It has now been 3 years since the von Willebrand factor (VWF)–cleaving protease implicated in thrombocytopenic purpura (TTP) pathogenesis was identified as ADAMTS13 (adisintegrin-like and metalloprotease with thrombospondin type 1 motif 13). More than 50 ADAMTS13 mutations resulting in familial TTP have been reported. Considerable progress has also been realized toward understanding the role of ADAMTS13 in normal hemostasis, as well as the mechanisms by which ADAMTS13 deficiency contributes to TTP pathogenesis. Measurement of ADAMTS13 activity in TTP and other pathologic conditions also remains a focus of a substantial clinical research effort. Building on these studies, continued investigation of ADAMTS13 and VWF holds considerable promise for advancing the understanding of TTP pathogenesis and should lead to improved diagnosis and treatment for this important hematologic disease.
Introduction
The disease thrombotic thrombocytopenic purpura (TTP) was first described in 1924 when Eli Moschcowitz reported a 16-year-old girl with sudden onset of fever and hemolytic anemia, followed rapidly by paralysis, coma, and death (recently reprinted).1 Since that report, TTP has intrigued hematologists and has been the focus of considerable clinical and basic science research. Much of the interest in TTP likely relates to its dramatic appearance in previously healthy individuals, its rapidly progressive and often fatal course, and to the fact that timely diagnosis can be life saving.
In 1960 Schulman et al2 reported a similar disorder in an 8-year-old girl who exhibited relapsing episodes of thrombocytopenia. This patient responded well to plasma infusion, and Schulman proposed that the disorder was due to deficiency of a platelet-stimulating factor. Upshaw3 later reported comparable findings in a 29-year-old woman whose first episode occurred at the age of 6 months. He suggested deficiency of a plasma factor that promotes platelet and red blood cell survival as the underlying pathogenic mechanism. It is now clear that the disorder described by Schulman and Upshaw, which now bears their name (Upshaw-Schulman syndrome, Online Mendelian Inheritance in Man [OMIM] no. 274150),4 represents a familial form of TTP.
TTP classically has been defined as the pentad of fever, thrombocytopenia, microangiopathic hemolytic anemia, renal dysfunction, and neurologic symptoms, although presentation of these symptoms is highly variable.5 Platelet-rich microthrombi in the small vessels of multiple organs are the pathologic hallmark of this disease.6 The annual incidence of TTP is estimated at approximately 4 cases per 1 000 000 in the United States and appears to be increasing.7,8 TTP is more frequent among women with a female-to-male ratio of 3:2.7
The term TTP refers to several different forms of the disorder, the most common being acquired (also referred to as “sporadic”) TTP, which typically occurs without an obvious inciting event in individuals between 20 and 60 years of age.5 Initial descriptions of TTP reported mortality greater than 90%. The advent of plasma exchange and modern supportive therapy have reduced mortality to less than 20%.9 Morbidity, however, remains high, and treatment carries significant associated risks, including exposure to blood products from multiple donors. Relapses following an initial episode of acquired TTP are common, with about one third of cases becoming chronic.10,11
Familial TTP usually manifests in the immediate postnatal period or during infancy, although some cases with onset as late as the second to third decade have been reported.12-19 Patients with familial TTP typically exhibit a chronic relapsing course and, in contrast to patients with acquired TTP, respond well to plasma infusion and usually do not require plasma exchange.18
TTP-like syndromes can also arise secondary to other conditions such as HIV,20 bone marrow transplantation,21,22 pregnancy,23 treatment with the antiplatelet agents ticlopidine24 and clopidogrel,25 and autoimmune disorders such as systemic lupus erythematosus.26 In addition, a number of other disorders are also associated with thrombotic microangiopathy (TMA). These diseases often exhibit clinical characteristics overlapping those of TTP and can complicate diagnosis. In particular, the presentation of the hemolytic uremic syndrome (HUS) can be very similar to that of TTP, and these 2 disorders are grouped by some experts into the single category TTP/HUS.27 However, most cases of acute HUS are caused by Shigatoxin-producing strains of Escherichia coli and Shigella dysenteriae, and the nature of the clots observed in TTP (platelet and von Willebrand factor [VWF] rich) are different than those observed in HUS (fibrin rich). Therefore, these 2 disorders are likely to differ at least partially in their mechanisms of pathogenesis.
TTP pathogenesis: altered VWF homeostasis
Considerable evidence now implicates the blood clotting protein VWF as a key component in TTP pathogenesis.28 VWF, an abundant plasma glycoprotein, serves as the primary adhesive link between platelets and the subendothelium, and it also carries and stabilizes coagulation factor VIII (FVIII) in the circulation. VWF is synthesized in endothelial cells and megakaryocytes as a precursor containing a signal peptide and large propeptide.29 Endothelial cell VWF is secreted via both constitutive and regulated pathways.30 VWF transported through the regulated pathway is first stored in a unique storage granule called the Weibel-Palade body.31 In response to a variety of stimuli, VWF is released from endothelial cells in the form of ultralarge multimers (UL-VWF), which can be up to approximately 20 000 kDa in size32,33 and are the most adhesive and reactive form of VWF. Circulating VWF analyzed by reducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) reveals mature monomeric VWF (∼300 kDa), as well as several smaller fragments.34 Two of these smaller forms (the 176-kDa and 140-kDa fragments) have been shown by amino acid sequence analysis to be the products of cleavage of the intact VWF subunit at the bond between tyrosine 1605 and methionine 1606.35 For a more detailed discussion of VWF biochemistry, the interested reader is referred to an in-depth review.36
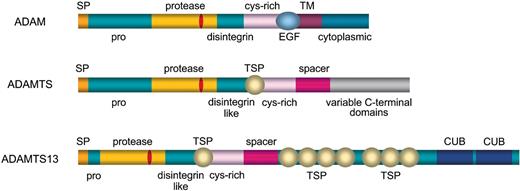
Schematic diagram of ADAM and ADAMTS metalloproteases and of ADAMTS13. SP indicates signal peptide; pro, propeptide; protease, metalloprotease (location of zinc-binding motif shown in red); disintegrin, disintegrin domain; dis-like, disintegrin-like domain; cys-rich, cysteine-rich domain; TSP, thrombospondin type-1 motif; EGF, epidermal growth factor-like repeat; TM, transmembrane domain; CUB, CUB domain.
Schematic diagram of ADAM and ADAMTS metalloproteases and of ADAMTS13. SP indicates signal peptide; pro, propeptide; protease, metalloprotease (location of zinc-binding motif shown in red); disintegrin, disintegrin domain; dis-like, disintegrin-like domain; cys-rich, cysteine-rich domain; TSP, thrombospondin type-1 motif; EGF, epidermal growth factor-like repeat; TM, transmembrane domain; CUB, CUB domain.
The first link between VWF proteolysis and TTP pathogenesis was suggested in 1982 by Moake et al37 who observed the presence of UL-VWF in the plasma of 4 patients with chronic relapsing TTP. These investigators hypothesized that UL-VWF formed the spontaneous platelet and VWF-rich thrombi characteristic of TTP and that deficiency of a VWF “depolymerase” is the underlying cause of TTP. In 1996, 2 groups independently described a proteolytic activity in plasma that was able to cleave VWF at the tyrosine 1605-methionine 1606 bond.38,39 In both studies, proteolysis of VWF required that it be in a denatured conformation, achieved by preincubation with either guanidine or urea, or be subjected to high shear stress in vitro.39
While the identity of the VWF-cleaving protease was still unknown, the suggestion that UL-VWF multimers play a role in TTP prompted the same investigators to examine this VWF-cleaving activity in patients with TTP. In 1997, deficiency of this proteolytic activity in the plasma of 4 patients with familial TTP was described and suggested to be inherited in an autosomal recessive fashion.40 Subsequently, deficient VWF-cleaving activity was described in patients with acquired TTP41 and was determined to be due to the presence of immunoglobulin G (IgG) autoantibody inhibitors.42,43 These results, therefore, implicated a common mechanism for the pathogenesis of both familial and acquired TTP and also provided a possible explanation for the differences in response to plasma infusion between these 2 forms of the disorder. It was hypothesized that efficacy of plasma infusion alone in familial cases is consistent with constitutional deficiency of the VWF-cleaving protease, while, in acquired cases, plasma exchange may serve the additional function of removing the IgG inhibitors.
Identification of the VWF-cleaving protease as ADAMTS13
In 2001, several groups identified the VWF-cleaving protease as ADAMTS13 (adisintegrin-like and metalloprotease with thrombospondin type 1 motif 13), a novel member of the ADAMTS family of metalloproteases. One line of investigation used a genetic positional cloning strategy that started with a collection of familial TTP pedigrees, with subsequent linkage analysis localizing the responsible gene to chromosome 9q34.44 Fine mapping of this region further localized the gene to a much smaller genetic interval. DNA sequence analysis of candidate genes in this region led to identification of mutations in 15 of 17 affected chromosomes within a predicted gene encoding a new member of the ADAMTS (ADisintegrin and Metalloprotease with Thrombospondin type-1 motifs) family, ADAMTS13. Independently, 3 other groups purified the VWF-cleaving protease activity from either human plasma or commercial factor VIII/VWF concentrate, with peptide sequence analysis identifying a protein encoded by the same ADAMTS13 gene.45-48
ADAMTS13 is the 13th member of the ADAMTS family of metalloproteases, which is related to the large ADAM (ADisintegrin and Metalloprotease) family (Figure 1). ADAM proteases are named for their characteristic domain structure that bears similarity to the snake venom metalloproteases. ADAM proteases are type I transmembrane proteins that possess a signal peptide and propeptide preceding a metalloprotease domain, disintegrin-like domain, EGF repeat, transmembrane domain, and cytoplasmic domain (Figure 1).49 To date, more than 30 ADAM family members have been described, and these molecules serve a variety of functions ranging from sperm-egg fusion (ADAMs 1 and 2) to shedding of extracellular ligands such as tumor necrosis factor-alpha (TNF-α; ADAM 17) and Notch (ADAM10).49
Similar to ADAM metalloproteases, members of the ADAMTS family (of which there are 19) exhibit a similar domain structure with a signal peptide, propeptide, metalloprotease domain, and disintegrin-like domain.50 In contrast to ADAM proteases, ADAMTSs lack EGF-like repeats and a transmembrane domain and, therefore, are secreted rather than membrane bound. In addition, all ADAMTS family members possess one or more thrombospondin type 1 (TSP1) motifs50 and variable additional C-terminal domains (Figure 1). ADAMTS2 cleaves the propeptide of collagen II, and mutations in this protein cause Ehlers-Danlos syndrome type VII C.51 Mutations in ADAMTS10 cause autosomal recessive Weill-Marchesani sydrome.52 Interestingly, some mutations in the fibrillin gene can cause the autosomal dominant form of this disease,53 suggesting that fibrillin may be a substrate of ADAMTS10. ADAMTS3 and ADAMTS14, which bear homology to ADAMTS2, can also cleave procollagen, suggesting possible redundant functions or as-yet-unrecognized differences in substrate specificity.54 ADAMTS1, ADAMTS4, and ADAMTS5/11 (also known as aggrecanases) cleave the cartilage proteoglycan aggrecan and may play a role in inflammatory joint disease.55-57 In mice, disruption of the Adamts1 gene results in growth retardation and several developmental defects, including adipose tissue malformation, histologic changes in the uterus and ovaries, enlarged renal calices with fibrotic changes, and abnormal adrenal medullary architecture.55
ADAMTS13 mutations identified in patients with familial TTP. Missense mutations are indicated below the diagram in blue, and mutations predicted to result in a truncated protein (nonsense, frameshift, and splice site) are indicated above the diagram in orange.
ADAMTS13 mutations identified in patients with familial TTP. Missense mutations are indicated below the diagram in blue, and mutations predicted to result in a truncated protein (nonsense, frameshift, and splice site) are indicated above the diagram in orange.
ADAMTS13 is one of the largest members of the ADAMTS family, and exhibits the characteristic domain structure (Figure 1). Unlike other family members, the ADAMTS13 sequence contains 2 C-terminal CUB domains of unknown function. CUB domains are found in many developmentally regulated proteins (eg, the dorsoventral patterning protein tolloid, bone morphogenetic protein 1, and spermadhesins) and may play a role in protein-protein interactions.58 TSP1 motifs are known also to mediate protein-protein interactions, especially among proteins in the extracellular matrix.59 Finally, disintegrin-like domains are known to bind integrins through an RGD sequence also present within the ADAMTS13 cysteine-rich domain.60 Thus, through its multiple domains, ADAMTS13 has the potential to interact with a wide range of molecules.
The human ADAMTS13 gene comprises 29 exons spanning approximately 37 kb of genomic sequence.44 The ADAMTS13 cDNA predicts a 1427 amino acid protein with a calculated molecular mass of 145 kDa, in contrast to the apparent molecular mass of approximately 190 kDa observed for ADAMTS13 purified from human plasma.46,47 This difference in calculated versus observed molecular mass is likely due to glycosylation.61 ADAMTS13 is synthesized predominantly in the liver,44,62 although variable expression has been observed in other tissues and cell types, including platelets.63
ADAMTS13 mutations in familial TTP
Considerable work from multiple laboratories has now identified more than 50 mutations in patients with familial TTP (Figure 2).44,61,64-71 Approximately 60% of these mutations are missense single amino acid substitutions, and 40% are nonsense, frameshift, or splice site mutations that are predicted to result in a truncated protein. There are no apparent clustering or mutation hot spots, although roughly 75% of the missense mutations are found in the first half of the molecule (protease domain through TSP2).
Several investigators have examined the effect of ADAMTS13 point mutations on synthesis and secretion of ADAMTS13 in cell culture. Most of these mutations appear to result in greatly reduced ADAMTS13 secretion into the cell culture medium.61,64,66 The few mutations that demonstrate a small amount of ADAMTS13 secretion also result in abrogation of enzymatic activity. These data suggest that most patients with familial ADAMTS13 deficiency are likely to be cross-reacting material (CRM) negative for ADAMTS13. Definitive demonstration of ADAMTS13 antigen status in patients with familial TTP awaits development of appropriate reagents and clinical testing. Interestingly, a common ADAMTS13 polymorphism (P475S) has been demonstrated in the Japanese population that results in greatly reduced ADAMTS13 activity in vitro.61 As many as 10% of the Japanese population may be heterozygous for P475S and, therefore, may exhibit significantly reduced ADAMTS13 activity. The significance of this finding for the development of TTP or other disorders remains to be determined.
To date, no clear genotype to phenotype correlations have yet been observed. It is tempting to speculate that mutations near the amino terminus of ADAMTS13 (eg, a mutation within the protease domain) may be more “severe” than mutations near the carboxy terminus. However, analysis to date has generally been restricted to patients already carrying a clear diagnosis of familial TTP, potentially presenting an ascertainment bias against the identification of mild mutations. Thus, the possibility that less severe mutations may be associated with milder forms of this disease cannot be excluded. The rarity of proximal truncating mutations or compound heterozygosity for mutations in the protease domain has led to the speculation that most or all patients with familial TTP may express low, residuals levels of ADAMTS13 activity, and that “complete” ADAMTS13 deficiency may be lethal. Further testing of this hypothesis will require the identification of patients who are compound heterozygous or homozygous for a null mutation or the development of a knock-out mouse model.
ADAMTS13 structure and function
Since the isolation and cloning of the ADAMTS13 cDNA, several laboratories have expressed recombinant ADAMTS13 in cell culture. Recombinant ADAMTS13 cleaves VWF in vitro, providing formal demonstration that ADAMTS13 is indeed the VWF-cleaving protease identified in earlier studies.61,72 Furthermore, several groups have gone on to perform structure/function analyses of ADAMTS13 to determine the structural requirements for its activity.
Two groups have investigated the effects of C-terminal truncations on ADAMTS13 secretion and VWF-cleaving activity.61,73 Truncation of ADAMTS13 within, or distal to, TSP1 results in proteins that retain VWF-cleaving activity in vitro, while truncations proximal to TSP1 (within the protease, TSP1, cysteine-rich, or spacer domains) result in an inactive protein. These results indicate that sequences within the region spanning the protease domain to the spacer domain of ADAMTS13 are sufficient for VWF-cleaving activity, at least in vitro. With regard to secretion of these mutants, truncation of ADAMTS13 within the cysteine-rich and spacer domains results in variable secretion from the cell. Additionally, mutation of the RGD sequence within the cysteine-rich region (to RGE) had no effect on secretion or VWF-cleaving activity in vitro.74
Recently, a truncated variant of murine ADAMTS13 was identified from several common inbred strains of laboratory mice.75 In these strains, including the commonly-studied strain C57BL/6, insertion of an intracisternal A particle sequence (a retrovirus-like element) results in ADAMTS13 protein truncated following TSP6. This truncated form of ADAMTS13, termed ADAMTS13S, exhibits in vitro VWF-cleaving activity identical to wild-type murine ADAMTS13. The normal phenotype of mouse strains that express ADAMTS13S nearly exclusively suggests that sequences distal to TSP6 may not be required for ADAMTS13 function in vivo. However, this observation is in contrast to the fact that disease-causing mutations distal to TSP6 have been identified in patients with familial TTP.
The propeptide of ADAMTS13 contains 41 amino acids, in contrast to the approximately 200 amino acids that comprise the propeptides of most other ADAM and ADAMTS family members.50,76 In addition, the propeptide of ADAMTS13 does not appear to contain a potential “cysteine-switch” motif. This motif functions in other metalloproteases to inhibit protease activity by coordinating with the active site Zn2+ in the protease domain to keep the molecule in an inactive zymogen form until cleavage of the propeptide (by a furin-like enzyme) coincident with secretion from the cell.77 In contrast to what has been observed for other metalloproteases, deletion of the ADAMTS13 propeptide does not impair secretion or enzymatic activity, demonstrating that the propeptide is not required for folding or secretion and likely does not function to maintainADAMTS13 in an inactive form.76 However, theADAMTS13 propeptide sequence has been conserved across wide evolutionary distance (from fish to mammals), suggesting that it may serve an as yet unknown function.
ADAMTS13 autoantibodies and acquired TTP
In 1998 it was demonstrated that acquired TTP is caused by inhibitory IgG autoantibodies to the VWF-cleaving protease.43,78 Isolation and cloning of the ADAMTS13 cDNA has now enabled several laboratories to begin to map the epitopes recognized by the anti-ADAMTS13 autoantibodies found in patients with acquired TTP.74,79 First, IgG antibodies purified from the plasma of 3 patients with acquired TTP were found to react with the cysteine-rich and spacer domains of recombinant ADAMTS13.74 Subsequently, plasma from 25 patients with acquired TTP were analyzed against ADAMTS13 fragments expressed in E coli. Antibodies recognizing the ADAMTS13 cysteine-rich and spacer domains were found in 100% of the plasma samples, and in 3 cases antibodies were only found against these domains.79 These data suggest that the cysteine-rich and spacer domains of ADAMTS13 may harbor the critical epitopes needed for the generation of inhibitory autoantibodies that can cause TTP. Varying combinations of antibodies against the propeptide, TSP motifs, and CUB domains of ADAMTS13 were also found, providing convincing evidence of a polyclonal autoantibody response in a majority of cases.79 The finding of antibodies directed against the ADAMTS13 propeptide suggests that unprocessed ADAMTS13 (or free propeptide) may be present in the circulation of some individuals. Finally, a recent report demonstrated the presence of anti-ADAMTS13 non-neutralizing antibodies (both IgG and IgM) in a patient with acquired TTP.80 It is hypothesized that these antibodies function by promoting ADAMTS13 clearance, or by interfering with its binding to the endothelium or other plasma proteins. Because of the success of these early studies, it should be possible to fine-map the critical individual epitopes involved in the autoimmune response leading to acquired TTP.
ADAMTS13 cleaves VWF under flowing conditions in an in vitro system
Resting platelets transiently adhere to stimulated endothelium in vivo in a VWF-dependent manner.81,82 Following initial binding, platelets are eventually released from the endothelial surface through a mechanism that has been hypothesized to be mediated by ADAMTS13 cleavage of VWF.83 Recent studies have addressed this hypothesis using an in vitro system of human umbilical vein endothelial cells (HUVECs) cultured in a parallel plate flow chamber.84,85 Perfusion of normal plasma containing ADAMTS13 (or partially purified ADAMTS13) results in almost immediate cleavage of platelet/UL-VWF strings that were released in response to endothelial agonists, and this effect is not seen when ADAMTS13-deficient plasma from patients with TTP is used.84 These results support the notion that ADAMTS13 indeed may function to cleave newly secreted UL-VWF from the surface of endothelial cells, which otherwise may have persisted long enough to induce platelet adhesion.
An interaction between ADAMTS13 and UL-VWF has also been demonstrated under both static and flowing conditions.84 ADAMTS13-coated beads were shown to adhere to UL-VWF released from stimulated HUVECs and to immobilize VWF in a cell-free system. Interestingly, the ADAMTS13 beads also bound to immobilized VWF A3 domain, suggesting that this domain may be a site of ADAMTS13 docking on VWF. Finally, the inflammatory cytokine interleukin 6 (IL-6; when present at levels considerably higher than typically observed in vivo) greatly abrogates the ability of ADAMTS13 to cleave UL-VWF under flowing conditions in the parallel plate chamber.86 This effect of IL-6 on ADAMTS13 activity has been speculated to represent another link between inflammation and thrombosis.
ADAMTS13 deficiency, TTP, and other thrombotic microangiopathies
Several laboratories have measured VWF-cleaving protease activity in large series of patients with the clinical diagnosis of TTP, with mixed and often conflicting results.87 The sensitivity of decreased VWF-cleaving protease activity for the diagnosis of acquired TTP ranges from 33% to 100%, with similarly variable data for specificity.42,88-94 Decreased VWF-cleaving activity has been reported in a wide variety of conditions, including liver cirrhosis, chronic uremia, idiopathic thrombocytopenic purpura (ITP), disseminated intravascular coagulation (DIC), systemic lupus erythematosus (SLE), leukemia, pregnancy, the postoperative state, the neonatal period, and with advancing age.95,96 In contrast, results regarding the specificity of severe ADAMTS13 deficiency for distinguishing TTP from acute HUS have been more convincing, as the incidence of severe ADAMTS13 deficiency in patients diagnosed with acute HUS has been reported at 0% in 3 studies (61 cases),89,90,92 3% (1 of 29 cases),97 9% (2 of 23),42 and 60% (3 of 5),93 for a total of 5% (6 of 118 cases).
Owing to the differences reported in these studies, the subject of the sensitivity and specificity of ADAMTS13 deficiency for the diagnosis of TTP remains controversial. Much of the variability in these studies likely relates to the following: (1) lack of a true “gold standard” laboratory test for TTP diagnosis; (2) difficulty in distinguishing TTP from HUS and other forms of TMA based solely on clinical manifestations (ie, is TTP a distinct clinical entity, or rather part of a TMA disease continuum?); (3) potential sources of error in the assays (eg, other proteases that can cleave VWF, or heterogeneity of the autoantibody response); and (4) multiple assays involving different principles, techniques, preparations, and operators, which make direct comparison among assays difficult.
In an effort to address the differences between the ADAMTS13 activity assays currently in use, a recent study compared several tests and found good interassay and interlaboratory agreement in their ability to identify severe ADAMTS13 deficiency and the presence of ADAMTS13 inhibitors.98 A follow-up study investigated the ability of 11 independent laboratories (each performing different ADAMTS13 activity assays) to determine the ADAMTS13 activity present in the same set of coded plasmas.99 The results showed that the majority of methods were able to correctly identify plasma with 0% activity, and several of the methods were able to discriminate between 0% and 10% ADAMTS13 activity. The best performance was observed for the methods measuring VWF cleavage by ristocetin cofactor, collagen binding, and immunoblotting of degraded VWF multimers.
Although the ability to determine ADAMTS13 activity and perform anti-ADAMTS13 antibody screening would be of great diagnostic value for the clinician, the use of these tests currently is restricted by limited availability and long turnaround times. Therefore, the clinical application of ADAMTS13 testing in the management of TTP is still evolving and continues to be the subject of considerable research activity. Although improved diagnostic tests based on recombinant VWF fragments as substrates are in development,94,100-103 at this time TTP still remains largely a clinical diagnosis.
Future
Identification of ADAMTS13 as the VWF-cleaving protease deficient in TTP has resulted in considerable progress toward understanding the pathogenesis of this disease. However, many questions remain. For example, it is becoming increasingly clear that loss of ADAMTS13 activity with an associated increase in circulating UL-VWF is necessary, but not sufficient, to initiate an episode of acute TTP.104 Patients in clinical remission can still demonstrate UL-VWF multimers in the plasma and absent ADAMTS13 activity in vitro,42,105 Similarly, regarding familial TTP, there is the question of incomplete penetrance (the likelihood that a gene mutation will cause clinical disease in a given individual) and variable expressivity (variation in disease severity among affected individuals). For example, most patients with familial TTP present in the neonatal period. However, some cases are not diagnosed until early adulthood, and there are rare individuals that have ADAMTS13 disease-causing mutations but still have not manifested clinical disease through the third decade of life.12 In addition, family members with the same ADAMTS13 mutation(s) can demonstrate great variability in disease severity.12 Whether these observations are due in part to different ADAMTS13 mutations or to other environmental “triggers” or genetic “modifying factors” remains to be determined. Regarding acquired TTP, the triggers that initiate anti-ADAMTS13 autoantibody production remain unknown, and questions remain concerning their mechanism of action (ie, in addition to blocking enzymatic function, do autoantibodies also promote accelerated ADAMTS13 clearance?). Furthermore, individual epitope targets need to be mapped. Building on these exciting recent advances, continued study of ADAMTS13 and VWF holds considerable promise for advancing the understanding of TTP pathogenesis and should lead to improved treatment for this important hematologic disease.
Prepublished online as Blood First Edition Paper, March 17, 2005; DOI 10.1182/blood-2004-10-4097.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal