Comment on Landrette et al, page 2900
Plag1 and Plagl2, identified as Cbfb-MYH11 candidate cooperating genes in genetic screens using retrovirus, are now found to have increased expression in human AML and are individually capable of rapidly producing AML in mice in combination with Cbfb-MYH11.
Retroviruses can uncover cooperative steps in leukemia development when used in combination with genetically engineered mice that are predisposed to disease. Following infection of genetically engineered mice, insertional mutagenic events caused by the virus can result in activation or inactivation of important disease genes and accelerate leukemia. Although such studies to investigate lymphomagenesis began in the 1980s, it was not until the last few years that retroviruses were used to model acute myeloid leukemia (AML) in genetically modified mice.1-3 The first of these studies to identify genes that synergize with a fusion gene important to human AML was initiated in the laboratory of Paul Liu at the National Institutes of Health.4 The group was carrying out research on the CBFB-MYH11 gene, which joins CBFB with the smooth myosin heavy chain (MYH11) and is found at chromosome 16 inversion (inv(16)(p13;q22)).4 They were interested in determining mechanisms by which CBFB-MYH11 blocks differentiation and induces leukemia.4 Although Cbfb+/MYH11 mice lack definitive hematopoiesis and die at about embryonic day 12.5, knock-in chimeras expressing the gene product core binding factor β–smooth muscle myosin heavy chain (Cbfβ-SMMHC) survive and develop leukemia after additional mutations.5 By application of the retrovirus genetic screen approach, it was possible to identify genes that potentially synergize with Cbfb-MYH11 in the AML pathogenesis.3 Included in this group of genes were Plag1 and Plagl2 (see figure).FIG1
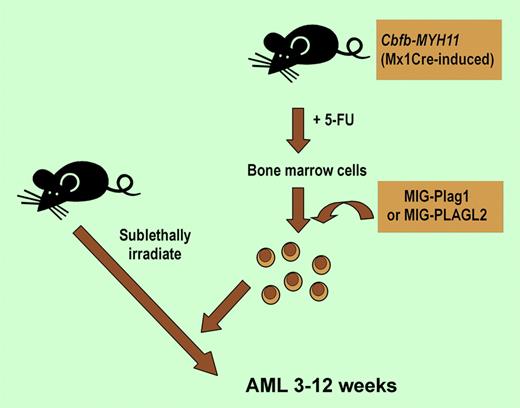
Mice with a Cbfb-MYH11 allele that was created using Cre-Lox technology. Following treatment with 5-fluorouracil for 6 days, bone marrow cells were harvested and infected with MIG retrovirus expressing Plag1 or PLAGL2 or MIG alone. Cells were transplanted into sublethally irradiated mice and observed for leukemia.
Mice with a Cbfb-MYH11 allele that was created using Cre-Lox technology. Following treatment with 5-fluorouracil for 6 days, bone marrow cells were harvested and infected with MIG retrovirus expressing Plag1 or PLAGL2 or MIG alone. Cells were transplanted into sublethally irradiated mice and observed for leukemia.
In this issue of Blood, Landrette and colleagues show that AML can be induced in 3 to 12 weeks with 100% penetrance by coexpressing either Plag1 or Plagl2, which were tagged in the retrovirus screen, and Cbfb-MYH11. The very short latency and full penetrance of the leukemia is indeed a striking finding and suggests very few genetic changes, perhaps as few as 2, are required to trigger leukemia. They further showed that Plag1 and PLAGL2 individually can increase proliferation of hematopoietic progenitors from Cbfb-MYH11–expressing bone marrow by inducing entry into S phase.
Evidence that the findings were very much relevant to human disease came when Landrette and colleagues, as reported here, discovered that the genes encoding these transcription factors are overexpressed in 20% of human AML. Incredibly, PLAGL2 is preferentially overexpressed in samples carrying inv(16). PLAG1 overexpression, on the other hand, did not correlate with a distinct cytogenetic subgroup. This Blood paper is the first to implicate PLAGL2 in any human cancer and the first to demonstrate that both Plag1 and PLAGL2 function to cause neoplastic disease in vivo. These investigations profoundly underscore the power of retroviruses as tools for exposing new cancer genes. ▪

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal