Abstract
We identified 13 new gene expression markers that were elevated and one marker, ANKRD15, that was down-regulated in patients with polycythemia vera (PV). These 14 markers, as well as the previously described PRV1 and NF-E2, exhibited the same gene expression alterations also in patients with exogenously activated granulocytes due to sepsis or granulocyte colony-stimulating factor (G-CSF) treatment. The recently described V617F mutation in the Janus kinase 2 (JAK2) gene allows defining subclasses of patients with myeloproliferative disorders based on the JAK2 genotype. Patients with PV who were homozygous or heterozygous for JAK2-V617F exhibited higher levels of expression of the 13 new markers, PRV1, and NF-E2 than patients without JAK2-V617F, whereas ANKRD15 was down-regulated in these patients. Our results suggest that the alterations in expression of the markers studied are due to the activation of the Jak/signal transducer and activator of transcription (STAT) pathway through exogenous stimuli (sepsis or G-CSF treatment), or endogenously through the JAK2-V617F mutation.
Introduction
Myeloproliferative disorders (MPDs) are a heterogeneous group of diseases characterized by increased hematopoiesis leading to elevated numbers of nonlymphoid cells or platelets in the peripheral blood. Diagnostic criteria for polycythemia vera (PV), essential thrombocythemia (ET), and idiopathic myelofibrosis (IMF) have been defined by the Polycythemia Vera Study Group (PVSG) and more recently by the World Health Organization (WHO).1-3 The Janus kinase (Jak) family of proteins mediate the effects of hematopoietic cytokines, for example, erythropoietin and granulocyte colony-stimulating factor (G-CSF), by phosphorylating cytoplasmic targets, including the signal transducers and activators of transcription (STATs).4 Recently, a mutation exchanging valine to phenylalanine at position 617 (V617F) of the Jak2 protein has been identified in patients with MPDs.5-8 This mutation results in the activation of the Jak2 kinase. Comparison of granulocyte mRNA expression by differential display or microarrays has been used to define biomarkers for MPDs.9-11 The value of markers, such as elevated expression of mRNAs for polycythemia rubra vera 1 (PRV1) and nuclear factor erythroid-derived 2 (NF-E2) in granulocytes,10,11 in the diagnostic workup of patients with suspected MPDs remains to be established. Here we analyzed 14 new gene expression markers as well as PRV1 and NF-E2 by quantitative polymerase chain reaction (PCR) in patients with MPDs classified according to the JAK2-V617F mutation status and in patients with exogenous stimulation of granulocytes by cytokines.
Study design
Patients
We studied 98 patients with MPDs (54 PV, 33 ET, 11 IMF); 30 patients with PV were from Italy, and the remaining 68 patients with MPD were from Switzerland. The PVSG criteria were applied for the Italian patients with PV, whereas the WHO diagnostic criteria were followed for all patients with MPD in the Swiss cohort.1-3 Four individuals with secondary erythrocytosis (SE) with elevated serum erythropoietin due to pulmonary dysfunction, 4 with secondary neutrophilia (3 due to sepsis and 1 postsurgical neutrophilia), and 28 healthy individuals were included. In addition, 3 healthy donors were examined before and after G-CSF treatment for stem-cell mobilization. The study was approved by the ethics committee of Basel (Ethikkommission Beider Basel), Switzerland. Patients provided informed consent in accordance with the Declaration of Helsinki.
RNA and DNA analyses
Isolation of granulocytes and RNA and cDNA preparation were performed as described.12 The primer sequences are listed in Table S1 (available at the Blood website; click on the Supplemental Table link at the top of the online article). Quantitative PCR was performed with the SYBR green master mix in duplicate (Applied Biosystems, Foster City, CA). The differences of threshold cycles (ΔCT) were derived by subtracting the CT value for the internal reference, ribosomal protein L19 (RPL19), from the CT values of the marker genes.13,14 One of the healthy control cDNAs was chosen as a calibrator for calculating the ΔΔCT values. The JAK2 genotyping and the quantification of the PRV1 mRNA were performed as described.13,14
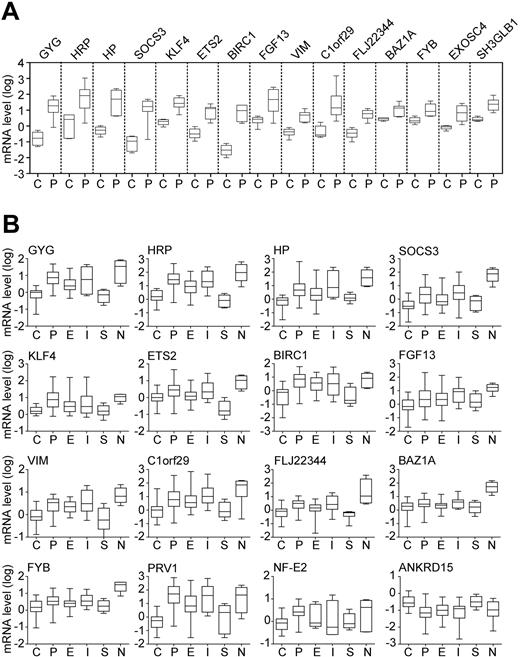
Validation of gene expression changes by quantitative PCR. (A) Marker expression in the learning cohort. Fifteen differentially expressed genes identified by microarray analysis were re-examined by real-time PCR in 4 healthy controls (C) and 7 patients with polycythemia vera (P). Boxes represent the interquartile range that contains 50% of the values; the horizontal line in the box marks the median and bars indicate the range of values. Relative gene expression is shown on a logarithmic scale (log10) and the values represent the mRNA expression relative to a healthy control calibrator sample, calculated by the ΔΔCT method. All 15 genes examined showed a statistically significant difference (P < .05; Mann-Whitney U test) between controls and PV. (B) Marker expression in the test cohort, consisting of 28 healthy controls (C), 47 patients with polycythemia vera (P), 33 patients with essential thrombocythemia (E), 11 patients with idiopathic myelofibrosis (I), 4 patients with secondary erythrocytosis (S), and 4 patients with secondary neutrophilia (N). Annotation as indicated in panel A.
Validation of gene expression changes by quantitative PCR. (A) Marker expression in the learning cohort. Fifteen differentially expressed genes identified by microarray analysis were re-examined by real-time PCR in 4 healthy controls (C) and 7 patients with polycythemia vera (P). Boxes represent the interquartile range that contains 50% of the values; the horizontal line in the box marks the median and bars indicate the range of values. Relative gene expression is shown on a logarithmic scale (log10) and the values represent the mRNA expression relative to a healthy control calibrator sample, calculated by the ΔΔCT method. All 15 genes examined showed a statistically significant difference (P < .05; Mann-Whitney U test) between controls and PV. (B) Marker expression in the test cohort, consisting of 28 healthy controls (C), 47 patients with polycythemia vera (P), 33 patients with essential thrombocythemia (E), 11 patients with idiopathic myelofibrosis (I), 4 patients with secondary erythrocytosis (S), and 4 patients with secondary neutrophilia (N). Annotation as indicated in panel A.
Microarray analysis
Total RNA from granulocytes was purified by the RNeasy total RNA isolation kit (Qiagen, Hilden, Germany). The U133A Human Genomic arrays (Affymetrix, Santa Clara, CA) were used for gene profiling as described.15
Results and discussion
Using microarray analysis, we screened for differentially expressed genes in granulocyte RNA from 7 patients with PV and 4 healthy controls. A total of 280 significant expression differences (P < .05) were observed (data not shown). The 15 most elevated genes in the microarrays were confirmed by quantitative PCR in the same 7 patients with PV and 4 controls, which constituted the learning cohort (Figure 1A). These genes encode transcription factors (ETS2, KLF4, BAZ1A), a cytokine (FGF13), proteins involved in signaling and apoptosis (SOCS3, FYB, SH3GLB1, BIRC1), a glycogen metabolism component (GYG), haptoglobin (HP) and haptoglobin-related protein (HRP), vimentin (VIM), the exosome component EXOSC4, and genes encoding proteins of unknown function (C1orf29, FLJ22344).
These results were validated in an independent test cohort consisting of 47 patients with PV and 28 healthy individuals. Thirteen of the 15 markers showed significant differences between controls and PV (Figure 1B). None of the observed changes were specific for PV, because the markers were also altered in some patients with IMF and ET. The marker expression in patients with SE and healthy controls was comparable (Figure 1B). We also examined the expression of PRV1 and NF-E2, 2 markers previously shown to be elevated in MPDs,10,11,14,16-18 and ANKRD15, a putative tumor suppressor gene located within the minimal 9pLOH region.7,19 Consistent with the previous reports,10,11,14,16-18 PRV1 and NF-E2 exhibited elevated expression in PV, IMF, and ET, whereas ANKRD15 mRNA was decreased in PV and IMF (Figure 1B).
Because granulocytes from patients with MPDs were shown to be functionally activated,20 we examined the possibility that the altered gene expression is caused by cytokine stimulation. Granulocytes from 4 patients with secondary neutrophilia showed elevated expression of all markers examined (Figure 1B), suggesting that the observed gene expression changes could be induced by cytokines. To examine the possibility that the Jak/STAT signaling pathway is involved, we determined the marker expression in granulocytes of healthy stem-cell donors before and after G-CSF administration. PRV1 was previously shown to increase after G-CSF treatment.10 We observed increased expression of all markers after G-CSF, whereas ANKRD15 mRNA was decreased (Figure 2A). The results obtained in neutrophilia and after G-CSF treatment indicate that exogenous cytokine stimulation can cause the same concerted alterations in gene expression as MPD.
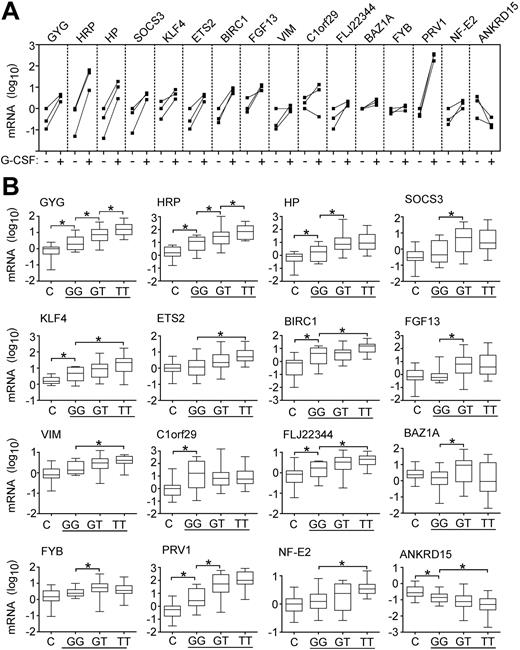
Effects of cytokine stimulation and JAK2 mutation on marker expression. (A) Expression of markers in granulocytes of 3 healthy donors before and after stem cells mobilization by G-CSF. For each individual, the gene expression is shown before (–) and after (+) G-CSF administration and the corresponding values are connected by solid lines. The mRNA levels of the markers were determined by quantitative PCR. Relative gene expression is shown on a logarithmic scale (log10) and the values represent the mRNA expression relative to a healthy control calibrator sample, calculated by the ΔΔCT method. (B) Marker expression in patients with the JAK2-V617F mutation. The mRNA level of individual markers was determined in 28 healthy controls (C) and in 54 patients with PV (horizontal line) grouped according to the JAK2 genotype. Ten patients with wild-type JAK2 (GG), 21 patients heterozygous for the JAK2-V617F mutation (GT), and 23 patients homozygous for the JAK2-V617F mutation (TT) were examined. Bars with asterisks indicate statistically significant differences between groups (P < .05; Mann-Whitney U test).
Effects of cytokine stimulation and JAK2 mutation on marker expression. (A) Expression of markers in granulocytes of 3 healthy donors before and after stem cells mobilization by G-CSF. For each individual, the gene expression is shown before (–) and after (+) G-CSF administration and the corresponding values are connected by solid lines. The mRNA levels of the markers were determined by quantitative PCR. Relative gene expression is shown on a logarithmic scale (log10) and the values represent the mRNA expression relative to a healthy control calibrator sample, calculated by the ΔΔCT method. (B) Marker expression in patients with the JAK2-V617F mutation. The mRNA level of individual markers was determined in 28 healthy controls (C) and in 54 patients with PV (horizontal line) grouped according to the JAK2 genotype. Ten patients with wild-type JAK2 (GG), 21 patients heterozygous for the JAK2-V617F mutation (GT), and 23 patients homozygous for the JAK2-V617F mutation (TT) were examined. Bars with asterisks indicate statistically significant differences between groups (P < .05; Mann-Whitney U test).
Because the JAK2-V617F mutation caused intrinsically activated Jak/STAT signaling in hematopoietic cell lines,5-7 the altered gene expression detected in patients with MPDs could be a consequence of the presence of the JAK2-V617F mutation. Therefore, we examined whether the expression of the markers correlates with the JAK2 mutation status in PV (Figure 2B). Six of the new markers and PRV1 exhibited significantly higher mRNA levels in patients heterozygous for the JAK2-V617F mutation compared to patients with PV who had wild-type JAK2. In the case of ETS2, KLF4, HRP, BIRC1, FLJ22344, VIM, and NF-E2, significant elevation of expression, and in the case of ANKRD15 significant decrease, were observed only when patients homozygous for JAK2-V617F were compared to patients with the wild-type JAK2. Although several markers exhibited a gradual alteration of expression dependent on the dosage of the mutated JAK2 gene, the gene expression differences between patients with PV who were heterozygous and homozygous for JAK2-V617F were statistically significant only in case of HRP and GYG (Figure 2B). Patients with PV who did not have mutation in JAK2 when compared to healthy controls showed significantly altered expression levels for HP, HRP, GYG, KLF4, BIRC1, C1orf29, FLJ22344, PRV1, and ANKRD15. This could be due to the presence of a small percentage of granulocytes with JAK2-V617F that contribute to the altered marker expression, which was not detected by sequencing. The presence of minor cell populations with JAK2-V617F in patients with PV was reported using allele-specific PCR.8 Alternatively, the patients with PV who did not have JAK2-V617F may carry a mutation in another component of the same signaling pathway. Interestingly, expression of SOCS3, a transcriptional target of STATs,21 was not increased in patients without JAK2-V617F.
Our results suggest that the alterations in expression of the 13 markers and ANKRD15 described in this report, as well as the 2 previously described markers PRV1 and NF-E2, are responsive to the activation of the Jak/STAT pathway through exogenous stimuli (secondary neutrophilia or G-CSF treatment), or endogenously through a gain-of-function mutation in JAK2. Thus, determining the presence or absence of the JAK2-V617F mutation will be the primary diagnostic assay and it remains to be determined how much additional information can be gained by measuring expression of biomarkers in MPDs.
Prepublished online as Blood First Edition Paper, August 4, 2005; DOI 10.1182/blood-2005-05-1889.
Supported by grants from the Swiss National Science Foundation (3100-066949.01), the Swiss Cancer League (OCS-01411-08-2003), the Lichtenstein Stiftung (R.C.S.), the Krebsliga beider Basel (R.K.), and by a grant from the Associazione Italiana per la Ricerca sul Cancro (AIRC), Milan, Italy (M.C.).
R.K. designed and performed research and wrote the article; S.-S.T., A.S.B., R.T., and D.P. performed research; M.B. contributed vital analytical tools and performed statistical analysis; A.T. contributed vital analytical tools and samples; F.P. performed research and contributed vital samples; M.C. designed research and contributed vital samples; and R.C.S. designed research and wrote the article.
An Inside Blood analysis of this article appears in the front of this issue.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Philippe Demougin for assistance with the microarray analysis and Jürg Schwaller for helpful comments on the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal