Abstract
Fibrinogen Philadelphia, a hypodysfibrinogenemia described in a family with a history of bleeding, is characterized by prolonged thrombin time, abnormal fibrin polymerization, and increased catabolism of the abnormal fibrinogen. Turbidity studies of polymerization of purified fibrinogen under different ionic conditions reveal a reduced lag period and lower final turbidity, indicating more rapid initial polymerization and impaired lateral aggregation. Consistent with this, scanning and transmission electron microscopy show fibers with substantially lower average fiber diameters. DNA sequence analysis of the fibrinogen genes A, B, and G revealed a T>C transition in exon 9 resulting in a serine-to-proline substitution near the γ chain C-terminus (S378P). The S378P mutation is associated with fibrinogen Philadelphia in this kindred and was not found in 10 controls. This region of the γ chain is involved in fibrin polymerization, supporting this as the polymerization defect causing the mutation. Thus, this abnormal fibrinogen is characterized by 2 unique features: (1) abnormal polymerization probably due to a major defect in lateral aggregation and (2) hypercatabolism of the mutant protein. The location, nature, and unusual characteristics of this mutation may add to our understanding of fibrinogen protein interactions necessary for normal catabolism and fibrin formation.
Introduction
Fibrinogen is a 340-kDa dimeric glycoprotein that is composed of 2 identical halves, each of which consists of 3 nonidentical polypeptide chains termed Aα, Bβ, and γ. The halves of the molecule are associated at the N-terminal regions by a series of disulfide bonds.1,2 Fibrinogen plays an important role in 3 major biologic processes: (1) soluble fibrinogen is converted into fibrin after activation of blood coagulation; (2) fibrinogen-derived fibrin serves as a template for the activation and localization of the fibrinolytic system that induces the dissolution of the fibrin clot; and (3) fibrinogen interacts with platelets, white cells, and endothelial cells to mediate several biologic processes, including hemostasis, inflammation, wound healing, and angiogenesis. Fibrin formation is initiated by the enzymatic activity of thrombin that cleaves fibrinopeptides A and B from the N-terminal region of the Aα and Bβ chains, resulting in exposure of polymerization sites “A” and “B,” respectively. Fibrin polymerization is initiated by binding of exposed site “A” with a complementary site, “a,” localized in the C-terminus of the γ chain of an adjacent fibrin molecule. This association organizes the initial fibrin polymer into a half-staggered overlap of 2-stranded protofibrils.1-3 Removal of fibrinopeptide B and exposure of the “B” site promotes the lateral aggregation of the protofibrils that produces fibrin fibers that aggregate with each other to yield fibrin bundles, which branch, resulting in a fibrin gel.1-4 Finally, the fibrin polymer is cross-linked by factor XIIIa that forms new peptide bonds between specific lysine and glutamine residues, resulting in the formation of γ-γ dimers and α polymers,5 which increases the strength of the clot and makes it more resistant to fibrinolysis.6-9 Fibrinogen is synthesized and assembled inside the hepatocyte and secreted into the circulation, where its plasma half-life ranges from 3 to 4 days.10,11
Genetic disorders involving fibrinogen have been classified as quantitative alterations (mainly afibrinogenemia and hypofibrinogenemia), alterations of function (dysfibrinogenemia), and combined defects (hypodysfibrinogenemia).12-14 Approximately 300 families with dysfibrinogenemia have been described and more than 50 molecular defects have been identified.12,15,16 The clinical expression of dysfibrinogenemias varies; roughly 60% are clinically silent, and the other 40% are almost equally associated with bleeding and thrombosis, and in few instances the abnormal fibrinogen is associated with both manifestations. Rarely, the abnormal fibrinogens have been associated with spontaneous abortions,12,13 delayed wound healing,13 or amyloidosis of the kidney.13 In some hypodysfibrinogenemia mutants, the structurally abnormal fibrinogen with altered functional activity is combined with a low fibrinogen concentration, arbitrarily defined as below 1.5 mg/mL, as measured by immunologic methods. About 15 families with hypodysfibrinogenemia have been fairly well characterized and in most of these, the functional defect is reflected in an abnormal polymerization phase, whereas a delay in fibrinopeptide release is the exception. Rarely, an abnormal interaction with platelets and endothelial cells has been described. Clinically, the individuals with hypodysfibrinogenemia may present with bleeding, thrombosis, or spontaneous abortions.12,13
Previously,10 fibrinogen isolation, chromatographic studies, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), fibrinopeptide release, fibrin polymerization, cross-linking, metabolic studies, and general coagulation tests, including euglobulin lysis time and presence of fibrinogen-fibrin degradation products in serum were examined in the fibrinogen Philadelphia kindred. Briefly, we showed that fibrinogen concentration, measured by immunologic methods, ranged between 0.45 and 0.78 mg/mL. The thrombin time of plasma or purified fibrinogen was very prolonged in affected members and improved after addition of calcium. The euglobulin lysis time and amounts of fibrinogen-fibrin degradation products in serum were normal. Sizes of the 3 fibrinogen chains, estimated by SDS-PAGE, were also normal. Functionally, although fibrinopeptide release was normal, the formed fibrin monomers showed a marked decrease in final turbidity. However, the formed fibrin monomers demonstrated a delay in polymerization, especially when monomers were incubated in high ionic strength medium. Metabolic studies done on the proband and her son, using autologous and homologous fibrinogen, showed a marked decrease in half-life in plasma of 46 and 38 hours of the autologous protein compared with 72 and 74 hours for the normal counterpart. These metabolic studies indicate that the hypercatabolism is due to an intrinsic molecular defect of the patients' fibrinogen because the normal protein had a normal metabolism. In this report, we examine the ability of the fibrinogen to polymerize and characterize clot structure made from the proband's purified fibrinogen, expand analysis of the kindred, define the molecular abnormality of fibrinogen Philadelphia, and describe a model to explain the altered polymerization.
Patients, materials, and methods
Study population
This study was approved by the Thomas Jefferson University Institutional Review Board and all patients gave their informed consent.
DNA isolation
Genomic DNA was isolated from peripheral blood using Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN) and from buccal brushings using BuccalAmp DNA extraction kit (Epicentre, Madison, WI) as per the manufacturers' instructions.
PCR amplification
Coding regions and exon/intron borders of the 3 fibrinogen genes (FGA, FGB, FGG) were amplified with gene-specific primer pairs (A. Fellowes, personal communication, May, 2002). Polymerase chain reactions (PCRs) were performed using 200 ng DNA, 200 ng forward and reverse primers, 0.2 mM deoxynucleotide triphosphate (dNTP), 1.5 mM MgCl2, and Jumpstart Taq polymerase (Sigma-Aldrich, St Louis, MO) in a 100-μL reaction. PCR conditions were 5 minutes at 95°C followed by 30 cycles of 94°C for 30 seconds, 52°C to 62°C for 30 seconds, and 72°C for 30 to 60 seconds followed by a final extension of 72°C for 5 minutes. Annealing temperature and extension times were varied. In family members where DNA was limited, PCR products were used as template in a second PCR amplification to generate sufficient material for DNA sequence analysis. PCR products were purified using, Montage PCR tubes (Millipore, Billerica, MA) and quantified by comparison to size markers on agarose gels.
Plasmid cloning of PCR products
PCR products encompassing exon 9 of the FG gene were cloned into the TA cloning vector, pCR2.1, and the plasmid transformed into OneShot Top10 competent bacteria as per the manufacturer's instructions (Invitrogen, Carlsbad, CA). Resulting colonies were screened for the presence of inserts using PCR colony screening with M13 forward and reverse primers. Clones containing inserts were further screened in 2 PCRs, containing one FGG exon 9 primer and either the M13 forward or reverse primer, because cloning was not unidirectional. Positive clones were grown and plasmid DNA was isolated using Qiagen miniprep kit (Qiagen, Valencia, CA). Resulting plasmid DNA was subjected to DNA sequence analysis. Ten clones from each individual were sequenced to confirm the presence of the 2 alleles (exon 9 T or C).
DNA sequence analysis
Amplified FGG fragments (∼100 ng) were sequenced using BigDye3.1 dye terminators (Applied Biosystems, Foster City, CA) as per the manufacturer's instructions and analyzed on an ABI 3100 capillary electrophoresis system using 50-cm capillaries with POP6 polymer and Data Collection and Sequencing Analysis v1.1 software packages (Applied Biosystems). A fragment containing exons 9 and 10 was amplified using FGG7501for (5′-ctg gca atg cac ttc gta at-3′) and FGG8376rev (5′-ctc tct gtt cag ata aag tcc-3′) and sequence analysis was performed with nested primers FGG7530for (5′-ttc ata gac ttg cag agg ta-3′) and FGG7737rev (5′-ggt ggt gtt gct gtc ctt ct-3′).
Genotyping of fibrinogen G promoter polymorphism,–597A>G
The 5′-flanking region of the FGG gene was amplified with FGG-827for (5′-agg gcc aaa cag aaa tga tg-3′) and FGG-101rev (5′-tct ttc ccg ttc ctt ttt cc-3′) primers, and the resulting 726–base pair (bp) product was digested with 10 U BseRI (New England Biolabs, Beverly, MA) for 4 hours followed by electrophoresis on a 3% (wt/vol) SeaKem agarose (Cambrex, East Rutherford, NJ) gel. Digestion of PCR products with an A at –597 results in 337-, 226-, 86-, and 77-bp products, whereas those with a G results in 233-, 226-, 10-, 86-, and 77-bp products. Alternatively, PCR products were subjected to DNA sequence analysis using a nested primer FGG-678 for (5′-cct ggc tct ttt ctc tgt gg-3′).
Contig assembly and analysis
Sequence chromatogram files were imported into Sequencher software (GeneCodes, Ann Arbor, MI) and assembled into contigs using FGA, FGB, FGG reference sequences (GenBank accession no. NT006258). Heterozygous nucleotide positions were identified by divergence from the reference sequence.
Fibrin polymerization: turbidity studies and factor XIII cross-linking
For all experiments, fibrinogen was dialyzed into the appropriate buffer for 18 hours at 4°C. Turbidity was measured using a protein concentration of 0.5 mg/mL with 0.5 National Institutes of Health (NIH) U/mL thrombin in the appropriate buffer. Approximately physiologic buffer conditions consisted of 0.20 M Cl buffer (0.05 M Tris [tris(hydroxymethyl) aminomethane] HCl, 0.15 M NaCl, pH 7.4), whereas low and high ionic strength conditions consisted of 0.05 M (0.05 M Tris HCl, pH 7.4) and 0.40 M (0.05 M Tris HCl, 0.35 M NaCl, pH 7.4), respectively. Citrate was used to bind free Ca++ without removing the Ca++ that is strongly bound to fibrinogen. Absorbance of polymerizing fibrin was measured at 350 nm in a Perkin-Elmer λ-4b spectrophotometer (Wellesley, MA) using quartz cuvettes at ambient temperature (22°C-25°C) for 30 minutes. Factor XIIIa-catalyzed cross-linking of fibrin was studied by polymerization of control and Philadelphia fibrinogen at a concentration of 1 mg/mL in 25 mM Tris, pH 7.4, 140 mM NaCl, 5 mM CaCl2, with 0.2 U/mL human thrombin and 10 μg/mL factor XIIIa (both from American Diagnostica, Stamford, CT). The reaction was stopped at each time point (0, 2, 3, 4, 6, 9, 15, 20, 30 minutes) by the addition of an equal volume of sample gel buffer and immediately boiling for 5 minutes. These clotting conditions were chosen to follow the time course of γ-chain cross-linking over time. Samples were run on 8% PAGE and gels were stained with Coomassie blue.
Scanning and transmission electron microscopy of fibrin clots
Mean fiber diameter and size distribution (mean ± SD; n = 500) were measured from transmission electron microscope samples prepared as described previously.17 Clots were formed as described for the turbidity measurements but were dispersed with a glass Pasteur pipet as they began to gel after the addition of thrombin (1.0-2.5 minutes). Sample volumes of 5 to 20 μL were pipetted onto freshly glow-discharged, 300-mesh carbon-coated Formvar grids, negatively contrasted with filtered 2% uranyl acetate, and allowed to air-dry. Samples were viewed at 80 kV in a JEOL-100B transmission electron microscope (JEOL, Peabody, MA) and fiber measurements were made from photographic enlargements of transmission electron microscope negatives.
Whole clots were examined by scanning electron microscopy and intermediate-voltage electron microscopy. Clots were formed under the same conditions for fiber diameter measurements, but the thrombin-activated fibrin solutions were allowed to clot for 2 hours at room temperature under humidified conditions. Some clots were stopped at 30 minutes and other times for comparison with turbidity measurements. Scanning electron microscopy samples were clotted in plastic sample chambers and intermediate-voltage electron microscope samples on 400-mesh carbon-coated Formvar grids. Both types of samples were fixed in buffered glutaraldehyde, dehydrated in ethanol, and critical-point dried from CO2. Scanning electron microscopy samples then were mounted on specimen stubs, sputter-coated with gold, and viewed in an Amray AMR 1400 scanning electron microscope (Bedford, MA).18 Intermediate-voltage electron microscope samples were rotary coated with carbon and viewed in a JEOL JEM-4000EX transmission electron microscope at 400 kV.19 Stereo pair micrographs were taken at 0°C, ± 2°C, and ± 5°C.
Electron microscope negatives were digitized and figures were prepared using Adobe Photoshop (Adobe Systems, San Jose, CA).
Protein structure prediction
Swiss-Pdb viewer20 (http://www.expasy.org/spdbv/) was used to generate a model of the C-terminal portion of the γ chain of fibrinogen Philadelphia containing the S378P mutation using the pdb file 1FIC derived from the crystal structure of the 30-kDa C-terminal fragment.21 The program determines the “best fit” structure, putative hydrogen bonds, and steric hindrances. Images were imported into Pov RAY v3.6 (http://www.povray.org/. Persistence of Vision Raytracer [Version 3.6]: Persistence of Vision Pty, 2004, computer software) and converted to graphics format for publication.
Results
Molecular defect determined
To determine the molecular defect of fibrinogen Philadelphia, DNA sequence analysis of the 3 fibrinogen genes, FGA, FGB and FGG, in the proband was performed. The DNA sequence of the FGA and FGB genes matched the reference sequence (GenBank accession no. NT006258). Sequence analysis of the FGG gene showed 2 nucleotide changes compared to the reference sequence: a TC change in exon 9 that is predicted to result in a serine to proline substitution (TCC to CCC) at amino acid 378 in the C-terminus of the γ chain (S378P) and an A to G change in the promoter 597 nucleotides upstream of the transcription start site (A–597). All members of the kindred tested were homozygous for the FGG promoter –597 A to G change. This nucleotide change is reported in the single nucleotide polymorphism database dbSNP (http://www.ncbi.nlm.nih.gov/SNP/; rs1800792), and has an allele frequency of 42%, similar to the 50% frequency seen when 10 white controls were screened using PCR-restriction fragment length polymorphism analysis.
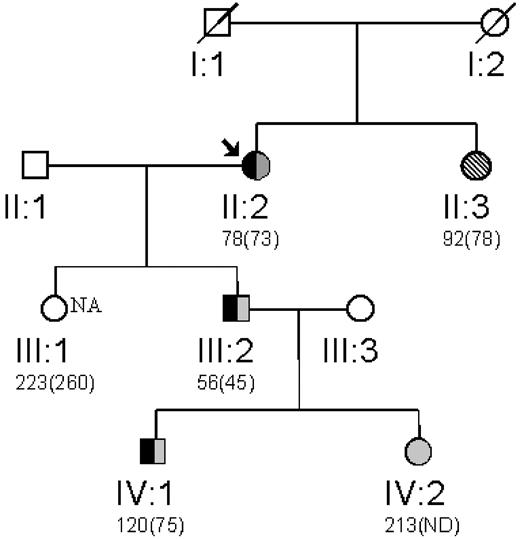
Exon 9 of the FGG gene was subsequently analyzed in the son (III-2) and grandson (IV-1), both of whom have hypodysfibrinogenemia as well as in the unaffected granddaughter (IV-2; Figure 1). To confirm the presence of 2 alleles at codon 378 in exon 9 in members of the family in whom DNA was limiting, PCR products were cloned into a shuttle vector, and resulting vectors subjected to DNA sequence analysis with a reverse primer. Sequence analysis of PCR products generated from the proband showed heterozygosity at this position and 2 representative clones showed each of the alleles (data not shown). The son and grandson, but not the unaffected granddaughter, were heterozygous for the T to C change resulting in S378P, as indicated in the pedigree in Figure l. This nucleotide change was not present in 10 control individuals screened by DNA sequence analysis.
Family pedigree. The propositus (arrow) with previously determined plasma fibrinogen levels assessed by thrombin-clottable protein shown below with immunodetectable fibrinogen (in parentheses). Inheritance of the fibrinogen G exon 9 T to C mutation resulting in a S378P amino acid substitution is shown in black. Gray-shaded objects represent the normal T allele and hatched bars represent the mutant C allele. The affected individual not genotyped is indicated by the shading.
Family pedigree. The propositus (arrow) with previously determined plasma fibrinogen levels assessed by thrombin-clottable protein shown below with immunodetectable fibrinogen (in parentheses). Inheritance of the fibrinogen G exon 9 T to C mutation resulting in a S378P amino acid substitution is shown in black. Gray-shaded objects represent the normal T allele and hatched bars represent the mutant C allele. The affected individual not genotyped is indicated by the shading.
Molecular modeling
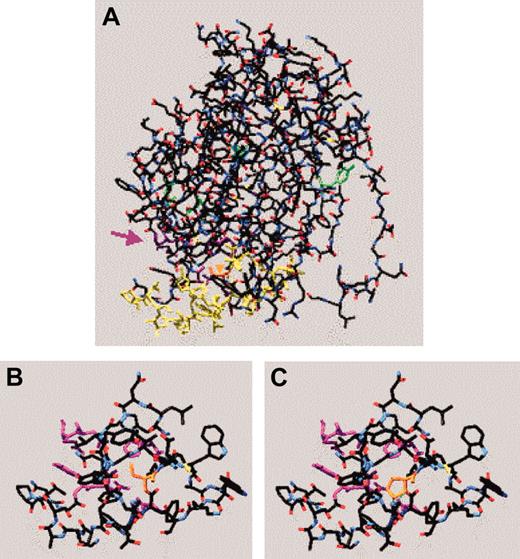
The effect of the fibrinogen Philadelphia mutation on the structure of the γ chain was investigated by using Swiss-Pdb viewer and entering the serine-to-proline change in the 1FIC of the 30 kDa C-terminal fragment.21 The location of the mutation in relation to some of the critical residues involved in calcium binding, D:D interactions, and lateral aggregation are shown in Figure 2A. The region surrounding residue 378 in the normal γ chain and in this model of fibrinogen Philadelphia are shown in Figures 2B and 2C, respectively. The amino acid change is predicted to result in formation of new hydrogen bonds between the hydroxyl group of proline 378 and glycine 352, residues likely to be involved in lateral aggregation.22 Interestingly, the functional studies presented are consistent with a defect in lateral aggregation.
Molecular model of fibrinogen Philadelphia. The γ-chain C-terminal 30-kDa fragment (1FIC) was examined using Swiss-Pdb viewer. The backbone is shown in red (oxygen), white (carbon), and blue (nitrogen) with amino acid residues of interest colored as follows: the fibrinogen Philadelphia mutation (378, orange), some of the residues thought to be critical to polymerization (329, 330, 340, 364, and 375, purple), some of the residues involved in lateral aggregation (350-360, 370-380, yellow), those involved in calcium binding (318, 320, green), and some of the residues involved in D:D interactions (275 and 280, green). (A) Complete C-terminal fragment of wild-type γ chain. The purple arrow indicates region of polymerization pocket and orange arrowhead (embedded in molecule) points to the location of Ser378, the residue affected in fibrinogen Philadelphia. (B) Amino acids 350 to 380 of the wild-type γ chain with residues critical to polymerization pocket visible. (C) Most favored conformation of amino acids 350 to 380 of fibrinogen Philadelphia with residues critical to polymerization pocket visible. Note the green dotted line (0.282 nm) indicating a newly created hydrogen bond between Gly352 (in red) and Pro378.
Molecular model of fibrinogen Philadelphia. The γ-chain C-terminal 30-kDa fragment (1FIC) was examined using Swiss-Pdb viewer. The backbone is shown in red (oxygen), white (carbon), and blue (nitrogen) with amino acid residues of interest colored as follows: the fibrinogen Philadelphia mutation (378, orange), some of the residues thought to be critical to polymerization (329, 330, 340, 364, and 375, purple), some of the residues involved in lateral aggregation (350-360, 370-380, yellow), those involved in calcium binding (318, 320, green), and some of the residues involved in D:D interactions (275 and 280, green). (A) Complete C-terminal fragment of wild-type γ chain. The purple arrow indicates region of polymerization pocket and orange arrowhead (embedded in molecule) points to the location of Ser378, the residue affected in fibrinogen Philadelphia. (B) Amino acids 350 to 380 of the wild-type γ chain with residues critical to polymerization pocket visible. (C) Most favored conformation of amino acids 350 to 380 of fibrinogen Philadelphia with residues critical to polymerization pocket visible. Note the green dotted line (0.282 nm) indicating a newly created hydrogen bond between Gly352 (in red) and Pro378.
Polymerization and cross-linking studies
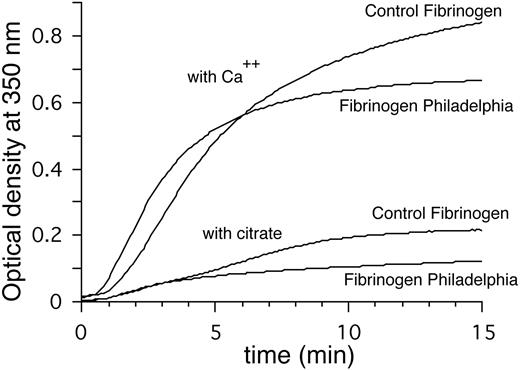
Measurements of turbidity as a function of time were used to compare the thrombin-activated polymerization of fibrinogen Philadelphia and control fibrinogen. Experiments were performed using 3 ionic strength buffers, with either 2 mM Ca++ or 0.02 M sodium citrate. Curves obtained using approximately physiologic (0.20 M NaCl buffer), low ionic strength (0.05 M NaCl), and high ionic strength (0.40 M NaCl) buffers were compared. In 0.20 M buffer with 2 mM Ca++, fibrinogen Philadelphia attained absorbance values nearly twice those of control fibrinogen during the first 2 minutes but was only 80% to 85% of normal values after 30 minutes (Figure 3; Table 1). Using 0.20 M buffer with 0.02 M citrate, both fibrinogens had absorbance values that were 75% to 80% less than 0.20 M buffer with Ca++ and a lag time of 0.8 minutes. Absorbance values were similar for the first 4 minutes; at 30 minutes fibrinogen Philadelphia reached absorbance values that were 66% of that for normal fibrinogen (Figure 3; Table 1). Turbidity experiments carried out in 0.40 M buffer with Ca++ resulted in severely depressed absorbance values, but fibrinogen Philadelphia attained maximum turbidity values that were nearly half of normal fibrinogen (0.020 ± 0.009 versus 0.035 ± 0.010, P < .0001, n = 10; Table 1). On the other hand, at 0.05 M ionic strength, the turbidity curves for fibrinogen Philadelphia were nearly normal but the lag period was shorter (Table 1). Fibrinogen Philadelphia clots form α and γ chain cross-links in the presence of factor XIIIa. The rate of cross-linking was similar to that of control clots (data not shown).
Turbidity curves. Representative turbidity curves of fibrinogen Philadelphia treated with thrombin in 0.05 M Tris HCl, 0.15 M NaCl, pH 7.4 either with 0.002 M CaCl2 or 0.02 M citrate. The presence of calcium ion (Ca++) has a profound effect on lateral aggregation, with a more pronounced difference for fibrinogen Philadelphia than for the control. In the presence of calcium, the lag period is shorter and the final turbidity is lower (Table 1).
Turbidity curves. Representative turbidity curves of fibrinogen Philadelphia treated with thrombin in 0.05 M Tris HCl, 0.15 M NaCl, pH 7.4 either with 0.002 M CaCl2 or 0.02 M citrate. The presence of calcium ion (Ca++) has a profound effect on lateral aggregation, with a more pronounced difference for fibrinogen Philadelphia than for the control. In the presence of calcium, the lag period is shorter and the final turbidity is lower (Table 1).
Polymerization of fibrinogen Philadelphia and clot structure as characterized by turbidity and electron microscopy
. | . | . | Maximum rate, × 10-5 sec-1 . | . | Turbidity, optical density at 350 nm . | . | . | . | . | . | Fiber diameter, nm . | . | . | . | . | . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Experimental condition . | Lag period, min . | . | . | . | Control . | . | . | Philadelphia . | . | . | Control . | . | . | Philadelphia . | . | . | |||||||||||
| . | Control . | Phil . | Control . | Phil . | 2 min . | 10 min . | 30 min . | 2 min . | 10 min . | 30 min . | 2 min . | 10 min . | 30 min . | 2 min . | 10 min . | 30 min . | |||||||||||
| 0.20 M Ca++ | 1.1 ± 0.3* | 0.6 ± 0.2* | 140 ± 18* | 180 ± 23* | 0.10 ± 0.05* | 0.73 ± 0.08* | 0.90 ± 0.09* | 0.19 ± 0.07* | 0.64 ± 0.09* | 0.75 ± 0.08* | 56 ± 14† | 75 ± 27*† | 111 ± 21*‡ | 57 ± 15† | 62 ± 20*† | 91 ± 19*‡ | |||||||||||
| 0.20 M citrate | 0.8 ± 0.2 | 0.8 ± 0.2 | 25 ± 5* | 17 ± 4* | 0.02 ± 0.01 | 0.20 ± 0.06* | 0.24 ± 0.07* | 0.02 ± 0.01 | 0.11 ± 0.05* | 0.16 ± 0.06* | — | 41 ± 11†§ | — | — | 45 ± 11†§ | — | |||||||||||
| 0.4 M Ca++ | 0 | 0 | 5 ± 1* | 1 ± 0.5* | 0.015 ± 0.003§ | 0.035 ± 0.010* | — | 0.018 ± 0.004§ | 0.020 ± 0.009* | — | — | 54 ± 15*† | — | — | 48 ± 14*† | — | |||||||||||
| 0.05 M citrate | 0.6 ± 0.2* | 0.3 ± 0.2* | 140 ± 20 | 140 ± 17 | 0.13 ± 0.07* | 0.74 ± 0.10 | 0.98 ± 0.10 | 0.21 ± 0.08* | 0.72 ± 0.09 | 0.98 ± 0.11 | — | — | — | — | — | — | |||||||||||
. | . | . | Maximum rate, × 10-5 sec-1 . | . | Turbidity, optical density at 350 nm . | . | . | . | . | . | Fiber diameter, nm . | . | . | . | . | . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Experimental condition . | Lag period, min . | . | . | . | Control . | . | . | Philadelphia . | . | . | Control . | . | . | Philadelphia . | . | . | |||||||||||
| . | Control . | Phil . | Control . | Phil . | 2 min . | 10 min . | 30 min . | 2 min . | 10 min . | 30 min . | 2 min . | 10 min . | 30 min . | 2 min . | 10 min . | 30 min . | |||||||||||
| 0.20 M Ca++ | 1.1 ± 0.3* | 0.6 ± 0.2* | 140 ± 18* | 180 ± 23* | 0.10 ± 0.05* | 0.73 ± 0.08* | 0.90 ± 0.09* | 0.19 ± 0.07* | 0.64 ± 0.09* | 0.75 ± 0.08* | 56 ± 14† | 75 ± 27*† | 111 ± 21*‡ | 57 ± 15† | 62 ± 20*† | 91 ± 19*‡ | |||||||||||
| 0.20 M citrate | 0.8 ± 0.2 | 0.8 ± 0.2 | 25 ± 5* | 17 ± 4* | 0.02 ± 0.01 | 0.20 ± 0.06* | 0.24 ± 0.07* | 0.02 ± 0.01 | 0.11 ± 0.05* | 0.16 ± 0.06* | — | 41 ± 11†§ | — | — | 45 ± 11†§ | — | |||||||||||
| 0.4 M Ca++ | 0 | 0 | 5 ± 1* | 1 ± 0.5* | 0.015 ± 0.003§ | 0.035 ± 0.010* | — | 0.018 ± 0.004§ | 0.020 ± 0.009* | — | — | 54 ± 15*† | — | — | 48 ± 14*† | — | |||||||||||
| 0.05 M citrate | 0.6 ± 0.2* | 0.3 ± 0.2* | 140 ± 20 | 140 ± 17 | 0.13 ± 0.07* | 0.74 ± 0.10 | 0.98 ± 0.10 | 0.21 ± 0.08* | 0.72 ± 0.09 | 0.98 ± 0.11 | — | — | — | — | — | — | |||||||||||
For turbidity and fiber diameters, the numbers of observations were 10 and 500, respectively.
Phil indicates fibrinogen Philadelphia; —, not determined.
P < .0001 versus control.
P < .001 versus control.
Indicates fiber diameter measurements from transmission electron micrographs of negatively contrasted fibrin.
Indicates fiber diameter measurements from scanning electron micrographs of fibrin clots.
Electron microscopy reveals abnormal clot structure
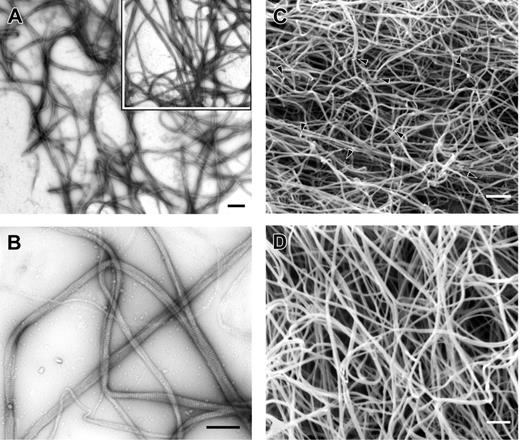
Transmission electron microscopy of negatively stained clots was performed to examine the details of clot structure under these same ionic conditions. Samples generally were taken every 2 minutes after the addition of thrombin, and fiber diameters and fiber densities were measured to correlate these observations with results seen in turbidity experiments. In most cases, the clots from fibrinogen Philadelphia appeared as a mesh of fibers of uniform size (Figure 4A). Fibers displayed the band pattern typical of normal fibrin (Figure 4B), indicating that assembly proceeded in the normal manner with half-staggered molecular packing.13 Diameters of fibers from fibrinogen Philadelphia clots made at 0.2 M buffer with Ca++ for 10 minutes were 80% of those of control clots (62 ± 20 versus 75 ± 27, P < .0001; Table 1). In contrast, under the same conditions the mean fiber diameter and size distribution after 2 minutes of polymerization were the same for both fibrinogens despite the fact that fibrin Philadelphia clots had reached absorbance values nearly twice those of normal fibrin at the same time point (0.19 versus 0.10, P < .0001; Table 1). However, the fiber density in transmission electron microscopy images of fibrinogen Philadelphia clots was considerably greater, accounting for the greater turbidity. Samples polymerized in 0.40 M buffer with Ca++ were poorly formed, with long, thin fibers with little bundling and irregularly spaced branch points (data not shown). Fiber diameters were greatly reduced at high ionic strength (Table 1), especially for fibrinogen Philadelphia clots, as predicted from the turbidity curves. A greater proportion of thin fibers and protofibrils were seen in the micrographs of fibrinogen Philadelphia clots prepared in 0.2 M NaCl, 0.02 M citrate buffer.
Transmission and scanning electron microscopy of Fg Philadelphia and normal control clot fibers. Clots were made under the same conditions as in Figure 2 with calcium. (A) Transmission electron micrograph of clot from fibrinogen Philadelphia at low magnification. Inset shows normal clot formed under similar conditions. Magnification bar = 1 μm. (B) Transmission electron micrograph of clot from fibrinogen Philadelphia at higher magnification, showing the details of fiber structure. Magnification bar = 0.5 μm. (C) Scanning electron micrograph of clot from fibrinogen Philadelphia showing the pattern of fiber branching, including fiber ends, with irregular branch points indicated by arrowheads. Magnification bar = 1 μm. (D) Scanning electron micrograph of control clot. Magnification bar = 1 μm.
Transmission and scanning electron microscopy of Fg Philadelphia and normal control clot fibers. Clots were made under the same conditions as in Figure 2 with calcium. (A) Transmission electron micrograph of clot from fibrinogen Philadelphia at low magnification. Inset shows normal clot formed under similar conditions. Magnification bar = 1 μm. (B) Transmission electron micrograph of clot from fibrinogen Philadelphia at higher magnification, showing the details of fiber structure. Magnification bar = 0.5 μm. (C) Scanning electron micrograph of clot from fibrinogen Philadelphia showing the pattern of fiber branching, including fiber ends, with irregular branch points indicated by arrowheads. Magnification bar = 1 μm. (D) Scanning electron micrograph of control clot. Magnification bar = 1 μm.
Scanning electron microscopy of whole clots was carried out to examine clot structure and fiber diameter after thrombin-induced polymerization was complete (Figure 4C-D). Measurements of mean fiber diameter in scanning electron microscopy images prepared under the same buffer conditions as described yielded differences comparable to those seen in transmission electron microscopy samples (Table 1). In other words, clots from fibrinogen Philadelphia had fiber diameters consistently lower than those of control clots. In addition, fiber ends were consistently seen in fibrin Philadelphia clots but not in controls (Figure 4C arrowheads).
Additional family members examined
Thrombin-clottable and immunodetectable fibrinogen levels were determined in 3 additional family members more than 20 years after the initial description of fibrinogen Philadelphia.10 Two children of the affected son (III-2) of the proband (II-2), and the proband's daughter were examined for immunologic levels of fibrinogen. Abnormally low levels were present in one grandson (IV-1), whereas the daughter (III-1) and granddaughter (IV-2) had normal levels (Figure 1). These additional findings further support autosomal dominant transmission of this genetic disorder.
Discussion
We have examined clot formation and structure and determined the molecular defect of fibrinogen Philadelphia, one of the first reported hypodysfibrinogenemias, an autosomal dominant condition functionally characterized by abnormal fibrin polymerization and about 30% of normal plasma fibrinogen levels. Thrombin-catalyzed polymerization of fibrinogen Philadelphia under a variety of conditions was characterized by a shorter lag period and lower maximum turbidity. On the other hand, although the initial characterization of mutant fibrin monomer had determined that clots formed from fibrin monomer at low ionic strength had lower final turbidity,10 this was not the case when fibrinogen was clotted with thrombin under low-salt conditions (Table 1). The reason for this difference is that polymerization is different with the sequential release of fibrinopeptides induced by thrombin than with fibrin monomer already missing both fibrinopeptides.4 With the polymerization of fibrinogen Philadelphia, the differences from control were most striking at high ionic strength or with low calcium at normal ionic strength. The results presented here reinforce the conclusion that lateral aggregation is sensitive to both ionic strength and calcium, suggesting that these interactions are ionic and involve the low-affinity calcium-binding sites. Consistent with these findings, transmission and scanning electron microscopy revealed clots composed of considerably thinner fibers. Although other dysfibrinogenemias are characterized by clots made up of thin fibers, the shortened lag period appears to be unique to fibrinogen Philadelphia. Approximately 91% of branch points in fibrinogen Philadelphia clots were trifunctional, similar to the 92% seen in control clots.23,24
The molecular defect γ S378P was present in 3 affected family members and was absent in one unaffected member, indicating that the nonconservative γ S378P change in exon 9 of the FGG gene is the cause of hypodysfibrinogenemia in this kindred. The 3 genes, FGA, FGB, and FGG, that encode the 3 fibrinogen chains (Aα,Bβ, and γ) are localized to a cluster on chromosome 4 (4q23-q32) are composed of 6, 8, and 10 exons, respectively,25,26 and are under coordinate control. Point mutations in the FGG gene are the most common cause of congenital dysfibrinogenemias, which are functionally expressed as defects in fibrin polymerization, although alterations in cross-linking also have been described.27
Our current knowledge of how fibrinogen is processed and interacts as well as the location of other mutations aid in understanding how the S378P mutation is associated with the biochemical, functional, and clinical phenotypes seen in the fibrinogen Philadelphia kindred. After the proteolytic cleavage of fibrinopeptide A by thrombin, the new N-terminal region exposes a polymerization site, named site “A,” that binds to a constitutively active complementary site termed “a” of adjacent fibrin molecules, localized in the C-terminal globular domain of the γ chain formed by several amino acids between G329 and R375.1-3,28,29 This association results in the formation of a double-stranded protofibril in which the fibrin monomers are arranged in a half-staggered overlap.3 Closely associated with the site “a” is the calcium-binding site pocket, where 2 aspartate residues, D318 and D320, play a fundamental role in calcium binding, and mutations of this segment may alter the proper folding of the molecule, resulting in defective fibrin polymerization.27 The C-terminal region of the γ chain also participates in the association D:D, where 2 molecules of fibrin associate through their C-terminal region, which is involved in the longitudinal growth of the protofibril30 and mutations of this area of the γ chain can cause delays in fibrin polymerization.31 The S378P mutation does not affect the D:D interface or factor XIII–induced cross-linking because it is not close to these regions (Figure 2). After the protofibrils reach a certain length they start to associate laterally to form fibrin fibers that branch to generate the fibrin gel. The molecular interactions involved in lateral aggregation are rather complex32-34 and not fully elucidated; our discussion will be limited to the role of the γ chain in this process. It appears that the γ chain may promote the lateral growth of fibrin fibers by direct interaction of the γ chains of different protofibrils via γ-chain segments 350 to 360 and γ 370 to 380.22 Because the S378P substitution in fibrinogen Philadelphia is in this region and we have observed striking effects of this mutation on the thickness of fibers, our results are consistent with this model. When the crystal structure of the C-terminal fragment of the γ chain21 is used to generate a molecular model, the S378 is located in a loop connecting 2 β sheets. One might speculate that substitution of serine with proline in a turn might not have a major effect on the tertiary structure of this region of the molecule. Whereas most dysfibrinogenemias that result in clots with low turbidity (thin fibers) also have an increased lag period, fibrinogen Philadelphia does not, suggesting that this abnormality is mainly due to a defect in lateral aggregation. Fibrinogen Paris I is structurally characterized by the insertion of 15 amino acids after γ Q350, and functionally manifests as a polymerization defect with normal cross-linking of the wild-type γ chains and no cross-linking of the γ Paris I chains.35 Analysis of the clot by electron microscopy also reveals abnormal clumps with irregular fibers and frequent terminations.36
Consistent with such conclusions is the low turbidity at high-salt concentration (Table 1), which has a characteristic effect on lateral aggregation. Interestingly, the lag period for fibrinogen Philadelphia is actually decreased under most conditions, indicating that this mutation also enhances the rate of oligomer formation. The observation that the number of fibers formed at 2 minutes after adding thrombin is higher for fibrinogen Philadelphia than for control (Table 1) is consistent with a higher rate of oligomer formation. These results are very similar to the effect of antibody 7E9 on fibrin polymerization, where this antibody enhanced fibrin oligomer formation by bringing monomers together and decreased lateral aggregation by capping the ends of protofibrils so that they did not get long enough to aggregate laterally.37 In both cases, fiber ends were observed, although this effect was more striking for 7E9 than for fibrinogen Philadelphia. Recent studies using a kinetic model for polymerization described previously38 support the scheme presented here. In conclusion, this unique mutation may provide evidence for the importance of γ378 in lateral aggregation, one of the most mysterious aspects of fibrin polymerization, although it may also affect protofibril initiation.
In other dysfibrinogenemias in which the molecular defect has been determined, the point mutations are close to the region identified in fibrinogen Philadelphia, yet their functional properties are different. Fibrinogen Osaka V, γ R375G, shows a delay in fibrin polymerization that is corrected in vitro by increasing the concentration of calcium. In this case, the defect in polymerization site “a” and calcium binding appear to be caused by the loss of the side chain of R375.39 In fibrinogen Kaiserslautern, γ K380N, the additional carbohydrate content is responsible for the polymerization defect.40 Thus, the structural and functional defect of fibrinogen Philadelphia is rather distinct.
A second abnormality associated with fibrinogen Philadelphia is the low plasma fibrinogen concentration, with a plasma level of approximately one third of normal. A unique property of this fibrinogen is the high catabolic rate compared to wild-type fibrinogen, indicative of the molecular defect being responsible for the rapid catabolism.10 In addition, the calculated rate of synthesis was normal in one affected family member and slightly decreased in the other,10 and the reason for the lack of compensatory synthesis is not known. A second hypodysfibrinogenemia, Bethesda III,41 exhibits properties similar to those of fibrinogen Philadelphia: no obvious truncations of the constitutive chains, a polymerization defect, a decrease in the concentration of fibrinogen to about one third to one fourth of normal, and a high catabolic rate of only the abnormal fibrinogen. In contrast, 2 other hypodysfibrinogenemias, fibrinogens Baltimore II42 and Chapel Hill I,43 manifest a normal catabolic rate of the autologous and homologous fibrinogens, indicating a defect in synthesis, assembly, or secretion. Two other hypodysfibrinogenemias, fibrinogens Marburg44 and Otago,45 are structurally characterized by truncations of the Aα chain that cause delay in fibrin polymerization and the low plasma fibrinogen could be due to impaired synthesis, intracellular assembly, or defects in secretion, as demonstrated with other hypofibrinogenemias.16,46 However, because the mechanism involved in the disposal of fibrinogen is not known, we cannot differentiate between the possibility that this mutation accelerates the regular mechanism of fibrinogen catabolism or that this mutation uncovers a new catabolic pathway.
The clinical expression of hypodysfibrinogenemias is similar to that of dysfibrinogenemias: bleeding, usually associated with trauma or surgical procedures, as seen in fibrinogen Philadelphia,10 bleeding and spontaneous abortions described in fibrinogen Bethesda III,41 postpartum bleeding followed by thrombosis in fibrinogen Marburg,44 or postsurgical bleeding and impaired wound healing as described in fibrinogen Otago.45
Our biophysical and molecular analyses of fibrinogen Philadelphia polymerization suggest that lateral aggregation is sensitive to both ionic strength and calcium concentration. The structural defect of fibrinogen Philadelphia localized at the C-terminus of the γ chain (S378P) is unique in the described mutations of fibrinogen and indicates that this region of the γ chain is functionally important in fibrin polymerization and plays a fundamental role in the formation of lateral aggregation of fibrin polymers. In addition, this substitution is likely also responsible for the high catabolic rate seen in this kindred, and further studies may uncover the pathways involved in fibrinogen catabolism.
Prepublished online as Blood First Edition Paper, January 4, 2005; DOI 10.1182/blood-2004-04-1621.
Supported in part by National Institutes of Health grants HL059227 (J.M.) and HL30954 (C.N., I.N.C., J.W.W.) and the Cardeza Foundation for Hematologic Research (M.A.K., M.K.B., S.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank A. Fellowes (Canterbury Health Laboratories, Christchurch, New Zealand) for supplying primer sequences for the human fibrinogen genes, Susan Miller-Samuel (Thomas Jefferson University, Philadelphia, PA) for creation of the pedigree figure, and Lillian Spadoni (Thomas Jefferson University, Philadelphia, PA) for assistance with molecular modeling.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal