Abstract
Cell-mediated immunity is essential for control of human cytomegalovirus (HCMV) infection. We used a pool of 138 synthetic overlapping pentadecapeptides overspanning the entire pp65 protein to generate polyclonal CMV-specific T-cell lines from 12 CMV-seropositive donors inheriting different HLA genotypes. Autologous monocyte-derived dendritic cells (DCs) pulsed with this complete pool consistently induced highly specific T cells that selectively recognized 1-3 pentadecapeptides identified by secondary responses to a mapping grid of pentadecapeptide subpools with single overlaps. Responses against peptide-loaded targets sharing single HLA class I or II alleles identified the restricting HLAalleles. HLA-A*0201+ donors consistently responded to pentadecapeptides containing HLA-A*0201-binding epitopeaa495-503NLVPMVATV. T-cell lines from other donors contained high frequencies of CD4 and/or CD8 T cells selectively reactive against peptides presented by other HLA alleles, including both known epitopes such as aa341-350QYDPVAALF (HLA-A*2402) as well as unreported epitopes such as aa267-275HERNGFTVL (HLA-B*4001 and B*4002) and aa513-523FFWDANDIYRI (HLA-DRB1*1301). These T cells consistently lysed CMV-infected target cells. Thus, this approach fosters expansion and selection of HLA-restricted CMV-pp65–reactive T-cell lines of high specificity that also lyse CMV-infected targets, and from a functional and regulatory perspective, may have advantages for generating virus-specific T cells for adoptive immunotherapy.
Introduction
Human cytomegalovirus (HCMV) infections continue to be a major complication of allogeneic hematopoietic stem cell transplants (SCTs).1-3 Prophylactic or preemptive administration of ganciclovir (GCV) has reduced the incidence of early CMV disease significantly, but prolonged antiviral treatment may result in a delayed immune reconstitution, favoring the onset of CMV disease after day 100.1,3-5
Cell-mediated immunity plays an essential role in the control of CMV infection and recovery from CMV disease.6,7 Riddell et al8 have shown that protective immunity could be successfully transferred by the infusion of donor-derived CMV-specific CD8+ cytotoxic T-cell clones. However, the approaches used to generate these clones were logistically complex, limiting their broad application. Furthermore, in the absence of CD4+ T-helper cells, CMV-specific CD8+ T-cell clones declined rapidly.9 Several alternative approaches to generate T cells for adoptive immunotherapy to prevent or treat CMV infections have been proposed. Sun et al10 demonstrated in in vitro studies that Epstein-Barr virus (EBV)–transformed B lymphoblastic cell lines (EBV-BLCLs) transduced with retrovirus encoding the immunodominant CMV protein pp65 could be used as antigen-presenting cells (APCs) to sensitize donor T lymphocytes against EBV and CMV epitopes simultaneously. However, these approaches necessitate gene transfer and selection of autologous transduced antigen-presenting cells in each individual case prior to the initiation of CMV-specific T-cell cultures. Recently, Einsele et al11 and Peggs et al12 have reported the use of donor-derived CMV-specific T cells generated by sensitization with CMV lysates loaded on either donor blood peripheral mononuclear cells (PBMCs) or monocyte-derived cytokine-activated dendritic cells, respectively. The former approach yields predominantly CD4+ CMV-specific T cells,11 while the latter fosters CD8+ cytotoxic T-cell populations.12 Nevertheless, the initial clinical results with each approach have been encouraging. However, concerns have been raised by regulatory agencies regarding the possibility that lysates of CMV-infected cells might contain live viral particles that could be transferred to the host. CMV-pp65 peptide–specific T cells sensitized with dendritic cells loaded with single immunogenic nonamers binding HLA-A*020113 or isolated from the blood on the basis of their capacity to bind HLA peptide tetramers14 also are being evaluated. However, T cells specific for single CMV-pp65–derived peptides may be unduly restricted in their antiviral activity. Furthermore, these techniques select for CD8+ T cells, excluding the CD4+ helper cell support required for expansion and persistence.9
To address these concerns, we have explored the use of pools of overlapping pentadecapeptides spanning the sequence of the major protein of CMV-pp65 for sensitization and generation of CMV-specific T cells. Such pentadecapeptides previously have been used to identify immunogenic viral epitopes recognized by T cells in the blood of healthy individuals and allograft recipients.15,16 In this report, we demonstrate that this methodology also permits the rapid generation of polyclonal CMV-specific T-cell lines that recognize immunodominant epitopes of CMV-pp65 presented by different HLA class I and class II alleles and can lyse CMV-infected targets. We further demonstrate that such cytotoxic T-cell lines can be regularly generated from CMV-seropositive individuals with uncommon HLA alleles for which immunogenic epitopes are not already identified. T cells could therefore potentially be generated from donors of any HLA type capable of presenting pp65 peptides.
Materials and methods
Donors
Blood samples were obtained from 12 consenting CMV-seropositive healthy donors according to protocols approved by our institutional review board of Memorial Sloan-Kettering Cancer Center (New York, NY). Of these donors, 5 expressed HLA-A*0201, 2 coexpressed HLA-A*2402, and 7 expressed other HLA alleles as noted in specific experiments. HLA typing of the blood donors and cell lines used in the experiments were performed by the Histocompatibility Testing Laboratory at Memorial Sloan Kettering Cancer Center. Analysis of specific nucleotide sequences was done using standard high-resolution typing techniques.
Generation, culture, and peptide pulsing of antigen-presenting cells
DCs were generated from PBMCs obtained after Ficoll-Hypaque centrifugation. CD14+ monocytes were enriched by negative selection using the monocyte isolation kit and Miltenyi microbead columns (Miltenyi Biotec, Auburn, CA). CD14+ monocytes were resuspended at a concentration of 1 × 106/mL in serum-free medium, supplemented with granulocyte-macrophage colony-stimulating factor (GM-CSF) (1500 IU/mL; Immunex, Seattle, WA) and interleukin-4 (1200 IU/mL; Biosource, Camarillo, CA). On days 2, 4, and 6 of culture, fresh cytokines were added. Fresh medium was added depending on cell growth. On day 7 of culture, 10 ng/mL tumor necrosis factor-α (TNF-α) (Biosource, Camarillo, CA) was added for the maturation of the dendritic cells.
Autologous EBV-BLCLs and a panel of allogeneic EBV-BLCLs, used to identify restricting HLA alleles, were generated as previously described.17,18 K562 cells transfected to express the HLA-A*0201 allele (K562/A*0201 transfectants) also were evaluated as APCs.19 Cells were fed RPMI 1640, 10% fetal calf serum (FCS), l-glutamine, penicillin, and streptomycin 3 times a week and expanded according to the growth and cell number.
Peptide pulsing of APCs was performed as previously described.17 In brief, 2.5 μg of each nonamer peptide (Research Genetics, Huntsville, AL) and single pentadecapeptide or a total of 24 μg pooled pentadecapeptides (2 μg/pentadecapeptide of subpools and 0.18 μg/pentadecapeptide of complete pool) was added to 1 × 106 APCs/mL in Iscove modified Dulbecco medium (IMDM) for 3 hours at room temperature prior to irradiation. Each of the 138 pentadecapeptides was synthesized by Natural and Medical Sciences Institute (NMI) (Tuebingen, Germany) to specifications of validated sequence, purity, sterility, and absence of endotoxin.
Generation of CMV-specific T-cell lines
T-cell lines were generated as previously described.17,18 In brief, CD3+ T-cell–enriched cell fractions were isolated by Ficoll-Hypaque gradient separation of PBMCs, followed by depletion of CD20+ B cells, CD14+ monocytes, and CD56+ natural killer (NK) cells with monoclonal antibody–coated immunomagnetic beads (Miltenyi Biotec). Thereafter, aliquots of enriched T cells, at a concentration of 1 × 106 cells/mL, were stimulated with 0.5 × 105/mL 6000 rad irradiated DCs and pulsed with single pentadecapeptide or with peptide pools in IMDM supplemented with 10% heat-inactivated human AB serum (Gemini, Calabasas, CA) for 7 days in 25 cm2 flasks. T cells were restimulated weekly at an effector-stimulator ratio of 20:1 and recultured at a concentration of 1 × 106/mL; 5 to 10 IU/mL of interleukin-2 (Collaborative Biomedical Products, Bedford, MA) was added at day 10 to the cultures and 2 to 3 times weekly thereafter.
The principle of the protein-spanning pools of overlapping pentadecapeptides
To generate polyclonal CMV-specific T-cell lines, we used pools of pentadecapeptides with 11–amino acid overlaps spanning the 561 amino acid sequence of pp65 protein. (Figure 1A). A total of 138 pentadecapeptides, spanning the pp65 protein, were purchased from NMI.
The pp65 protein and the overlapping pentadecapeptides. (A) The sequence of the pp65 protein, consisting of 561 amino acids and the principle of 11 amino acids overlapping pentadecapeptides are illustrated. The sequence of the first 3 pentadecapeptides with 11aa overlap each is highlighted. A total of 138 pentadecapeptides are required for overspanning the entire pp65 protein. As a result of the 11aa overlap, HLA-A*0201-restricted peptide NLVPMVATV (aa 495-503) is contained within the pentadecapeptides 123 (AGILA RNLVP MVATV) and 124 (ARNLV PMVAT VQGQN). The HLA-A*2402-restricted peptide QYDPVAALF (aa 341-350) is represented by the pentadecapeptides 85 (VELRQ YDPVA ALFFF) and 86 (QYDPV AALFF FDIDL). (B) The principle of pp65-derived peptide pools of overlapping pentadecapeptides. A total of 12 pentadecapeptides are contained within one peptide-pool (eg, pentadecapeptides 3, 15, 27, 39, 51, 63, 75, 87, 99, 111, 123, and 135 forming peptide pool 3) requiring a total of 24 pools overspanning the pp65 protein. Pentadecapeptides 123 and 124, containing the A*0201-restricted nonamer sequence NLVP MVATV, are found in peptide pools 3, 4, and 23.
The pp65 protein and the overlapping pentadecapeptides. (A) The sequence of the pp65 protein, consisting of 561 amino acids and the principle of 11 amino acids overlapping pentadecapeptides are illustrated. The sequence of the first 3 pentadecapeptides with 11aa overlap each is highlighted. A total of 138 pentadecapeptides are required for overspanning the entire pp65 protein. As a result of the 11aa overlap, HLA-A*0201-restricted peptide NLVPMVATV (aa 495-503) is contained within the pentadecapeptides 123 (AGILA RNLVP MVATV) and 124 (ARNLV PMVAT VQGQN). The HLA-A*2402-restricted peptide QYDPVAALF (aa 341-350) is represented by the pentadecapeptides 85 (VELRQ YDPVA ALFFF) and 86 (QYDPV AALFF FDIDL). (B) The principle of pp65-derived peptide pools of overlapping pentadecapeptides. A total of 12 pentadecapeptides are contained within one peptide-pool (eg, pentadecapeptides 3, 15, 27, 39, 51, 63, 75, 87, 99, 111, 123, and 135 forming peptide pool 3) requiring a total of 24 pools overspanning the pp65 protein. Pentadecapeptides 123 and 124, containing the A*0201-restricted nonamer sequence NLVP MVATV, are found in peptide pools 3, 4, and 23.
To identify the sensitizing pentadecapeptide in the complete pool, an analytic grid composed of 24 subpools of CMV pentadecapeptides was constructed. (Figure 1B). Each subpool contains 12 pentadecapeptides (eg, pool 3 contains pentadecapeptides 3, 15, 27, 39, 51, 63, 75, 87, 99, 111, 123, and 135; pool 15 contains pentadecapeptides 25 through 36). To identify the immunogenic epitope, T cells sensitized with the complete pool were washed and aliquots restimulated with each of the 24 subpools. T cells accumulating interferon-γ (IFN-γ) were quantitated by fluorescence-activated cell-sorter scanner (FACS) as described in the next paragraph. Immunogenic peptides were identified by the responses of T cells to intercepting subpools in the grid. For example, T cells from an HLA-A*0201+-seropositive donor might be expected to respond to pentadecapeptides 123 (AGILARNLVPMVATV) and 124 (ARNLVPMVATQGQN), each of which contain NLVP MVATV, a known A*0201-restricted antigenic nonamer. These pentadecapeptides would be identified as immunogenic if the T cells responded to pools 3, 4, and 23, which share pentadecapeptides 123 and 124 that contain this nonamer peptide.
Quantitation of antigen-specific T cells by intracellular IFN-γ–production assay
The intracellular IFN-γ–production assay was performed as previously described.17 T cells at a concentration of 1 × 106/mL were mixed with peptide-loaded or unmodified PBMCs or BLCLs at an effector-stimulator cell ratio of 5:1. Control tubes containing effector cells, or APCs only, were incubated separately until the staining procedure. Brefeldin A (Sigma, St Louis, MO) was added to both nonstimulated and stimulated samples at a concentration of 10 μg/mL of cells. Tubes were incubated overnight for 16 hours in a humidified 5% CO2 incubator at 37°C. Staining and analyses were performed as previously described.17
Determination of peptide-specific CD8+ T cells by tetramer analyses
Major histocompatibility complex (MHC)–tetramer complexes were generated in the MSKCC Tetramer Core Facility and used as previously described.17,20,21 The T cells were initially incubated with 25 μg/mL phycoerythrin (PE)–labeled tetrameric complex on ice for 30 minutes, washed, and then stained with 10 μL of monoclonal anti-CD3 allophycocyanin, 10 μL of anti-CD8 peridin chlorophyll protein (PerCP), and 10 μLof anti-CD62L fluorescein isothiocyanate (FITC) (BD Bioscience, San Jose, CA) for 20 minutes on ice. The stained cells were subsequently analyzed and quantitated with a FACSCalibur flow cytometer with dual lasers for 4-color capability (BD Biosciences).
Antigen-specific cytotoxic activity of CMV-specific T-cell lines
T-cell lines generated by sensitization with the entire pp65 peptide pool, selected peptide pools, or single pentadecapeptide also were assessed for their capacity to lyse peptide-pulsed or CMV-infected target cells using a standard 51Cr cytotoxicity assay as previously described.17,18 Target cells included BLCLs, K562/A*0201 transfectants, HLA-A*0201+ MRC-5 fibroblasts, or donor-derived skin fibroblasts, which were either pulsed with the relevant peptide or peptide pools or infected with CMV viral supernatant derived from the AD-169 strain. Targets either uninfected or pulsed with irrelevant peptides were included as controls.
Virus and fibroblast cell lines
MRC-5 fibroblasts (CCL-171; American Type Culture Collection, Manassas, VA) and fibroblasts from donor-derived skin biopsies were propagated in modified minimum essential media-α (MEM-α) supplemented with nonessential amino acids (NEAA), 10% FCS, and antibiotics. AD-169 CMV strain (VR-538; American Type Culture Collection) was propagated in MRC-5 fibroblasts, and infected cultures were harvested when a cytopathic effect was evident. Cells were spun at 400g (1500 rpm) for 10 minutes, and aliquots of supernatant were stocked at –80°C until used. Infectivity of viral supernatant was measured by standard CMV antigen tests of infected fibroblasts.
Results
Pentadecapeptide-loaded autologous APCs can induce CMV-pp65 epitope-specific T-cell responses
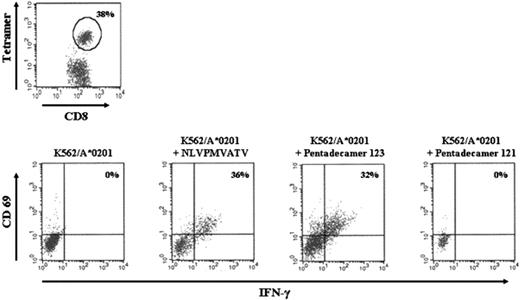
We sensitized T cells from HLA-A*0201+-seropositive donors with DCs pulsed with pentadecapeptide 123, containing the known immunogenic HLA-A*0201-binding epitope sequence NLVPMVATV. After 2 weeks, we analyzed the percentage of NLVPMVATV-specific T cells by MHC tetramer analyses and by intracellular IFN-γ production in response to 2°C stimulation with NLVPMVATV peptide or pentadecapeptides containing or not containing this specific amino acid sequence. For restimulations, we initially used HLA-A*0201-transfected K562 cells as APCs. In a representative experiment illustrated in Figure 2, 38% of CD8+ T cells were tetramer positive. In addition, 36% and 32% of the CD8+ T cells produced intracellular IFN-γ in response to restimulation with K562/A*0201 cells pulsed with NLVPMVATV peptide or pentadecapeptide 123, respectively. In contrast, nonpulsed K562/A*0201 cells or cells pulsed with pentadecapeptide 121, not containing the amino-acid sequence of the epitope, did not exhibit positive T-cell responses.
MHC tetramer analysis and intracellular IFN-γ analyses of pp65-derived CMV-specific T cells. CMV-specific T-cell lines of HLA-A*0201+ individuals were generated by stimulation of purified CD3+ cells with DCs pulsed with pentadecapeptide 123, containing the NLVPMVATV sequence. (A) Staining with HLA-A*0201–restricted MHC tetramer complexed with NLVPMVATV peptide. (B) Intracellular IFN-γ production after overnight stimulation with K562/A*0201 pulsed with NLVPMVATV peptide or the pentadecapeptides 123 or 121. The percentages of CD8+ CD69+ IFN-γ+ cells or CD8+ tetramer+ cells are shown in the right upper quadrant.
MHC tetramer analysis and intracellular IFN-γ analyses of pp65-derived CMV-specific T cells. CMV-specific T-cell lines of HLA-A*0201+ individuals were generated by stimulation of purified CD3+ cells with DCs pulsed with pentadecapeptide 123, containing the NLVPMVATV sequence. (A) Staining with HLA-A*0201–restricted MHC tetramer complexed with NLVPMVATV peptide. (B) Intracellular IFN-γ production after overnight stimulation with K562/A*0201 pulsed with NLVPMVATV peptide or the pentadecapeptides 123 or 121. The percentages of CD8+ CD69+ IFN-γ+ cells or CD8+ tetramer+ cells are shown in the right upper quadrant.
Peptide pools of pentadecapeptides permit the generation of CMV epitope-specific T-cell lines
We tested whether pools containing up to 12 pentadecapeptides, which included 1 pentadecapeptide containing a known immunogenic nonamer, could also be used to generate CMV-specific T cells and whether the responses to the pool would be comparable to responses to the single pentadecapeptide containing the immunogenic nonamer.
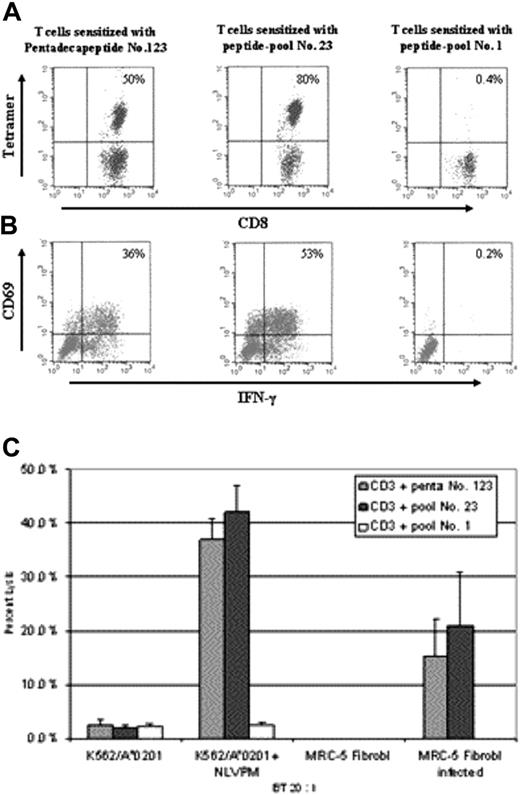
Three T-cell lines were generated from HLA-A*0201+ individuals using DCs pulsed with pentadecapeptide 123, peptide pool 23 containing pentadecapeptide 123, or peptide pool 1, which served as negative control. After 2 restimulations, aliquots of T cells from each line were tested. As seen in Figure 3A, 50% of CD8+ T cells sensitized with pentadecapeptide 123 and 80% of CD8+ T cells sensitized with peptide pool 23 bound NLVPMVATV-HLA-A*0201 tetramers. T cells sensitized with peptide pool 1 did not have tetramer-positive cells. Intracellular IFN-γ production in response to NLVPMVATV nonamer-pulsed K562/A*0201 target cells was detected in 36% and 53% of the CD8+ T cells sensitized with pentadecapeptide 123 or peptide pool 23, respectively. In contrast, no IFN-γ was noted in T cells sensitized with peptide pool 1 (Figure 3B). Furthermore, as demonstrated in Figure 3C, T cells generated by sensitization with pentadecapeptide 123 or with peptide pool 23 containing the epitope effectively lysed peptide-pulsed K562/A*0201 cells and CMV-infected MRC-5 fibroblasts. In contrast, nonpulsed or noninfected controls were not lysed. T cells sensitized with peptide pool 1 did not induce lysis of either the infected or the peptide-pulsed target cells, indicating the absence of CMV peptides immunogenic for this individual in this pool.
Comparative MHC tetramer staining, intracellular IFN-γ, and cytotoxicity analyses of CMV-specific T cells after sensitization with pentadecapeptides or peptide pools of pentadecapeptides. T lymphocytes of HLA-A*0201+ individuals were sensitized with pentadecapeptide 123 or peptide-pools 23 or 1. (A) T cells were analyzed for their capacity to bind MHC tetramer complexed with NLVPMVATV peptide. (B) Intracellular IFN-γ production by T lymphocytes was assessed in response to peptide-pulsed K562/A*0201 cells. 50% of the CD8+ cells of T lymphocytes sensitized with pentadecapeptide 123 bound MHC tetramer and 36% produced intracellular IFN-γ. 80% of the CD8+ cells of the T-cell line sensitized with the peptide pool 23 stained with the tetramer and 53% produced IFN-γ. T cells sensitized with pool 1 failed to stain with the MHC tetramer or to produce intracellular IFN-γ. Irrelevant MHC tetramers and target cells pulsed with irrelevant peptides did not produce results above background (data not shown). (C) T lymphocytes sensitized with either pentadecapeptide 123 (CD3 + penta 123), peptide pool 23 (CD3 + pool 23), or pool 1 (CD3 + pool 1) were analyzed for their capacity to induce lysis of peptide-pulsed K561/A*0201 cells or CMV-infected MRC-5 fibroblasts. Target cells were plated in triplicates. Results represent percentage of lysis ± standard deviation.
Comparative MHC tetramer staining, intracellular IFN-γ, and cytotoxicity analyses of CMV-specific T cells after sensitization with pentadecapeptides or peptide pools of pentadecapeptides. T lymphocytes of HLA-A*0201+ individuals were sensitized with pentadecapeptide 123 or peptide-pools 23 or 1. (A) T cells were analyzed for their capacity to bind MHC tetramer complexed with NLVPMVATV peptide. (B) Intracellular IFN-γ production by T lymphocytes was assessed in response to peptide-pulsed K562/A*0201 cells. 50% of the CD8+ cells of T lymphocytes sensitized with pentadecapeptide 123 bound MHC tetramer and 36% produced intracellular IFN-γ. 80% of the CD8+ cells of the T-cell line sensitized with the peptide pool 23 stained with the tetramer and 53% produced IFN-γ. T cells sensitized with pool 1 failed to stain with the MHC tetramer or to produce intracellular IFN-γ. Irrelevant MHC tetramers and target cells pulsed with irrelevant peptides did not produce results above background (data not shown). (C) T lymphocytes sensitized with either pentadecapeptide 123 (CD3 + penta 123), peptide pool 23 (CD3 + pool 23), or pool 1 (CD3 + pool 1) were analyzed for their capacity to induce lysis of peptide-pulsed K561/A*0201 cells or CMV-infected MRC-5 fibroblasts. Target cells were plated in triplicates. Results represent percentage of lysis ± standard deviation.
Sensitization with overlapping peptides spanning the pp65 protein selectively induces HLA-restricted T-cell lines specific for immunogenic epitopes of CMV-pp65
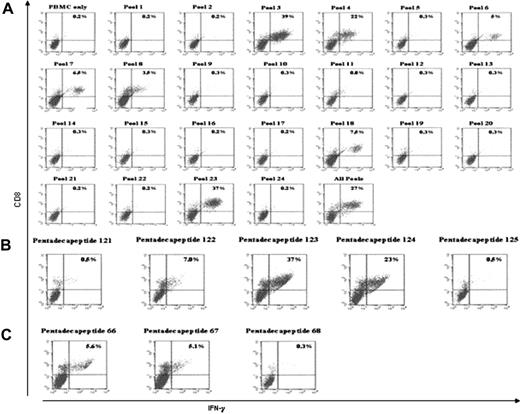
We subsequently sensitized T cells from 12 CMV-seropositive individuals with autologous DCs pulsed with a pool containing all 138 pentadecapeptides overspanning the entire pp65 protein. The T cells were restimulated on day 7 and 14 of culture. On day 18, these T cells were washed and restimulated with PBMCs pulsed with 1 of each of the 24 pentadecapeptide pools or the complete pool. In Table 1, we summarize these results. As can be seen, each donor had low but detectable frequencies of IFN-γ+ T cells at initiation of culture (0.15%-0.4%). By day 18, the proportion and absolute number of CMV-pp65–specific IFN-γ+ T cells had increased 60- to 110-fold. We subsequently evaluated each of these cultures for their responses to the analytic grid of pentadecapeptide subpools, identified the specific pentadecapeptide eliciting responses, and thereafter defined the immunogenic epitopes and their presenting HLA alleles. A representative experiment is shown in Figure 4A. When CD3+ T cells from this donor were analyzed, only pools 3, 4, 6, 7, 18, 23, and the complete pool stimulated T cells to produce IFN-γ production above background. The IFN-γ+ cells also were uniformly CD69+. The majority of pentadecapeptide pools (pools 1, 2, 5, 8-17, 19-22, 24) did not induce any T-cell response. Pools 3, 4, and 23, which contain the known HLA-A*0201-binding epitope NLVPMVATV, induced intracellular IFN-γ production in 22% to 39% of the CD3+ T cells. These T cells were CD8+. In subsequent analyses of responses to each of the single pentadecapeptides (121-132) contained within peptide pool 23, pentadecapeptides 123 and 124, each of which contains the NLVPMVATV epitope, induced 37% and 23% IFN-γ+ CD8+ cells, respectively (Figure 4B). Pentadecapeptide 122, containing 5 amino acids of the nonamer, induced 7% IFN-γ+ CD8+ T cells, but the remaining pentadecapeptides of this pool did not induce IFN-γ production above background. A similar pattern of response to subpools 3, 4, and 23 and to pentadecapeptides 123 and 124 was elicited from 5 HLA-A0201+ donors (Table 1).
T-cell lines from 12 donors showing donor HLA type, yield of pp65 peptide-specific IFNγ+ T cells, the pentadecapeptides eliciting responses, specific epitopes, and restricting HLA alleles
. | HLA type of donor . | . | . | . | . | T-cell response to CMV-pp65 peptide pool, total cell no. (% IFN+ T cells) . | . | . | Subsets on day 18, % . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. . | A . | B . | C . | DRB*1 . | DQB*1 . | Day 0 . | Day 18 . | Fold increase . | CD4 . | CD8 . | HLA restriction . | Immunogenic pentadecapeptide . | Sequence eliciting response . | ||||||
| 1 | 0201 | 3901 | 0702 | 0401 | 0302 | ND | ND | ND | ND | ND | A*0201 | 123, 124 | NLVPMVATV495-503 | ||||||
| 0301 | 1302 | 06BC | 07AD | 02MN | ND | ND | ND | ND | ND | NA | NA | NA | |||||||
| 2 | 0201 | 4402 | 05MN | 1101 | 03AF | 20 (0.18) | 49 (7.6) | 103.4 | 73 | 21 | A*0201 | 123, 124 | NLVPMVATV495-503 | ||||||
| 2402 | 3901 | 12NP | 1101 | 03AF | 20 (0.18) | 49 (7.6) | 103.4 | 73 | 21 | A*2402 | 85, 86 | QYDPVAALF341-349 | |||||||
| 3 | 0201 | 4002 | 0202 | 1101 | 03AF | 20 (0.15) | 79.5 (5.3) | 79.5 | 65 | 32 | A*0201 | 123, 124 | NLVPMVATV495-503 | ||||||
| 2402 | 27FFW | 0202 | 12AG | 03AF | 20 (0.15) | 79.5 (5.3) | 79.5 | 65 | 32 | A*2402 | 85, 86 | QYDPVAALF341-349 | |||||||
| 4 | 0201 | 40CCP | 0304 | 1302 | 06CUK | ND | ND | ND | ND | ND | A*0201 | 123, 124 | NLVPMVATV495-503 | ||||||
| 31AB | 1509 | 07ET | 0801 | 0402 | ND | ND | ND | ND | ND | NA | NA | NA | |||||||
| 5 | 0206 | 46AB | 1202 | 1501 | 0401 | 20 (0.20) | 48 (5.0) | 60 | 55 | 41 | A*0206 | 123, 124 | NLVPMVATV495-503 | ||||||
| 0207 | 1518 | 0801 | 0405 | 06WG | 20 (0.20) | 48 (5.0) | 60 | 55 | 41 | NA | NA | NA | |||||||
| 6 | 0201 | 1501 | 0300 | 1302 | 0302 | 20 (0.30) | 52 (7.94) | 68 | 66 | 31 | A*0201 | 123, 124 | NLVPMVATV495-503 | ||||||
| 0301 | 4001 | 0304 | 0401 | 0604 | 20 (0.30) | 52 (7.94) | 68 | 66 | 31 | B*4001 | 66, 67 | HERNGFTVL267-275 | |||||||
| 7 | 2402 | 27FFW | 0102 | 1501 | 06AAC | 20 (0.14) | 59 (4.52) | 95.2 | 55 | 41 | A*2402 | 85, 86 | QYDPVAALF341-349 | ||||||
| 68FPY | 27FFW | 0102 | 0402 | 06WG | 20 (0.14) | 59 (4.52) | 95.2 | 55 | 41 | NA | NA | NA | |||||||
| 8 | 6801 | 4002 | 04APA | 0804 | 03AF | 20 (0.15) | 62 (6.82) | 70.4 | 60 | 38 | B*4002 | 66, 67 | HERNGFTVL267-275 | ||||||
| 6601 | 5301 | 0305 | 1303 | 03AF | 20 (0.15) | 62 (6.82) | 70.4 | 60 | 38 | NA | NA | NA | |||||||
| 9 | 3002 | 4201 | 0202 | 0302 | 02MN | 20 (0.4) | 70 (5.9) | 51.6 | 55 | 42 | B*4201 | 66, 67 | RPHERNGFTVL265-275 | ||||||
| 3001 | 1503 | 17MN | 1101 | 0402 | 20 (0.4) | 70 (5.9) | 51.6 | 55 | 42 | DRB1*1101 | 91, 92 | EHPTFTSQYRI513-523 | |||||||
| 10 | 03AD | 0702 | 0702 | 0701 | 02XX | 20 (0.23) | 60 (4.8) | 62.6 | 65 | 32 | B*0702 | 66, 67 | RPHERNGFTV265-274 | ||||||
| 31AB | 0702 | 0702 | 1501 | 06XX | 20 (0.23) | 60 (4.8) | 62.6 | 65 | 32 | NA | NA | NA | |||||||
| 11 | 26JV | 44CU | 1601 | 1301 | 06AAC | 20 (0.2) | 56 (7.91) | 110 | 73 | 24 | DRB1*1301 | 128, 129 | FFWDANDIYRI513-523 | ||||||
| 29BD | 08DKG | 07AG | 1501 | 06WG | 20 (0.2) | 56 (7.91) | 110 | 73 | 24 | NA | NA | NA | |||||||
| 12 | 0101 | 4403 | 03XX | 0702 | 05XX | 20 (0.14) | 48 (4.7) | 75.2 | 75 | 22 | DQBI*05XX | 128, 129 | FFWDANDIYRI513-523 | ||||||
| 2301 | 5501 | 04XX | 1602 | 02XX | 20 (0.14) | 48 (4.7) | 75.2 | 75 | 22 | NA | NA | NA | |||||||
. | HLA type of donor . | . | . | . | . | T-cell response to CMV-pp65 peptide pool, total cell no. (% IFN+ T cells) . | . | . | Subsets on day 18, % . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. . | A . | B . | C . | DRB*1 . | DQB*1 . | Day 0 . | Day 18 . | Fold increase . | CD4 . | CD8 . | HLA restriction . | Immunogenic pentadecapeptide . | Sequence eliciting response . | ||||||
| 1 | 0201 | 3901 | 0702 | 0401 | 0302 | ND | ND | ND | ND | ND | A*0201 | 123, 124 | NLVPMVATV495-503 | ||||||
| 0301 | 1302 | 06BC | 07AD | 02MN | ND | ND | ND | ND | ND | NA | NA | NA | |||||||
| 2 | 0201 | 4402 | 05MN | 1101 | 03AF | 20 (0.18) | 49 (7.6) | 103.4 | 73 | 21 | A*0201 | 123, 124 | NLVPMVATV495-503 | ||||||
| 2402 | 3901 | 12NP | 1101 | 03AF | 20 (0.18) | 49 (7.6) | 103.4 | 73 | 21 | A*2402 | 85, 86 | QYDPVAALF341-349 | |||||||
| 3 | 0201 | 4002 | 0202 | 1101 | 03AF | 20 (0.15) | 79.5 (5.3) | 79.5 | 65 | 32 | A*0201 | 123, 124 | NLVPMVATV495-503 | ||||||
| 2402 | 27FFW | 0202 | 12AG | 03AF | 20 (0.15) | 79.5 (5.3) | 79.5 | 65 | 32 | A*2402 | 85, 86 | QYDPVAALF341-349 | |||||||
| 4 | 0201 | 40CCP | 0304 | 1302 | 06CUK | ND | ND | ND | ND | ND | A*0201 | 123, 124 | NLVPMVATV495-503 | ||||||
| 31AB | 1509 | 07ET | 0801 | 0402 | ND | ND | ND | ND | ND | NA | NA | NA | |||||||
| 5 | 0206 | 46AB | 1202 | 1501 | 0401 | 20 (0.20) | 48 (5.0) | 60 | 55 | 41 | A*0206 | 123, 124 | NLVPMVATV495-503 | ||||||
| 0207 | 1518 | 0801 | 0405 | 06WG | 20 (0.20) | 48 (5.0) | 60 | 55 | 41 | NA | NA | NA | |||||||
| 6 | 0201 | 1501 | 0300 | 1302 | 0302 | 20 (0.30) | 52 (7.94) | 68 | 66 | 31 | A*0201 | 123, 124 | NLVPMVATV495-503 | ||||||
| 0301 | 4001 | 0304 | 0401 | 0604 | 20 (0.30) | 52 (7.94) | 68 | 66 | 31 | B*4001 | 66, 67 | HERNGFTVL267-275 | |||||||
| 7 | 2402 | 27FFW | 0102 | 1501 | 06AAC | 20 (0.14) | 59 (4.52) | 95.2 | 55 | 41 | A*2402 | 85, 86 | QYDPVAALF341-349 | ||||||
| 68FPY | 27FFW | 0102 | 0402 | 06WG | 20 (0.14) | 59 (4.52) | 95.2 | 55 | 41 | NA | NA | NA | |||||||
| 8 | 6801 | 4002 | 04APA | 0804 | 03AF | 20 (0.15) | 62 (6.82) | 70.4 | 60 | 38 | B*4002 | 66, 67 | HERNGFTVL267-275 | ||||||
| 6601 | 5301 | 0305 | 1303 | 03AF | 20 (0.15) | 62 (6.82) | 70.4 | 60 | 38 | NA | NA | NA | |||||||
| 9 | 3002 | 4201 | 0202 | 0302 | 02MN | 20 (0.4) | 70 (5.9) | 51.6 | 55 | 42 | B*4201 | 66, 67 | RPHERNGFTVL265-275 | ||||||
| 3001 | 1503 | 17MN | 1101 | 0402 | 20 (0.4) | 70 (5.9) | 51.6 | 55 | 42 | DRB1*1101 | 91, 92 | EHPTFTSQYRI513-523 | |||||||
| 10 | 03AD | 0702 | 0702 | 0701 | 02XX | 20 (0.23) | 60 (4.8) | 62.6 | 65 | 32 | B*0702 | 66, 67 | RPHERNGFTV265-274 | ||||||
| 31AB | 0702 | 0702 | 1501 | 06XX | 20 (0.23) | 60 (4.8) | 62.6 | 65 | 32 | NA | NA | NA | |||||||
| 11 | 26JV | 44CU | 1601 | 1301 | 06AAC | 20 (0.2) | 56 (7.91) | 110 | 73 | 24 | DRB1*1301 | 128, 129 | FFWDANDIYRI513-523 | ||||||
| 29BD | 08DKG | 07AG | 1501 | 06WG | 20 (0.2) | 56 (7.91) | 110 | 73 | 24 | NA | NA | NA | |||||||
| 12 | 0101 | 4403 | 03XX | 0702 | 05XX | 20 (0.14) | 48 (4.7) | 75.2 | 75 | 22 | DQBI*05XX | 128, 129 | FFWDANDIYRI513-523 | ||||||
| 2301 | 5501 | 04XX | 1602 | 02XX | 20 (0.14) | 48 (4.7) | 75.2 | 75 | 22 | NA | NA | NA | |||||||
ND indicates not determined; NA, not applicable.
Analyses of CMV-specific T-cell line of an HLA-A*0201+healthy seropositive donor after sensitization with the complete pool of pentadecapeptides overspanning the entire pp65 protein. (A) Capacity of T cells to produce intracellular IFN-γ in response to PBMCs pulsed with 1 of each of the 24 pentadecapeptide pools was analyzed. Percentages of CD8+ CD69+ IFN-γ+ cells are shown in the right upper quadrant. Nonpulsed PBMCs and the majority of pentadecapeptide pools (pool 1, 2, 5, 9-17, 19-22, 24) did not induce intracellular IFN-γ production above background. In contrast, pools 3, 4, and 23, containing the known HLA-A*0201 binding epitope NLVPMVATV, induced intracellular IFN-γ production of 22% to 39% of CD8+ cells. In addition, pools 6, 7, and 18 induced intracellular IFN-γ production in 5% to 7.5% of CD8+ cells. (B) IFN-γ production in response to single pentadecapeptides (121-132) of peptide-pool 23. Pentadecapeptides 123 and 124, containing the full length of the NLVPMVATV epitope, induced intracellular IFN-γ production in 37% and 23% of the CD8+ cells, respectively. Pentadecapeptide 122, containing 5 amino acids of the nonamer, induced IFN-γ production by 7% of CD8+ cells. IFN-γ response was negative for the remaining pentadecapeptides of this pool. (C) Intracellular IFN-γ production in response to single pentadecapeptides (61-72) of peptide pool 18. Pentadecapeptides 66 and 67 induced intracellular IFN-γ production by 5.6% and 5.1% of CD8+ cells, respectively. The remaining pentadecapeptides did not induce IFN-γ production above background.
Analyses of CMV-specific T-cell line of an HLA-A*0201+healthy seropositive donor after sensitization with the complete pool of pentadecapeptides overspanning the entire pp65 protein. (A) Capacity of T cells to produce intracellular IFN-γ in response to PBMCs pulsed with 1 of each of the 24 pentadecapeptide pools was analyzed. Percentages of CD8+ CD69+ IFN-γ+ cells are shown in the right upper quadrant. Nonpulsed PBMCs and the majority of pentadecapeptide pools (pool 1, 2, 5, 9-17, 19-22, 24) did not induce intracellular IFN-γ production above background. In contrast, pools 3, 4, and 23, containing the known HLA-A*0201 binding epitope NLVPMVATV, induced intracellular IFN-γ production of 22% to 39% of CD8+ cells. In addition, pools 6, 7, and 18 induced intracellular IFN-γ production in 5% to 7.5% of CD8+ cells. (B) IFN-γ production in response to single pentadecapeptides (121-132) of peptide-pool 23. Pentadecapeptides 123 and 124, containing the full length of the NLVPMVATV epitope, induced intracellular IFN-γ production in 37% and 23% of the CD8+ cells, respectively. Pentadecapeptide 122, containing 5 amino acids of the nonamer, induced IFN-γ production by 7% of CD8+ cells. IFN-γ response was negative for the remaining pentadecapeptides of this pool. (C) Intracellular IFN-γ production in response to single pentadecapeptides (61-72) of peptide pool 18. Pentadecapeptides 66 and 67 induced intracellular IFN-γ production by 5.6% and 5.1% of CD8+ cells, respectively. The remaining pentadecapeptides did not induce IFN-γ production above background.
In addition to the intense responses to pools 3, 4, and 23, 5% to 7.5% IFN-γ+ CD8+ cells were detected in T cells from the donor depicted in Figure 4A when restimulated with PBMCs pulsed with pools 6, 7, and 18. From the analytical grid illustrated in Figure 1B, it was noted that pentadecapeptides 66 and 67 are shared by these pools. When these T cells were analyzed after secondary stimulation with individual pentadecapeptides 61 through 72 (pool 18), 5.6% and 5.1% of CD8+ T cells produced IFN-γ in response to pentadecapeptide 66 and 67, respectively. The remaining pentadecapeptides of pool 18 did not induce IFN-γ response above background (Figure 4C). EBV-BLCL sharing single HLA alleles with this donor (HLA-type A*0201, A*0301, B*4001, B*1501) were pulsed with pentadecapeptide 67, and T-cell responses were evaluated that identified HLA-B*4001 as the presenting allele. The pattern of response further implicates the peptide HERNG FTVL, which is shared by pentadecapeptides 66 and 67 and can be presented by HLA-B*4001 as the sensitizing antigen as further illustrated in Figure 6D.
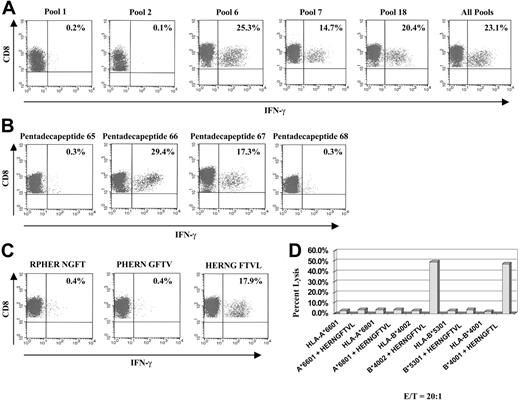
Determination of sequence and HLA restriction of a pp65-derived epitope in a seropositive individual with uncommon HLA type after sensitization of T cells with the complete pool. T cells of an individual with the HLA-type A*6801, A*6601 and B*4002, B*5301 were generated as described. (A) Intracellular IFN-γ production in response to PBMCs pulsed with pools 6, 7, 18 and complete pool was significant. Pools 1 and 2, representative for all other pentadecapeptide pools, were negative for intracellular IFN-γ production. (B) Only pentadecapeptides 66 and 67 of pool 18 stimulated T cells effectively. IFN-γ response of neighboring pentadecapeptides 65 and 68, as well as all other pentadepeptides contained within the pool 18 (not shown), was negative. (C) Based on the overlapping principle, the amino acid sequence RPHER NGFTVL, represented within pentadecapeptide 66 and 67, likely contains the candidate nonameric peptide being recognized by the T cells. Three candidate nonamers were synthesized and tested. Only the peptide HERNG FTVL induced a significant T-cell response. (D) The HLA restriction of this nonamer was tested by pulsing BLCLs, each expressing one of the individual's HLA allele only. BLCLs encoding the HLA-B*4002 allele were effectively lysed after pulsing with the HERNG FTVL peptide, whereas BLCLs expressing other alleles, pulsed with the peptide, did not induce cytotoxic T-cell responses. T cells of an individual with HLA-type A*0201, A*0301 and B*4001, B*1501 demonstrated a response to HLA-A*0201-restricted NLVPMVATV peptide and a response to the pentadecapeptides 66 and 67 of the pool 18. (D) B*4001-expressing BLCLs of this individual pulsed with HERNG FTVL peptide also were effectively lysed by T cells of the B*4002-expressing individual above.
Determination of sequence and HLA restriction of a pp65-derived epitope in a seropositive individual with uncommon HLA type after sensitization of T cells with the complete pool. T cells of an individual with the HLA-type A*6801, A*6601 and B*4002, B*5301 were generated as described. (A) Intracellular IFN-γ production in response to PBMCs pulsed with pools 6, 7, 18 and complete pool was significant. Pools 1 and 2, representative for all other pentadecapeptide pools, were negative for intracellular IFN-γ production. (B) Only pentadecapeptides 66 and 67 of pool 18 stimulated T cells effectively. IFN-γ response of neighboring pentadecapeptides 65 and 68, as well as all other pentadepeptides contained within the pool 18 (not shown), was negative. (C) Based on the overlapping principle, the amino acid sequence RPHER NGFTVL, represented within pentadecapeptide 66 and 67, likely contains the candidate nonameric peptide being recognized by the T cells. Three candidate nonamers were synthesized and tested. Only the peptide HERNG FTVL induced a significant T-cell response. (D) The HLA restriction of this nonamer was tested by pulsing BLCLs, each expressing one of the individual's HLA allele only. BLCLs encoding the HLA-B*4002 allele were effectively lysed after pulsing with the HERNG FTVL peptide, whereas BLCLs expressing other alleles, pulsed with the peptide, did not induce cytotoxic T-cell responses. T cells of an individual with HLA-type A*0201, A*0301 and B*4001, B*1501 demonstrated a response to HLA-A*0201-restricted NLVPMVATV peptide and a response to the pentadecapeptides 66 and 67 of the pool 18. (D) B*4001-expressing BLCLs of this individual pulsed with HERNG FTVL peptide also were effectively lysed by T cells of the B*4002-expressing individual above.
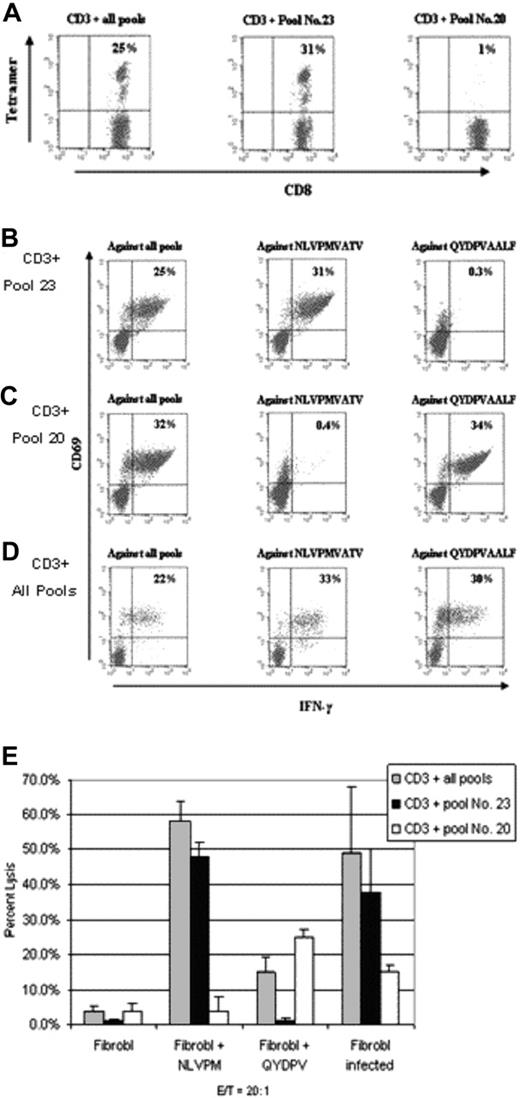
We also generated T cells from 2 seropositive HLA-A*0201- and HLA-A*2402-expressing individuals so as to determine whether the use of the complete pool of pentadecapeptides would induce T cells recognizing both the epitope presented by HLA-A*2402 QYDPVAALF22 as well as the NLVPMVATV epitope presented by HLA-A*0201. In these experiments, separate T-cell lines from the same donor also were generated simultaneously by stimulation with DCs pulsed with either peptide pool 23, which includes peptides with the epitope sequence NLVPMVATV, or peptide pool 20, which includes peptides containing the sequence QYDPVAALF. After 3 weekly stimulations intracellular IFN-γ production and tetramer binding by these T cells were tested. Results were similar for both donors. As shown in Figure 5A, 25%, 31%, and 1% of CD8+ T cells from one of these donors sensitized with the complete pool, pool 23 and pool 20, respectively, bound to the NLVPMVATV-HLA-A*0201 tetramers. Of the CD8+ T cells sensitized with pool 23, 34% produced IFN-γ after secondary stimulation with PBMCs loaded with NLVPMVATV peptide. PBMCs loaded with the QYDPVAALF peptide did not induce any response (0.3%). Conversely, 34% of the CD8+ T cells sensitized with pool 20 produced IFN-γ in response to PBMCs loaded with peptide QYDPVAALF but had no response to NLVPMVATV peptide (0.4%). In contrast, of the T cells sensitized with the complete pool, 22% produced IFN-γ in response to PBMCs loaded with the complete pool, 33% in response to PBMCs loaded with NVLPMVATV, and 30% to PBMCs loaded with QYDPVAALF (Figure 5B-D).
Analyses of T cells of an HLA-A*0201+- and A*2402+-expressing seropositive donor after sensitization with peptide pools of pentadecapeptides or with the complete pool. T-cell cultures were generated simultaneously by stimulation with either peptide pool 23 (CD3 + pool 23), containing HLA-A*0201–restricted peptide NLVPMVATV, or with peptide pool 20 (CD3 + pool 20), containing HLA-A*2402–restricted peptide QYDPVAALF. A third T-cell line was generated with DCs pulsed with the complete pool (CD3 + all pools). (A) Staining with A*0201-restricted MHC tetramers, complexed with NLVPMVATV peptide. (B-D) Intracellular IFN-γ responses to A*0201-restricted NLVPMVATV, A*2402-restricted QYDPVAALF, or to the complete pool. (E) T cells sensitized with the complete pool effectively lysed targets pulsed with either NLVPMVATV or QYDPVAALF peptide. T cells sensitized with peptide pool 23 or peptide pool 20 selectively lysed the targets after pulsing with the corresponding A*0201 or A*2402-restricted peptide. All 3 T-cell lines recognized autologous CMV-infected fibroblasts but failed to lyse nonpulsed, noninfected target cells. All target cells were plated in triplicates. Results represent percentage of lysis ± standard deviation.
Analyses of T cells of an HLA-A*0201+- and A*2402+-expressing seropositive donor after sensitization with peptide pools of pentadecapeptides or with the complete pool. T-cell cultures were generated simultaneously by stimulation with either peptide pool 23 (CD3 + pool 23), containing HLA-A*0201–restricted peptide NLVPMVATV, or with peptide pool 20 (CD3 + pool 20), containing HLA-A*2402–restricted peptide QYDPVAALF. A third T-cell line was generated with DCs pulsed with the complete pool (CD3 + all pools). (A) Staining with A*0201-restricted MHC tetramers, complexed with NLVPMVATV peptide. (B-D) Intracellular IFN-γ responses to A*0201-restricted NLVPMVATV, A*2402-restricted QYDPVAALF, or to the complete pool. (E) T cells sensitized with the complete pool effectively lysed targets pulsed with either NLVPMVATV or QYDPVAALF peptide. T cells sensitized with peptide pool 23 or peptide pool 20 selectively lysed the targets after pulsing with the corresponding A*0201 or A*2402-restricted peptide. All 3 T-cell lines recognized autologous CMV-infected fibroblasts but failed to lyse nonpulsed, noninfected target cells. All target cells were plated in triplicates. Results represent percentage of lysis ± standard deviation.
As illustrated in Figure 5E, T cells from pool 23 and pool 20 lines selectively lysed only the targets pulsed with the appropriate A*0201- or A*2402-restricted peptide, respectively. In contrast, T cells sensitized with the complete pool effectively lysed autologous targets pulsed with either NLVPMVATV or QYDPVAALF peptide. In addition, all 3 T-cell lines lysed CMV-infected autologous fibroblasts, whereas nonpulsed and noninfected targets were not recognized. Thus, T cells specific for immunogenic epitopes presented by both HLA-A*0201 and HLA-A*2402 could be generated within a single culture by stimulating donor T cells with the complete pool.
Peptide pools overspanning the pp65 protein permit generation of CMV-specific T-cell lines in individuals with uncommon HLA alleles and are useful for determination of sequence and HLA restriction of previously undescribed class I epitopes
T-cell lines from an individual with HLA-type A*6801/A*6601 and B*4002/B*5301 were generated by pulsing DCs with the complete pool. After 3 weeks of culture, CD8+ T cells producing intracellular IFN-γ were quantitated after restimulation with PBMCs pulsed with individual pentadecapeptides pools 1 to 24 or with the complete pool. Only peptide pools 6, 7, 18, and the complete pool induced significant frequencies of IFN-γ–producing T cells. T cells sensitized with the other pools were negative for IFN-γ production (Figure 6A). When the single pentadecapeptides from pool 18 were tested for their capacity to induce intracellular IFN-γ production, only pentadecapeptides 66 and 67, which also are contained in pools 6 and 7, induced IFN-γ+ CD8+ T cells. The neighboring pentadecapeptides 65 and 68, as well as all other pentadecapeptides contained within pool 18 (not shown), did not induce IFN-γ+ T cells (Figure 6B).
The amino acid sequence RPHER NGFTVL is represented within both pentadecapeptides 66 and 67. We therefore hypothesized that the nonameric peptide being recognized by the T cells of this individual was contained within this sequence. Three candidate nonamers were generated, and their capacity to induce intracellular IFN-γ was assessed. Only the nonameric peptide HERNG FTVL induced a significant response (Figure 6C). The HLA allele presenting this nonamer was analyzed by loading the peptide on a panel of EBV-BLCLs, each expressing only one of the individual's HLA alleles. As illustrated in Figure 6D, BLCLs encoding the HLA-B*4002 allele were effectively lysed after pulsing with the HERNG FTVL peptide, whereas peptide-pulsed BLCLs expressing other alleles did not induce any cytotoxic T-cell responses. Strikingly, these T cells also effectively lysed HERNG FTVL peptide-pulsed BLCLs from a donor with HLA type A*0201/A*0301 and B*4001/B*1501, strongly suggesting that the peptide can be recognized when presented by either the HLA-B*4002 or the HLA-B*4001 micro variant allele (see “Note added in proof”).
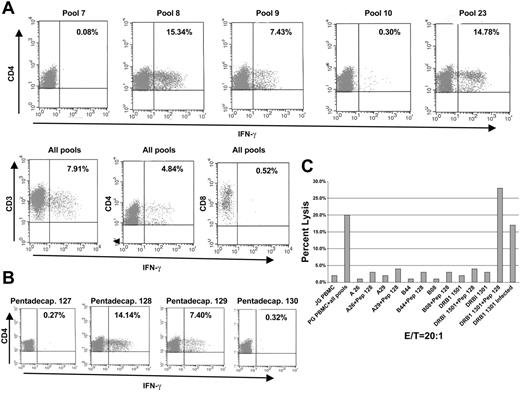
HLA class II restriction pattern and peptide sequence of an epitope inducing CMVpp65-specific CD4+ T cells determined using the complete pool
T cells of an individual with the HLA-type A*26JV, A*29BD, and B*44CU, B*08DKG, DRB1*-1301, and DRB1*-1501 were generated by pulsing autologous DCs with the complete pool. After 3 weeks of culture these cells were tested for their capacity to produce IFN-γ in response to secondary stimulation with PBMCs pulsed with individual pools 1 to 24 and to the complete pool. As shown in Figure 7A there was a predominant CD4+ T-cell response to pools 8, 9, 23 and to the complete pool. There was some CD8+ T-cell response noted to the same pools. All the other pools did not induce IFN-γ production. When the individual pentadecapeptides in pool 23 were tested, only pentadecapeptides 128 and 129 induced a significant response. Based on the overlapping principle, the amino acid sequence F FWDAN DIYRI represented within pentadecapeptide 128 and 129, likely contains the candidate peptide being recognized by the T cells of this individual (Figure 7B). We analyzed the HLA restriction of this pentadecapeptide by pulsing EBV-BLCLs each expressing one of the individual's HLA allele only. As illustrated in Figure 7C, only BLCL encoding the HLA-DRB1*-1301 allele were lysed after pulsing with the KYQEF FWDAN DIRYI peptide or after infection with CMV virus, whereas the EBV-BLCLs expressing the other alleles pulsed with the same peptide did not induce any cytotoxic T-cell responses.
HLA class II restriction pattern and peptide sequence of a pp65-derived epitope in a seropositive individual with uncommon HLA type after generation of CMV-specific T cells with complete pool. T cells of an individual with HLA-type A*26JV, A*29BD and B*44CU, B*O8DKG, DRB1*1301, DRB1*1501 were generated as previously described. (A) PBMCs pulsed with pentadecapeptide pools 8, 9, 23 and complete pool induced significant intracellular IFN-γ production of CD4+ cells in this individual. All other pentadecapeptide pools failed to induce a response. (B) Only pentadecapeptides 128 and 129, encoded in pool 23, stimulated T cells effectively. The neighboring pentadecapeptide pools 127 and 130, as well as all other pentadepeptides contained within pool 23 (not shown), did not induce intracellular IFN-γ production. Based on the overlapping principle, the amino acid sequence F FWDAN DIYRI represented within pentadecapeptide 128 and 129 likely contains the candidate peptide being recognized by the T cells of this individual. (C) HLA restriction of this pentadecapeptide was analyzed by pulsing EBV-BLCLs, each expressing one of the individual's HLA allele only. BLCLs encoding the HLA-DRB1*1301 allele were lysed after pulsing with the KYQEF FWDAN DIYRI peptide or after infection with CMV virus, whereas the EBV-BLCLs expressing the other alleles pulsed with the same peptide did not induce any cytotoxic T-cell responses.
HLA class II restriction pattern and peptide sequence of a pp65-derived epitope in a seropositive individual with uncommon HLA type after generation of CMV-specific T cells with complete pool. T cells of an individual with HLA-type A*26JV, A*29BD and B*44CU, B*O8DKG, DRB1*1301, DRB1*1501 were generated as previously described. (A) PBMCs pulsed with pentadecapeptide pools 8, 9, 23 and complete pool induced significant intracellular IFN-γ production of CD4+ cells in this individual. All other pentadecapeptide pools failed to induce a response. (B) Only pentadecapeptides 128 and 129, encoded in pool 23, stimulated T cells effectively. The neighboring pentadecapeptide pools 127 and 130, as well as all other pentadepeptides contained within pool 23 (not shown), did not induce intracellular IFN-γ production. Based on the overlapping principle, the amino acid sequence F FWDAN DIYRI represented within pentadecapeptide 128 and 129 likely contains the candidate peptide being recognized by the T cells of this individual. (C) HLA restriction of this pentadecapeptide was analyzed by pulsing EBV-BLCLs, each expressing one of the individual's HLA allele only. BLCLs encoding the HLA-DRB1*1301 allele were lysed after pulsing with the KYQEF FWDAN DIYRI peptide or after infection with CMV virus, whereas the EBV-BLCLs expressing the other alleles pulsed with the same peptide did not induce any cytotoxic T-cell responses.
Discussion
In the last 5 to 10 years, results from several phase 1 and small phase 2 trials have provided evidence that adoptive transfer of virus-specific T cells can prevent and effectively treat chemotherapy refractory monoclonal EBV lymphomas and prevent the development of clinically significant CMV infections.8,9,11,23 However, the techniques used to sensitize virus-specific T cells in vitro have entailed culture of the T cells with virus-infected autologous cells, such as CMV-virus–infected skin fibroblasts,9 EBV-transformed B cells,10 or DCs transduced with retroviral or adenoviral vectors to express specific viral proteins or immunogenic peptides.24 Such approaches have proved to be both time- and labor-intensive and difficult to implement or sustain. Alternative sensitization strategies using DCs pulsed with lysates of CMV-infected cells have shown promise, but such lysates are difficult to standardize and have raised regulatory concerns that they might contain and transmit live virus.12
Recently, Einsele et al13 have initiated trials of T cells sensitized with DCs loaded with a limited number of immunogenic nonapeptides of CMV-pp65 selected on the basis of computerized estimates of their capacity to bind to and be presented by common HLA class I allele such as HLA-A*0201 inherited by both the donor and transplant recipient. However, a proportion of patients, such as individuals co-inheriting HLA-B*0701 and HLA-A*0201, may preferentially respond to epitope presented by an allele such as HLA-B*0701 and not respond to an immunogenic peptides presented by HLA-A0201.25 Also, current strategies for selecting potentially immunogenic epitopes based on predicted binding affinities26,27 are useful for only a limited number of HLA alleles. Furthermore, as demonstrated by Elkington et al,28 even for those peptides of CMV-pp65 predicted to bind to common HLA alleles, only 40% elicited cytokine-producing T cells detected by enzyme-linked immunospot (ELISPOT) assay, and only a subset of the T-cell lines generated from HLA-A*0201+-seropositive donors in response to these peptides actually lysed CMV-infected cells.
To address several of these limitations, we have explored the use of pools of overlapping pentadecapeptides spanning the entire sequence of CMV-pp65 for the generation of T cells for adoptive cell therapy. These peptides are synthetic, sterile, and can be manufactured under good manufacturing practice (GMP) conditions. In prior studies, several investigators have used pools of overlapping 11-20 mers to measure antigen-specific T-cell responses in the blood and to map viral epitopes recognized by T cells in vivo or in vitro following sensitization with virus-infected autologous fibroblasts or antigen-presenting dendritic cells and other cell lines transduced to express a specific viral protein.29,30 In particular, Kern et al16 analyzed frequencies of CMV-specific CD8+ T cells in CMV-seropositive healthy individuals by quantitating T cells producing IFN-γ in isolated PBMCs stimulated for 6 hours with overlapping pentadecapeptides spanning the entire HCMV-pp65 protein. In their studies of randomly selected CMV-seropositive donors, 83% had CD8+, and 63% had detectable CD4+ T-cell responses. IFN-γ+ cells detected ranged from 0.03% to 2.7%. In a subset of these donors, T cells responding to the complete pool were subsequently stimulated with a set of subpools of the pentadecapeptides arranged in the matrix design described in Figure 1B. By analysis of responses to intercepting subpools, specific pentadecapeptides containing immunogenic epitopes could be identified and the epitopes subsequently defined by testing responses to individual nonamer sequences contained in that pentadecapeptide.
In the present study, we tested whether repeated sensitization of T cells from seropositive donors over 14 to 28 days in vitro with autologous DCs loaded with the complete pool of overlapping pentadecapeptides spanning the sequence of CMV-pp65 could induce the generation of large populations of CMV-pp65–reactive CD8+ and CD4+ T cells of sufficient specificity and activity against CMV-infected targets to be considered for adoptive cell therapy.
T-cell lines containing high frequencies of CMV-pp65–reactive IFN-γ+ T cells were generated in response to the complete pool of pentadecapeptides from each of the 12 CMV-seropositive donors tested, including individuals inheriting uncommon HLA alleles. Initially, our principal concern was the possibility that T cells, after extended culture in vitro, would react to many peptides in the pool, including peptides not normally presented by CMV-infected cells or peptides not specific to CMV, which might induce nonspecific background in vitro or unwanted tissue cross-reactions in vivo. In each case, however, significant populations of IFN-γ+ CD3+ T cells were generated in response to only a limited number of pentadecapeptides in the complete pool, which could be easily identified by analysis of responses to the grid of overlapping pentadecapeptide subpools (Figure 4). Responses to other pentadecapeptide subpools or individual pentadecapeptides were similar to background responses. Further analysis of responses to individual peptides contained in the pentadecapeptide subpools demonstrated that the CD8+ T cells generated from each of the 12 patients studied were responding to, at most, 1 to 2 epitopes, each presented by a different class I HLA allele. Similarly, CD4+ IFN-γ+ T-cell responses thus far studied have been principally directed against one pentadecapeptide presented by a single HLA class II allele. These studies thus suggest that T-cell lines generated in response to the complete pool of pentadecapeptides spanning the sequence of CMVpp65 selectively react against a very limited number of immunodominant epitopes. The limited spectrum of reactivity exhibited by these lines likely reflects the repertoire already selected in the CMV-seropositive donor rather than an artifact of in vitro selection, since studies by Kern et al16 and others who have used overlapping peptides to test for CMV-specific T cells in the blood of such donors also have documented immediate CD8+ and CD4+ IFN-γ+ T-cell responses at frequencies of less than 1%, specific for at most 2 to 4 class I binding and 1 to 2 class II binding peptide epitopes in any given donor.
The specific epitopes presented by a given HLA allele that elicit a response appear to be also highly selected. For example, when T cells from 5 donors expressing the common HLA-A*0201 allele were sensitized with the complete pool, the T-cell lines from each donor responded to pentadecapeptides 123 and 124 containing the immunogenic peptide NLVPMVATV, known to bind to and be presented by this allele (Figure 4A-B). These T cells specifically lysed both HLA-A*0201+ targets loaded with the NLVPMATV peptide as well as CMV-infected HLA-A*0201+ cell targets (Figure 5E). None of these T-cell lines responded to other pentadecapeptides containing peptides predicted to bind to HLA-A*0201 and reported to elicit T-cell responses detectable by ELISPOT analyses when used individually to sensitize T cells in vitro such as aa14-22VLGPISGHV (pentadecapeptide 3, pool 13), aa522-530RIFAELEGV (pentadecapeptide 130, pool 10), or aa20-128MLNIPSINV (pentadecapeptide 30, pool 15).28 It is possible that pentadecapeptides containing these potentially immunogenic HLA-A*0201-binding sequences cannot be adequately or appropriately digested by the ectopeptidases in the cell cultures, which have been shown to trim off amino acid sequences from the pentadecapeptides not fitting in the groove of the presenting HLA allele.31,32 Experiments to address this possibility are in progress. However, it also is possible that these other sequences of CMVpp65 are not efficiently processed and presented by CMV-infected cells and therefore do not elicit a major cytotoxic T-cell response. In this regard, it is of interest that of 6 HLA-A0201-binding nonamer epitopes of CMVpp65 shown by Elkington et al28 to induce significant frequencies of IFN-γ+ T cells in ELISPOT assays, only the aa495-503NLVPMVATV peptide (contained in pentadecapeptides 123 and 124) and the 5 amino acid–overlapping sequence aa491-499ILARNLVPM (contained in pentadecapeptide 123) elicited T cells capable of lysing CMV-infected targets.
We also analyzed the specificity and function of specific pentadecapeptides presented by other HLA class I alleles, including HLA-A*2402, HLA-B*4001, HLA-B*4002, HLA-B*4201, and HLA-B*0701. In each instance, the pentadecapeptide-eliciting responses could be rapidly ascertained by analysis of responses to the grid of pentadecapeptide subpools. In each instance, we also were able to identify a specific nonamer contained in the pentadecapeptide as the sensitizing epitope, including an unreported epitope presented by HLA-B*4001 and HLA-B*4002 as very recently described by Kondo et al33 (see “Note added in proof”). Importantly, these T cells only lysed targets loaded with the specific pentadecapeptide (or nonamer therein) that also expressed the restricting HLA allele. Furthermore, each of these T-cell lines also specifically lysed CMV-infected autologous fibroblasts. Thus, these studies further support the possibility that T-cell lines generated by this approach select for T cells reactive against epitopes that can be processed and presented by CMV-infected autologous targets. The potential for selection of functional T cells responsive to immunodominant epitopes from seropositive donors of diverse HLA genotypes provided by this approach may thus improve the probabilities of generating clinically active CMV-specific CD8+ T cells for adoptive immunotherapy.
Sensitization of T cells with DCs loaded with the complete pool of pentadecapeptides also stimulates expansion of HLA class II–restricted CD4+ T cells. Indeed, while the APCs and culture conditions used have focused the expansion of CMV-pp65 specific CD8+ T cells, significant populations of specific CD4+ T cells also are regularly detected. In some donors, such as the donor described in Figure 7, the peptide-specific IFN-γ+ T cells generated have been predominantly CD4+ T cells. Like the peptide-specific CD8+ T cells, these CD4+ T cells have consistently exhibited cytotoxic activity against infected targets as well as peptide-loaded cells expressing the restricting HLA class II allele. However, since most of the CMV-infected cells encountered in vivo are likely not to express HLA class II, the principal contribution of these CD4+ T cells would be expected to be based on their capacity to recruit and maintain CD8+ T-cell effectors through their helper functions.
In conclusion, synthetic overlapping pentadecapeptides spanning the sequence of the immunodominant protein of CMV-pp65, when loaded on DCs, can consistently stimulate the in vitro generation of CD8+ and CD4+ T-cell lines from seropositive donors of diverse HLA genotypes. These cell lines are selectively enriched for T cells specific for a limited number of immunodominant epitopes, each presented by a single HLA class I or class II allele. The CMV-pp65–specific T cells generated by this approach are consistently able to lyse autologous CMV-infected targets. This approach, which exposes donor T cells to the gamut of potentially immunogenic peptides from CMV-pp65, thus may promote expansion of precommitted T cells recognizing epitopes that have been previously presented by CMV-infected cells of the donors in the context of class I and II alleles selected as immunodominant by the donor. Thus, from both functional and regulatory perspectives, this approach may therefore have advantages for generation of clinically active T cells for adoptive immunotherapy.
Note added in proof. During the review process of this manuscript, the HERNGFTVL epitope and the HLA restriction of this peptide to HLA-B*4001 and HLA-B*4002 have been published by Kondo et al.33
Prepublished online as Blood First Edition Paper, October 28, 2004; DOI 10.1182/blood-2003-05-1433.
Supported in part by the National Cancer Institute (grants CA59350 and CA23766), The Larry H. Smead Fund, The Aubrey Fund for Pediatric Cancer Research, The Vincent Astor Chair Clinical Research Fund, and a Translational Research Grant from the Leukemia and Lymphoma Society (G.K.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Dr Eric Pamer and Ingrid Leiner (MSKCC Tetramer Core Facility) for the generation and supply of the MHC tetramers used in these studies; Dr Ekaterina Doubrovina for supplying EBV-transformed B-cell lines; and Dr B. Dupont for HLA typing of donor cells and BLCLs. We thank Drs W. Herr and C. Britten (Mainz, Germany) for providing the K562/A*0201-transfected cell line.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal