 1).
The ICAM-1–binding domain of JDP8Icvar has been mapped to its DBLβC2 domain.22 The C2 domain, which contains 4 conserved cysteines, is commonly found in association with DBLβ domains of PfEMP-1.30 It has been previously shown that DBLβ and C2 domains of JDP8Icvar do not bind ICAM-1 when expressed separately indicating that both regions are required for binding.22 However, it was not clear if the C2 region plays a structural role and is required for correct folding of the binding pocket or if C2 contains binding residues for ICAM-1. Here, we have used chimeric constructs that contain regions from the ICAM-1–binding JDP8-DBLβC2 domain fused to regions from FCR3-DBLβC2, which does not bind ICAM-1, to distinguish between these possibilities. The FCR3-DBLβ region contains the 12 conserved cysteines (C1-C12) found in JDP8-DBLβ and FCR-C2 contains the 4 conserved cysteines (C13-C16) found in JDP8-C2. The chimeric constructs, CH1 and CH2, should thus be able to form the correct disulfide linkages and fold correctly. The inability of CH1 and CH2 to bind ICAM-1 indicates that both DBLβ and C2 regions of JDP8Icvar contain contact residues for ICAM-1. Deletion constructs demonstrated that as observed for other DBL domains, the N-terminal amino acid stretch spanning cysteines 1 to 4 of JDP8-DBLβC2 is not necessary for binding to ICAM-1.
In conclusion, we have mapped receptor-binding residues of DBL domains from EBPs that bind the Duffy blood group antigen and sialic acids on glycophorin A to mediate red cell invasion, as well as DBL domains from PfEMP-1 that bind diverse cytoadherence receptors such as CR1 to mediate rosetting, a binding phenotype associated with severe malaria, and CSA, a cytoadherence phenotype implicated in placental malaria. In each case, the binding residues lie in the central regions of the DBL domains. The central regions of DBL domain F2, R29-DBLα, and FCR3-DBLγ retain binding function when expressed on their own suggesting that they constitute functional modules that can fold independently. The flanking amino- and carboxyl-terminal regions of these DBL domains are not essential for binding but may play a role in modulating the fine specificity or affinity of the binding interaction. A multidomain architecture with a central functional, receptor-binding module was previously proposed for DBL domains based on studies on the prototype DBL domains PvRII and PkαRII from PvDBP and PkDBP that bind the Duffy antigen.24 The studies presented here suggest that the model is valid for a wide range of DBL domains from EBPs as well as from PfEMP-1 with diverse binding specificity. The ICAM-1–binding DBLβC2 domain appears to be a variation on this theme in that amino acids from both the central region of DBLβ as well as the associated C2 region are required for binding to ICAM-1.
Identification of the regions within DBL domains that contain critical binding residues opens up the path toward understanding the structure-function bases for their interaction with host receptors. Efforts to determine the structures of DBL domains can now focus on these regions, which contain the receptor-binding sites. In addition, these regions may be targeted for site-directed mutagenesis to identify the contact residues that form the receptor-binding pocket. A clear understanding of the receptor-ligand interactions mediated by DBL domains may enable the development of novel therapeutic or prophylactic strategies to block these interactions and inhibit red cell invasion or reverse cytoadherence and protect against malaria.
Prepublished online as Blood First Edition Paper, September 2, 2004; DOI 10.1182/blood-2004-05-1722.
Supported by grants from the Wellcome Trust (A.S. and C.E.C.) and from the Howard Hughes Medical Institute (C.E.C.). A.S. and C.E.C. are Wellcome Trust International Senior Research Fellows. C.E.C. is a Howard Hughes International Research Scholar.A.M. was supported by a Fellowship from the Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica (SEIMC).
A.M., N.B., R.S., and S.S. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr B. K. L. Sim for providing a plasmid containing DNA encoding F2 region of EBA-175, Dr Alex Rowe for providing a plasmid containing DNA encoding DBLα region of R29var1, Dr Artur Scherf for providing a plasmid containing the gene for FCR3varCSA, Dr Roselyn Eisenberg and Dr Gary Cohen for providing plasmid pRE4 and monoclonal antibody DL6, Dr Karina Yazdanbaksh for providing CR-1 deficient erythrocytes, Dr Suman Dhar for providing 293T cells, and Dr Tarvinder Taneja for constructing DBLβC2 plasmids.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal