Abstract
Reticuloendotheliosis viral oncogene homolog/nuclear factor of kappa light polypeptide gene enhancer in B cells 1 (Rel/NF-κB) activation is a ubiquitous outcome of engaging Toll-like receptors (TLRs), yet the cell-type–specific functions of this pathway in response to particular microbial signals remain poorly defined. Here we show that NF-κB1 and C-Rel, Rel/NF-κB proteins induced in conventional dendritic cells (cDCs) and plasmacytoid dendritic cells (pDCs) by cytosine-phosphate-guanosine (CpG) DNA, a TLR-9 ligand, serve markedly different functions in these DC subsets. With the exception of impaired interleukin-12 (IL-12) production, cultured Nfkb1–/–C-Rel–/– cDCs responded relatively normally to CpG DNA. In contrast, CpG-treated Nfkb1–/–C-Rel–/– pDCs, which were still able to produce type I interferon and regulated on activation normal T-cell expressed and secreted (RANTES), but not IL-6 or IL-12, failed to acquire an activated dendritic phenotype and underwent apoptosis. Although the TLR-9–mediated death of Nfkb1–/–C-Rel–/– pDCs, which coincided with a failure to up-regulate the prosurvival proteins B-cell lymphoma apoptosis regulator xL (Bcl-xL) and A1, was blocked by Bcl-2 transgene expression, this inhibition of apoptosis still failed to rescue the differentiation defects. This indicated that these NF-κB transcription factors independently regulate TLR-9–mediated pDC morphogenesis and survival. Collectively, these findings establish that NF-κB1 and c-Rel, while largely dispensable for TLR-9–induced cDC activation, are critical for regulating differentiation and survival programs during pDC activation.
Introduction
Plasmacytoid dendritic cells (pDCs) represent a dendritic-cell (DC) subset that differs from the CD11chi–major histocompatibility complex II (MHCII)hi conventional DC (cDC), commonly viewed as the classic stimulators of naive T cells.1 A distinctive feature of activated pDCs is their capacity to rapidly produce high levels of type I interferon (IFN) in response to infectious agents, highlighting the importance of these DCs in innate and adaptive immune responses.2 Prior to activation, pDCs lack the dendrites and ruffled cytoplasm characteristic of mature cDCs, instead displaying a plasma-cell–like morphology and expressing lymphocyte-related cell-surface markers.3,4 When activated, pDCs differentiate into cells of typical dendritic appearance and acquire a cell-surface phenotype resembling cDCs. In addition to producing type I IFN, activated pDCs secrete the cytokines interleukin 6 (IL-6), IL-12, and regulated on activation normal T-cell expressed and secreted (RANTES).3,4 Stimuli reported to activate pDCs include viruses, single-stranded RNA, or synthetic imiquimod ligands for Toll-like receptor 7(TLR-7), plus cytosine-phosphate-guanosine (CpG) oligonucleotides (ODNs) for TLR-9.3
Various signaling pathways have been implicated in cDC development and function, among which the Reticuloendotheliosis viral oncogene homolog/nuclear factor of kappa light polypeptide gene enhancer in B cells 1 (Rel/NF-κB) pathway features prominently.5-9 The transcriptional activators of this pathway comprise dimers of related subunits (c-Rel, RelA, RelB, NF-κB1, and NF-κB2), some of which possess intrinsic transactivating properties.10 In most cells, the bulk of Rel/NF-κB dimers are retained in the cytoplasm by inhibitory (IκB) proteins.11 Diverse stimuli, including pathogen-associated molecular patterns (PAMPs), activate an IκB kinase (IKK) complex, which in turn phosphorylates IκB proteins,12 targeting them for ubiquitin-dependent proteosome-mediated degradation.11 Upon release from IκB, Rel/NF-κB dimers translocate to the nucleus, controlling transcription by binding to decameric sequences (κB elements) located within the regulatory regions of target genes.10
Nuclear up-regulation of Rel/NF-κB appears to be a universal response to engaging TLRs in many cell types, including DCs,13,14 yet little is known about the functional importance of specific family members in DC subsets activated by a particular TLR signal. Given TLR-9 expression is common to pDCs and cDCs, we focused on the function of Rel/NF-κB dimers activated in these DCs by TLR-9 ligation. Although NF-κB1/c-Rel heterodimers and NF-κB1 homodimers represented a major component of the Rel/NF-κB complexes up-regulated in both CpG1668-ODN–stimulated cDCs and pDCs, studies using DCs from mice lacking NF-κB1 and c-Rel revealed the roles of these transcription factors differed markedly during the activation of these 2 types of DCs. Cultured Nfkb1–/–C-Rel–/– cDCs displayed a typical DC morphology and cell-surface phenotype, which was enhanced upon CpG1668-ODN–induced activation. Moreover, the viability of activated Nfkb1–/–C-Rel–/– cDCs was normal. In contrast, CpG1668-ODN–treated Nfkb1–/–C-Rel–/– pDCs, while able to produce normal levels of IFN-α, failed to acquire a DC morphology or up-regulate activation markers, exhibited diminished IL-6 production, and cells underwent apoptosis. Although enforced Bcl-2 transgene expression inhibited CpG-ODN–induced Nfkb1–/–C-Rel–/– pDC apoptosis, differentiation, activation marker expression, and IL-6 production remained impaired. Collectively, these findings establish that during CpG1668-ODN–induced DC activation, NF-κB1 and c-Rel regulate multiple aspects of pDC activation, which in cDCs are controlled independently of these transcription factors.
Materials and methods
Mice
Mice were bred under special pathogen-free (SPF) conditions at the Walter and Eliza Hall Institute and used at 6 to 10 weeks of age. C-Rel–/–,15 Nfkb1–/–,16 and Nfkb1–/–C-Rel–/–17 mice were on a C57BL/6 genetic background (N:9-10 backcross generations). Vav–bcl-2 transgenic mice have been described previously.18 T cells used in allostimulatory mixed leukocyte reactions were from CBA/CaH mice. Age- and sex-matched C57BL/6 Ly-5.1 mice were used as recipients in bone marrow chimera experiments. All animal experiments were conducted with the approval of and in accordance with the guidelines of the Royal Melbourne Hospital (RMH) Animal Ethics Committee, RMH Research Foundation, Royal Melbourne Hospital, Melbourne, Victoria, Australia.
Bone marrow chimeras
Radiation chimeras generated by injecting 5 × 105Nfkb1–/–C-Rel–/– bone marrow cells into the tail veins of C57BL/6-Ly5.1 mice that had received 1100 rad of γ-irradiation were maintained on water containing 100 mg/mL neomycin sulfate. Flow cytometric analysis 6 weeks after reconstitution confirmed that more than 95% of recipient hemopoietic cells were donor-cell derived.
Purification of DCs from mouse lymphoid organs
The mAbs, fluorescent conjugates, and multicolor labeling procedures for sorting and analyses of pDCs and cDCs from lymphoid organs were exactly as described.4 DC preparations containing cDCs and pDCs were isolated from the spleen and thymus as described.4,19 For purification of cDC and pDC populations, the presorted DC preparation was labeled with anti-CD11c (N418)–fluorescein isothiocyanate (FITC) conjugate and anti-CD45RA (clone 14.8)–phycoerythrin (PE)–conjugate (BD-PharMingen, San Diego, CA) and the distinct CD11cintCD45RAhi (pDC) and CD11c+CD45RA– (cDC) populations sorted using the MoFlo instrument.
Generation of pDCs from bone marrow cultures
Cultures of Flt3 ligand supplemented bone marrow (FLBM) cells were generated as described.20 Briefly, bone marrow cells were cultured after red blood cell (RBC) lysis at a density of 1.5 × 106 cells/mL in RPMI-1640, 10% fetal calf serum (FCS), and 50 to 200 ng/mL Flt3L for 9 to 10 days. pDCs and cDCs within the FLBM cultures were sorted using the MoFlo instrument.
Cytokines and stimulants of DCs
Murine recombinant granulocyte macrophage–colony stimulating factor (rGM-CSF; 200 U/mL) and murine rIL-4 (100 U/mL) were from Immunex (Seattle, WA). Recombinant rat IFN-γ (20 ng/mL) was purchased from PeproTech (Rocky Hill, NJ). Murine rIL-12 p70 was purchased from R&D Systems (Minneapolis, MN). Oligonucleotides containing a CpG motif and fully phosphorothioated backbone (CpG1668)21 or a partially phosphorothioated backbone (CpG2216)22 were synthesized by GeneWorks Pty (Adelaide, Australia) and, unless otherwise stated, used at a concentration of 500 nM (CpG1168) or 1000 nM (CpG2216). The TLR agonists Pam-3-Cys and R848 were purchased from InvivoGen (San Diego, CA). Lipopolysaccharide (LPS) and polyinosinic-polycytidylic acid (poly I:C) were purchased from Sigma-Aldrich (Poole, United Kingdom). Beta propiolactone-inactivated (BPL) influenza A virus (A/Guangdong/25/93) was provided by Michael Hocart (Commonwealth Serum Laboratory [CSL], Melbourne, Australia). Herpes simplex virus (HSV)–1disc23 was provided by Dr Mark Suter (University of Zurich, Switzerland).
Differentiation and activation of DCs in culture
DCs were cultured at 0.5 × 106 cells/mL in round-bottomed 96-well tissue culture plates or in flat-bottomed 96- or 24-well tissue culture plates in 10% CO2 at 37°C for 2 to 72 hours. Mouse tonicity RPMI-1640 medium was used, together with the appropriate stimulants as specified in the text. For the analysis of IL-6, IL-12p70, and RANTES production, cells were incubated in a cocktail of rGM-CSF, rIL-4, and rIFN-γ previously shown to enhance IL-12p70 production in mouse cDC24 and to enhance IL-12p70 plus IL-6 production in pDCs (M. O'Keeffe, unpublished results, January 2004).
Phase-contrast microscopy
Cells cultured for 16 to 24 hours in 24- or 96-well flat-bottom culture plates were visualized (× 20 or × 40) using a Nikon ELWD03 Phase Contrast inverted microscope equipped with a 20×/0.4 objective lens (Figures 2A, 3A), and were photographed with a connected Nikon SLR camera (Nikon, Tokyo, Japan).
Mixed leukocyte reaction cultures
CD4+ T cells were purified from the pooled lymph nodes of CBA/CaH mice as described.25 Freshly isolated CD11cintCD45RAhi pDCs (5 × 103) were added to round-bottom 96-well plates containing 2 × 104 T cells in 100 μL RPMI 1640 medium in the absence or presence of 0.15 μM CpG1668-ODN. Replicate cultures were incubated at 37°C in 10% CO2 for 3.5 to 6.5 days, and at days 3.5, 4.5, 5.5, and 6.5, were pulsed with 3H-thymidine (1 μCi [0.037 MBq]/well) for 8 hours. Cells were then harvested onto glass fiber filters and thymidine incorporation determined by liquid scintillation counting. All cultures were performed in triplicate and controls, with T cells or pre-DCs only, in the absence or presence of stimulants included at each time point.
Quantitation of cytokine production
Extraction of nuclear proteins and electrophoretic mobility shift assays
A 32P-dATP end-labeled probe encompassing the κB3 site (5′-GCGGGAAATCCCCC-3′) from the murine C-Rel promoter was incubated with 1 to 2 μg nuclear extracts prepared from unstimulated or activated (2 and 12 hours) FLBM pDCs or FLBM cDCs as described.26 For supershift analysis, antibodies that specifically recognize NF-κB1, RelA, c-Rel,26 or NF-κB2 (Santa Cruz Biotechnology, Santa Cruz, CA) were incubated with nuclear extracts on ice for 30 minutes before adding radiolabeled probe. Reactions were incubated for 20 minutes at room temperature (RT), 2 μL Ficoll dye added, and the reactions fractionated on 5% nondenaturing polyacrylamide gels. Gels were then dried and exposed to autoradiography at –70°C.
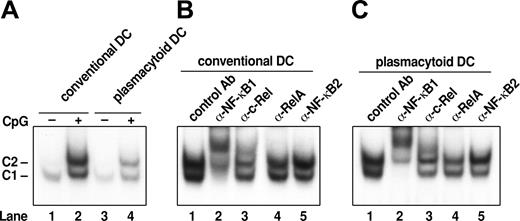
Similar patterns of Rel/NF-κB complexes are induced in cDCs and pDCs by CpG DNA. (A) Rel/NF-κB complexes induced in response to CpG. Nuclear extracts isolated from cDCs (lanes 1 and 2) and pDCs (lanes 3 and 4) prior to (lanes 1 and 3) and after CpG1668-ODN–induced activation (2 hours; lanes 2 and 4) were subjected to electrophoretic mobility shift assays (EMSA). (B, C) Antibody shifts of Rel/NF-κB complexes in CpG-activated cDCs and pDCs. Preimmune sera (lane 1) or antibodies specific for NF-κB1 (lane 2), c-Rel (lane 3), RelA (lane 4), and p52NF-κB2 (lane 5) were first incubated with nuclear extracts from wt cDCs and pDCs stimulated for 2 hours with CpG prior to performing EMSA.
Similar patterns of Rel/NF-κB complexes are induced in cDCs and pDCs by CpG DNA. (A) Rel/NF-κB complexes induced in response to CpG. Nuclear extracts isolated from cDCs (lanes 1 and 2) and pDCs (lanes 3 and 4) prior to (lanes 1 and 3) and after CpG1668-ODN–induced activation (2 hours; lanes 2 and 4) were subjected to electrophoretic mobility shift assays (EMSA). (B, C) Antibody shifts of Rel/NF-κB complexes in CpG-activated cDCs and pDCs. Preimmune sera (lane 1) or antibodies specific for NF-κB1 (lane 2), c-Rel (lane 3), RelA (lane 4), and p52NF-κB2 (lane 5) were first incubated with nuclear extracts from wt cDCs and pDCs stimulated for 2 hours with CpG prior to performing EMSA.
Western blotting
Protein blots were performed as described27 using total cell extracts isolated from 106 wt or Nfkb1–/–C-Rel–/– pDCs that were unstimulated or activated in culture for 6 hours with CpG1668-ODN (0.5 μM). Filters were probed with anti-A1 rat mAb MAB8516 (R&D Systems, Minneapolis, MN), anti–B-cell lymphoma apoptosis regulator xL (Bcl-xL) mAb clone 44 (BD Transduction Labs, San Diego, CA), purified hamster anti–mouse Bcl-2 mAb 3F11 (BD Pharmingen, San Diego, CA), or anti-HSP70 mAb (gift of D. Huang, Walter and Eliza Hall Institute) and bound antibody revealed by horseradish peroxidase (HRPO)–conjugated anti-Ig antibodies using enhanced chemoluminescence (Amersham Pharmacia Biotech, Piscataway, NJ).
Results
NF-κB1, c-Rel, and RelA are recruited to the nucleus of TLR-9–stimulated pDCs and cDCs
To identify Rel/NF-κB family members recruited to the nucleus of cDCs and pDCs in response to a common TLR signal, electrophoretic mobility gel shift assays were performed on nuclear extracts isolated from both DC populations stimulated with CpG1668-ODN (Figure 1). A single complex (C1) was present in unstimulated cDCs and pDCs (Figure 1A, lanes 1 and 3). Within 2 hours of CpG1668-ODN treatment, C1 was up-regulated, and a novel nuclear complex (C2) was induced in both types of DCs (Figure 1A, lanes 2 and 4). The composition of C1 and C2 in activated cDCs (Figure 1B) and pDCs (Figure 1C) was determined using antibodies specific for different Rel/NF-κB proteins. NF-κB1–specific antibodies supershifted C1 and almost all of C2 in both DCs (Figure 1B,C, lane 2). In cDCs, a majority of C2 also was supershifted by anti–c-Rel antibodies (Figure 1B, lane 3); the C2 complex in cDCs also was diminished by anti-RelA antibodies, albeit to a lesser extent (Figure 1B, lane 4). Antibody experiments indicated that qualitatively the C2 complex appeared to be equivalent in pDCs and cDCs, but unlike cDCs in which c-Rel predominated, in activated pDCs, c-Rel and RelA were present at similar levels in the C2 complex (Figure 1C; compare 1, 3, and 4). Neither complex was recognized by antibodies specific for NF-κB2 (lane 5) or RelB (not shown). These findings demonstrate that the C1 complex rapidly induced in cDCs and pDCs by CpG1668-ODN comprise NF-κB1 homodimers, whereas the C2 complex in both cell types consisted of NF-κB1/c-Rel and NF-κB1/RelA heterodimers.
To ascertain if Rel/NF-κB complexes change in abundance or composition during CpG-ODN stimulation of cDCs and pDCs, DNA binding studies were performed on nuclear extracts isolated from both DC subsets 12 hours after initiating activation. These experiments (Supplemental Figure S1, available at the Blood website; click on the Supplemental Figures link at the top of the online article) establish that the C1 complex still comprises NF-κB1 homodimers, but the C2 complex in cDCs and pDCs now contains less RelA than seen after 2 hours of stimulation and consists mainly of NF-κB1/c-Rel heterodimers. This indicates that the relative nuclear levels of RelA and c-Rel appear to change during the course of CpG-ODN–mediated DC activation and differentiation.
NF-κB1 and c-Rel are required to generate normal numbers of splenic cDCs and pDCs
Although nuclear expression of NF-κB1, c-Rel, and RelA all were induced in CpG1668-ODN–activated cDCs and pDCs, initially we examined the individual and combined functions of NF-κB1 and c-Rel in both DC subsets using mice lacking these transcription factors. Possible roles for RelA in TLR-9–activated DCs revealed through the use of mice lacking this transcription factor were not pursued in this study due to the complications that arise from the embryonic lethality associated with an absence of RelA.28 First, possible roles for these transcription factors in cDC and pDC development or maintenance was assessed by purifying and enumerating both DC populations in wild-type (wt), Nfkb1–/–, c-Rel–/–, and Nfkb1–/–C-Rel–/– mice. Splenic cDC (CD11chi CD45RA–) and pDC (CD11cintCD45RAhi) populations were present in each Rel/NF-κB mutant, but absolute cell numbers within each DC population varied between the different mouse strains (Table 1). In the absence of NF-κB1, cDC and pDC numbers were reduced approximately 2- and 3-fold, respectively. In the C-Rel–/– mutant, cDC numbers were normal, but pDC numbers were 2-fold lower than in C57BL/6 mice. The combined absence of NF-κB1 and c-Rel further reduced cell numbers in both the cDC and pDC populations (5- and 6-fold, respectively, compared to wt mice). Staining with antibodies to CD4 and CD8, which further delineates DC populations, established that the loss of NF-κB1 selectively diminished CD4+ cDC numbers, whereas all conventional DC subsets were significantly reduced in the combined absence of NF-κB1 and c-Rel (Table 1).
Enumeration of splenic cDC and pDC populations
. | Spleen DC populations (× 106) . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
. | Wild type . | Nfkb1-/- . | C-Rel-/- . | Nfkb1-/- C-Rel-/- . | Nfkb1-/- C-Rel-/- BM chimeras . | ||||
| Plasmacytoid | 0.93 ± 0.11 | 0.31 ± 0.08 | 0.47 ± 0.02 | 0.15 ± 0.06 | 0.43 ± 0.05 | ||||
| Conventional | 2.61 ± 0.21 | 1.19 ± 0.18 | 2.56 ± 0.21 | 0.48 ± 0.03 | 1.93 ± 0.24 | ||||
| CD4+ | 1.28 ± 0.06 | 0.48 ± 0.06 | 1.49 ± 0.24 | 0.19 ± 0.01 | ND | ||||
| CD8+ | 0.50 ± 0.09 | 0.39 ± 0.07 | 0.45 ± 0.04 | 0.23 ± 0.08 | ND | ||||
| CD4-CD8- | 0.48 ± 0.03 | 0.36 ± 0.03 | 0.54 ± 0.08 | 0.08 ± 0.01 | ND | ||||
. | Spleen DC populations (× 106) . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
. | Wild type . | Nfkb1-/- . | C-Rel-/- . | Nfkb1-/- C-Rel-/- . | Nfkb1-/- C-Rel-/- BM chimeras . | ||||
| Plasmacytoid | 0.93 ± 0.11 | 0.31 ± 0.08 | 0.47 ± 0.02 | 0.15 ± 0.06 | 0.43 ± 0.05 | ||||
| Conventional | 2.61 ± 0.21 | 1.19 ± 0.18 | 2.56 ± 0.21 | 0.48 ± 0.03 | 1.93 ± 0.24 | ||||
| CD4+ | 1.28 ± 0.06 | 0.48 ± 0.06 | 1.49 ± 0.24 | 0.19 ± 0.01 | ND | ||||
| CD8+ | 0.50 ± 0.09 | 0.39 ± 0.07 | 0.45 ± 0.04 | 0.23 ± 0.08 | ND | ||||
| CD4-CD8- | 0.48 ± 0.03 | 0.36 ± 0.03 | 0.54 ± 0.08 | 0.08 ± 0.01 | ND | ||||
Numbers of purified cDCs (total and CD4+, CD8+, and CD4-CD8- subpopulations) and pDCs isolated from wild-type, Nfkb1-/-, C-Rel-/-, and Nfkb1-/- C-Rel-/- mice or irradiated C57BL6Ly5. 1 mice engrafted with Nfkb1-/- C-Rel-/- bone marrow were determined. The results shown are the range within either five independent experiments (for total pDCs and cDCs) or three independent experiments (for cDC subsets) or two independent experiments for the chimera results.
ND indicates not determined.
To determine if reduced numbers of DC in Nfkb1–/–C-Rel–/– mice were due to a DC intrinsic defect or an abnormal environment, lethally irradiated Ly5.1+ C57BL/6 mice were reconstituted with Ly5.2+Nfkb1–/–C-Rel–/– bone marrow. Donor-derived DC populations in these chimeras were examined 5 to 7 weeks later (Table 1). The Nfkb1–/–C-Rel–/– splenic pDC numbers in engrafted recipients were higher than those in Nfkb1–/–C-Rel–/– mice, but still only approximately 50% of wild-type levels. In contrast, Nfkb1–/–C-Rel–/– cDC numbers were relatively normal in reconstituted mice. This indicated that the roles of NF-κB1 and c-Rel in generating a normal-sized cDC population were non-DC intrinsic, whereas autonomous and environmental cues controlled by these transcription factors regulate pDC numbers.
The activated phenotype of plasmacytoid but not conventional DCs is dependent on NF-κB1 and c-Rel
The roles served by NF-κB1 and c-Rel during TLR-9–mediated DC activation were assessed by stimulating wt, Nfkb1–/–, C-Rel–/–, and Nfkb1–/–C-Rel–/– splenic cDCs and pDCs in culture with CpG1668-ODN. In the absence of microbial stimuli, cultured cDCs normally form clusters of cells with protruding dendrites. All mutant cDCs behaved like wt cells, acquiring a dendritic phenotype after culture that included increased expression of MHCII, CD40, CD80, and CD86 cell-surface markers, all of which were further up-regulated by stimulation with TLR-9 ligand (Supplemental Figure S2 and data not shown).
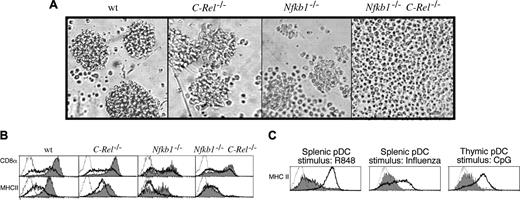
In response to CpG1668-ODN, C-Rel–/– pDCs behaved like wt cells, whereas Nfkb1–/– pDCs displayed fewer and smaller clusters of activated cells (Figure 2A). Most striking, however, were the CpG1668-ODN–treated Nfkb1–/–C-Rel–/– pDC cultures. Whereas the viability of activated wt, C-Rel–/–, and Nfkb1–/– pDCs was approximately 40% to 50% after overnight stimulation (∼16 hours), activated Nfkb1–/–C-Rel–/– pDC cultures displayed high levels (> 95%) of cell death (Supplemental Figure S3A), with increased apoptosis in CpG1668-ODN–treated Nfkb1–/–C-Rel–/– pDC cultures evident within 8 hours of stimulation. The few viable CpG1668-ODN–stimulated Nfkb1–/–C-Rel–/– pDCs were morphologically indistinguishable from untreated cells, with no sign of cluster or dendrite formation (Figure 2A).
Nfkb1–/– C-Rel–/– pDCs do not undergo morphologic or phenotypic activation in response to CpG-ODN stimulation. Purified splenic pDCs isolated from wt, Nfkb1–/–, C-Rel–/–, and Nfkb1–/–C-Rel–/– mice were incubated for 16 hours in culture in the absence or presence of CpG1668-ODN. (A) The morphology of CpG-stimulated CD11cintCD45RAhi pDCs. These data are representative of 5 independent experiments. (B) Impaired induction of the activation markers CD8 and MHCII in Nfkb1–/–C-Rel–/– pDCs. The expression of CD8 and MHCII are shown on viable unstimulated (heavy dark line) or CpG-stimulated (gray shaded) pDCs from mice of different genotypes. Unstained controls are represented by dotted histograms. This data are representative of results obtained from 5 independent experiments. (C) The Nfkb1–/–C-Rel–/– pDC activation defect is common to TLR-7 and different TLR-9 ligands. MHCII expression was monitored on wt (heavy line) and Nfkb1–/–C-Rel–/– (gray shaded) pDC stimulated with the TLR-7 ligand R848 (splenic pDCs), influenza virus (splenic pDCs), or CpG (thymic pDCs). Unstained controls are represented by dotted histograms. These data are representative of 5 and 2 experiments done with the splenic and thymic pDC populations, respectively.
Nfkb1–/– C-Rel–/– pDCs do not undergo morphologic or phenotypic activation in response to CpG-ODN stimulation. Purified splenic pDCs isolated from wt, Nfkb1–/–, C-Rel–/–, and Nfkb1–/–C-Rel–/– mice were incubated for 16 hours in culture in the absence or presence of CpG1668-ODN. (A) The morphology of CpG-stimulated CD11cintCD45RAhi pDCs. These data are representative of 5 independent experiments. (B) Impaired induction of the activation markers CD8 and MHCII in Nfkb1–/–C-Rel–/– pDCs. The expression of CD8 and MHCII are shown on viable unstimulated (heavy dark line) or CpG-stimulated (gray shaded) pDCs from mice of different genotypes. Unstained controls are represented by dotted histograms. This data are representative of results obtained from 5 independent experiments. (C) The Nfkb1–/–C-Rel–/– pDC activation defect is common to TLR-7 and different TLR-9 ligands. MHCII expression was monitored on wt (heavy line) and Nfkb1–/–C-Rel–/– (gray shaded) pDC stimulated with the TLR-7 ligand R848 (splenic pDCs), influenza virus (splenic pDCs), or CpG (thymic pDCs). Unstained controls are represented by dotted histograms. These data are representative of 5 and 2 experiments done with the splenic and thymic pDC populations, respectively.
The extent to which the different mutant pDCs underwent CpG1668-ODN–induced differentiation also was reflected in the levels of activation marker expression (Figure 2B). Activated C-Rel–/– pDCs expressed relatively normal levels of CD8 and MHCII, whereas MHCII levels that were slightly reduced in unstimulated Nfkb1–/– pDCs were up-regulated to normal levels in 50% of these cells. CD8 was only slightly up-regulated on Nfkb1–/– pDCs. Those CpG1668-ODN–treated Nfkb1–/–C-Rel–/– pDCs that remained viable displayed no up-regulation of MHCII and only a slight induction of CD8. The enhanced apoptosis of activated Nfkb1–/–C-Rel–/– pDCs, their failure to undergo morphologic transformation or to up-regulate MHCII, CD40, and CD8, appeared to be pDC intrinsic, since Nfkb1–/–C-Rel–/– pDCs derived from FLBM cultures or wt mice reconstituted with Nfkb1–/–C-Rel–/– BM cells exhibited the same defects as pDCs isolated from Nfkb1–/–C-Rel–/– mice when stimulated with CpG1668-ODN (data not shown).
The failure of CpG1668-ODN to activate Nfkb1–/–C-Rel–/– pDCs was not restricted to this oligonucleotide, nor was it TLR-9 specific, as a different immunostimulatory CpG oligonucleotide (CpG2216, Supplemental Figure S3B), beta-propialactone inactivated influenza virus, and R848 (TLR-7 ligand), all potent stimulators of pDCs, failed to activate splenic Nfkb1–/–C-Rel–/– pDCs (Figure 2C). Furthermore, thymic Nfkb1–/–C-Rel–/– pDCs were refractory to CpG1668-ODN–mediated activation (Figure 2C), indicating this defect is not specific to pDCs isolated from a particular tissue. Finally, TLR-2, TLR-3, and TLR-4 ligands, not normally stimulatory for pDCs, were still unable to activate Nfkb1–/–C-Rel–/– pDCs, establishing that these cells still followed the normal rules for TLR-mediated activation (data not shown).
Enforced Bcl-2 expression inhibits the increased apoptosis but fails to rescue other activation defects in TLR-9–stimulated Nfkb1–/–C-Rel–/– pDCs
Rel/NF-κB transcription factors control antiapoptotic pathways in part by regulating the expression of Bcl-2 and its prosurvival relatives, A1 and Bcl-xL.29 The findings that Nfkb1–/–C-Rel–/– mice had fewer pDCs and that TLR-9 stimulated Nfkb1–/–C-Rel–/– pDCs exhibited high levels of apoptosis prompted us to investigate whether these defects arose from impaired Bcl-2–like prosurvival function. Initially, this was examined by introducing a bcl-2 transgene (bcl-2T) under the transcriptional control of the hemopoietic restricted vav promoter18 onto the Nfkb1–/–C-Rel–/– background. Enumeration of cDC and pDC populations in wt.bcl-2T and Nfkb1–/–C-Rel–/–.bcl-2T mice is summarized in Table 2. Enforced Bcl-2 expression in the hemopoietic compartment normalized splenic cDC numbers on the Nfkb1–/–C-Rel–/– background, but the pDC population in Nfkb1–/–C-Rel–/–.bcl-2T mice (25% of wt.bcl-2T numbers) was only slightly larger than that of Nfkb1–/–C-Rel–/– nontransgenic littermates (15% of wt levels).
Enumeration of splenic cDC and pDC populations in wt and Nfkb1-/- C-Rel-/- mice expressing a bcl-2 transgene
. | Spleen DC populations (× 106) of bcl-2T mice . | . | |
|---|---|---|---|
| Genotype . | cDCs . | pDCs . | |
| wt.bcl-2T | 2.67 ± 0.66 | 1.95 ± 0.07 | |
| Nfkb1-/- C-Rel-/- .bcl-2T | 2.81 ± 0.21 | 0.43 ± 0.03 | |
. | Spleen DC populations (× 106) of bcl-2T mice . | . | |
|---|---|---|---|
| Genotype . | cDCs . | pDCs . | |
| wt.bcl-2T | 2.67 ± 0.66 | 1.95 ± 0.07 | |
| Nfkb1-/- C-Rel-/- .bcl-2T | 2.81 ± 0.21 | 0.43 ± 0.03 | |
Numbers of purified cDCs and pDCs isolated from wt.bcl-2T and Nfkb1-/- C-Rel-/- .bcl-2T mice were determined from 3 independent experiments. The range of values obtained between experiments is shown.
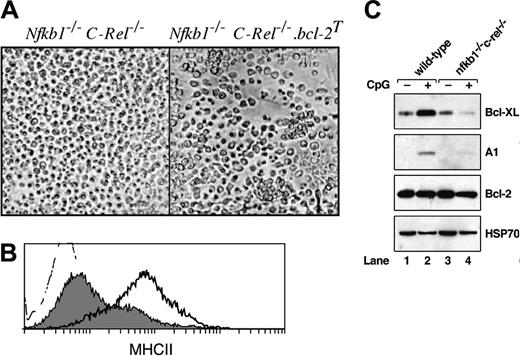
Next we examined the consequences of enforced Bcl-2 expression for TLR-9–stimulated Nfkb1–/–C-Rel–/– pDC survival and activation. pDCs isolated from wt, Nfkb1–/–C-Rel–/–, wt.bcl-2T, and Nfkb1–/–C-Rel–/–.bcl-2T mice were stimulated for 16 to 40 hours with CpG1668-ODN. The viabilities of stimulated wt.bcl-2T and Nfkb1–/–C-Rel–/–.bcl-2T pDCs each were more than 80% (results not shown), indicating that Bcl-2 transgene expression inhibited the increased TLR-9–mediated pDC death observed in the combined absence of NF-κB1 and c-Rel. Whereas activated wt.bcl-2T pDCs behaved identically to nontransgenic wt controls, forming large clusters and up-regulating MHCII and co-stimulation markers (data not shown), stimulated Nfkb1–/–C-Rel–/–.bcl-2T pDCs, while forming a few small clusters, lacked dendrites (Figure 3A), and showed no other signs of phenotypic activation, including a failure to up-regulate MHCII (Figure 3B), CD40, or CD8 (data not shown). Similar results were obtained using pDCs from FLBM cultures (Supplemental Figure S4). This indicated that morphologic transformation and survival of pDC in response to TLR-9 signals are independently regulated by NF-κB1 and c-Rel.
The capacity of transgenic Bcl-2 expression to overcome the increased apoptosis of TLR-9–activated Nfkb1–/–C-Rel–/– pDCs prompted an examination of endogenous BH3 prosurvival protein expression in wt and Nfkb1–/–C-Rel–/– pDCs (Figure 3C). In unstimulated wt (lane 1) and Nfkb1–/–C-Rel–/– (lane 3) pDCs, A1 was undetectable and Bcl-xL was expressed at the same low levels. In response to 6-hour treatment with CpG1668-ODN, a stimulation period that precedes detectable death in activated Nfkb1–/–C-Rel–/– pDC cultures, both Bcl-xL and A1 were up-regulated in wt pDCs (lane 2), but in Nfkb1–/–C-Rel–/– pDCs, A1 was barely induced and Bcl-xL expression was less than preactivation levels (lane 4). Bcl-2 expression in unstimulated wt and Nfkb1–/–C-Rel–/– pDCs was equivalent and did not change in response to CpG1668-ODN stimulation.
Activated Nfkb1–/–C-Rel–/– pDCs synthesize normal levels of IFN-α, but IL-6 and IL-12 production is reduced
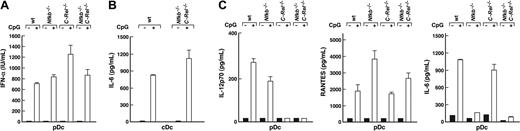
Since TLR-dependent synthesis of IFN-α is a hallmark of pDC activation, IFN-α production was examined in CpG1668-ODN–stimulated wt, Nfkb1–/–, C-Rel–/–, and Nfkb1–/–C-Rel–/– pDC cultures (Figure 4A). Activated pDC from Nfkb1–/– and Nfkb1–/–C-Rel–/– mice produced relatively normal levels of IFN-α whereas C-Rel–/– cells consistently secreted higher amounts of IFN-α than cells from wt mice. As expected, Pam-3-Cys, LPS, and polyI:C did not induce IFN-α production by pDCs (results not shown).
Other cytokines produced by CpG1668-ODN–stimulated pDCs, namely IL-12p70, IL-6, and RANTES also were assessed in wt and Rel/NF-κB mutant pDC cultures (Figure 4C). Consistent with published findings for C-Rel–/– cDCs,8 IL-12p70 production by activated C-Rel–/– and Nfkb1–/–C-Rel–/– pDCs also was abrogated. For IL-6, normal amounts were produced by activated C-Rel–/– pDCs, but IL-6 levels in the corresponding Nfkb1–/– and Nfkb1–/–C-Rel–/– pDC cultures were only approximately 10% of that produced by wt cells. This contrasted with normal IL-6 production by CpG1668-ODN–stimulated Nfkb1–/–C-Rel–/– cDCs (Figure 4B). RANTES was expressed by all Rel/NF-κB mutant pDCs, with Nfkb1–/– and Nfkb1–/–C-Rel–/– pDC consistently producing more of this cytokine than wt cells. The reduced production of IL-12p70 and IL-6 by CpG1668-ODN–activated Nfkb1–/–C-Rel–/– pDCs was not simply a result of reduced pDC viability, as wt.bcl-2T and Nfkb1–/–C-Rel–/–.bcl-2T pDC still produced IL-12p70 and IL-6 at levels equivalent to that made by their nontransgenic counterparts (Supplemental Figure S5).
Transgenic Bcl-2 expression does not rescue the morphologic or phenotypic activation defects seen in Nfkb1–/– C-Rel–/– pDC. Purified splenic pDCs isolated from bcl-2T and Nfkb1–/–C-Rel–/–.bcl-2T mice were incubated in culture for approximately 16 hours in the absence or presence of CpG1668-ODN. (A) Cell morphology in response to CpG-mediated activation. (B) Histograms of MHCII expression on CpG-stimulated wt.bcl-2T (heavy dark line) and Nfkb1–/–C-Rel–/–.bcl-2T (gray shaded) pDCs. The unstained control is depicted as the dotted histogram. Data are representative of 3 independent experiments. (C) Western blot analysis of endogenous BH3 prosurvival protein expression in pDCs. Cellular extracts isolated from equivalent numbers (106) of wild-type (lanes 1 and 2) and Nfkb1–/–C-Rel–/– (lanes 3 and 4) pDCs prior to (lanes 1 and 3) and following CpG activation for 6 hours (lanes 2 and 4) were subjected to Western blot analysis. Filters were sequentially probed with antibodies specific for Bcl-XL, A1, Bcl-2, and HSP70. These data are representative of 3 independent experiments.
Transgenic Bcl-2 expression does not rescue the morphologic or phenotypic activation defects seen in Nfkb1–/– C-Rel–/– pDC. Purified splenic pDCs isolated from bcl-2T and Nfkb1–/–C-Rel–/–.bcl-2T mice were incubated in culture for approximately 16 hours in the absence or presence of CpG1668-ODN. (A) Cell morphology in response to CpG-mediated activation. (B) Histograms of MHCII expression on CpG-stimulated wt.bcl-2T (heavy dark line) and Nfkb1–/–C-Rel–/–.bcl-2T (gray shaded) pDCs. The unstained control is depicted as the dotted histogram. Data are representative of 3 independent experiments. (C) Western blot analysis of endogenous BH3 prosurvival protein expression in pDCs. Cellular extracts isolated from equivalent numbers (106) of wild-type (lanes 1 and 2) and Nfkb1–/–C-Rel–/– (lanes 3 and 4) pDCs prior to (lanes 1 and 3) and following CpG activation for 6 hours (lanes 2 and 4) were subjected to Western blot analysis. Filters were sequentially probed with antibodies specific for Bcl-XL, A1, Bcl-2, and HSP70. These data are representative of 3 independent experiments.
IL-6 and IL-12, but not IFN-α, production by pDCs is regulated by NF-κB1 and c-Rel. Wild-type, C-Rel–/–, Nfkb1–/–, and Nfkb1–/–C-Rel–/– splenic cDCs and pDCs were stimulated in culture for approximately 18 hours in a cytokine cocktail in the absence (–, closed bars) or presence (+, open bars) of CpG1668-ODN. Culture supernatants were harvested and examined for cytokine expression by ELISA. Data are representative of 3 independent experiments. Error bars represent the variation among replicate samples within an experiment. (A) IFN-α production by pDCs. (B) IL-6 production by wt and Nfkb1–/–C-Rel–/– cDCs. (C) IL-12p70, RANTES, and IL-6 production by pDCs.
IL-6 and IL-12, but not IFN-α, production by pDCs is regulated by NF-κB1 and c-Rel. Wild-type, C-Rel–/–, Nfkb1–/–, and Nfkb1–/–C-Rel–/– splenic cDCs and pDCs were stimulated in culture for approximately 18 hours in a cytokine cocktail in the absence (–, closed bars) or presence (+, open bars) of CpG1668-ODN. Culture supernatants were harvested and examined for cytokine expression by ELISA. Data are representative of 3 independent experiments. Error bars represent the variation among replicate samples within an experiment. (A) IFN-α production by pDCs. (B) IL-6 production by wt and Nfkb1–/–C-Rel–/– cDCs. (C) IL-12p70, RANTES, and IL-6 production by pDCs.
Activated Nfkb1–/–C-Rel–/– pDCs are poor stimulators of naive T cells
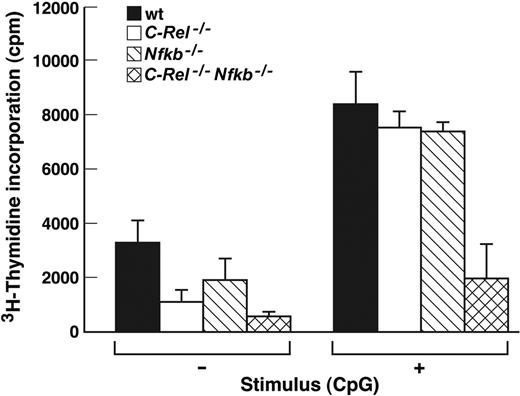
cDC activation of naive T cells in an allotypic mixed leukocyte reaction (MLR) is a hallmark of DC function. Activated pDCs also can stimulate naive T cells, albeit less efficiently than cDCs.4 To date, it remains unclear whether pDC stimulation of naive T cells depends on cell contact initiated through costimulatory molecules up-regulated during activation or by soluble mediators. Given the broad scope of the activation defects exhibited by Nfkb1–/–C-Rel–/– pDCs, we tested if this extended to T-cell activation. The relative ability of wt, C-Rel–/–, Nfkb1–/–, and Nfkb1–/–C-Rel–/– pDC to activate naive T cells into cycle in an MLR is summarized in Figure 5. Whereas CpG1668-ODN–treated wt, Nfkb1–/–, and C-Rel–/– pDCs induced similar, albeit low, levels of T-cell proliferation, CpG1668-ODN–treated Nfkb1–/–C-Rel–/– pDCs were markedly impaired in their ability to activate naive T cells. Specifically, T-cell proliferation induced by CpG1668-ODN–treated Nfkb1–/–C-Rel–/– pDCs was approximately 25% of that induced by the CpG1668-ODN–treated pDCs of all other genotypes and was within the range obtained when non–CpG1668-ODN–treated pDCs from wt, Nfkb1–/–, or C-Rel–/– mice were used. This conclusion was supported by microscopic examination of the cultures. Although T-cell proliferation and activation was evident in wells using CpG1668-ODN–treated wt, Nfkb1–/–, and C-Rel–/– pDCs, T-cell proliferation or activation was not detected in wells containing CpG1668-ODN–treated Nfkb1–/–C-Rel–/– pDCs or non–CpG1668-ODN–treated pDCs of any genotype (data not shown).
Nfkb1–/–C-Rel–/– pDCs fail to stimulate naive T cells in a mixed lymphocyte reaction. pDCs isolated from wt or Rel/NF-κB mutant mice were incubated with CBA/CaH CD4+ T cells and incubated for 4.5 days in the absence (–) or presence (+) of CpG1668-ODN. Cultures were then pulsed for 8 hours with 3H-thymidine, harvested, and incorporated radioactivity measured by scintillation counting. The data are representative of 2 independent experiments, and error bars represent variation among triplicate samples within an experiment.
Nfkb1–/–C-Rel–/– pDCs fail to stimulate naive T cells in a mixed lymphocyte reaction. pDCs isolated from wt or Rel/NF-κB mutant mice were incubated with CBA/CaH CD4+ T cells and incubated for 4.5 days in the absence (–) or presence (+) of CpG1668-ODN. Cultures were then pulsed for 8 hours with 3H-thymidine, harvested, and incorporated radioactivity measured by scintillation counting. The data are representative of 2 independent experiments, and error bars represent variation among triplicate samples within an experiment.
Discussion
Rel/NF-κB transcription factors contribute to the development, activation, and survival of diverse cell types.10,30 Here we show that NF-κB1 and c-Rel, both of which are up-regulated in cDCs and pDCs by the TLR-9 agonist CpG1668-ODN, serve markedly different roles in the morphologic transformation, activation, and function of these DC subsets. Our findings also establish that the production of IFN-α by pDCs does not require the acquisition of an activated DC phenotype, the latter of which is dependent on NF-κB1 and c-Rel.
Despite reduced numbers of cDC and pDC in mice lacking NF-κB1 and/or c-Rel, cell-surface–marker analysis shows that prior to being activated, these Rel/NF-κB–deficient DCs display a normal phenotype. This indicates these transcription factors, while necessary for generating or maintaining normal-sized DC populations, are dispensable for the differentiation of DCs from hemopoietic progenitors, a conclusion reinforced by the ability to generate cDC and pDC in Nfkb1–/–C-Rel–/– FLBM cultures. Restoration of Nfkb1–/–C-Rel–/– cDC numbers to normal levels in vivo by allowing DC development to occur in the wild-type environment of a radiation chimera or by expressing a Bcl-2 transgene in the hemopoietic compartment indicates that NF-κB1 and c-Rel regulate cDC homeostasis through stromal promotion of cDC survival. This conclusion agrees with the recent finding that the expression of endogenous Bcl-2 in secondary lymphoid organs is important for DC homeostasis.31 NF-κB1/c-Rel generation of a normal-sized pDC population appears more complex, involving a combination of DC intrinsic and environmental cues linked to survival-dependent and -independent mechanisms. Although the exact nature of the NF-κB1/c-Rel–dependent stromal signals regulating pDC and cDC numbers in vivo remains to be determined, consistent with the importance of NF-κB1/c-Rel–regulated environmental factor(s) in generating both splenic DC populations is that these defects coincide with a disruption of the splenic architecture in Nfkb1–/–C-Rel–/– mice.17
NF-κB1/c-Rel heterodimers and NF-κB1 homodimers were shown to be the major nuclear complexes expressed by CpG1668-ODN–activated cDCs and pDCs. Coupled with previous findings, this indicates the pattern of Rel/NF-κB transcription factors induced through TLR-9 may be cell-type specific. Whereas NF-κB1/c-Rel and NF-κB1 homodimers are mainly induced in B cells by CpG-ODN,32 NF-κB1/RelA is the predominant complex induced in macrophages by this stimulus.33 With CpG DNA signaling being TLR-9/MyD88 dependent,14 the induced cell-type–specific patterns of Rel/NF-κB complexes appear to be regulated independently of TLR/adaptor combinations, a conclusion supported by the finding that the same Rel/NF-κB dimers are induced in cDC by CpG1668-ODN (this paper) and LPS.7
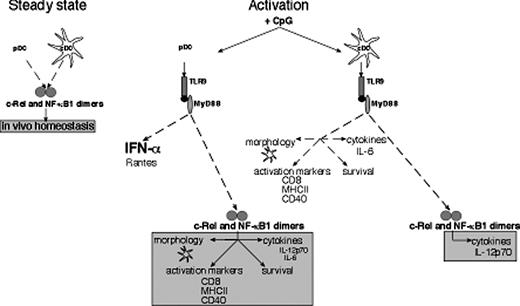
The transcription factors NF-κB1 and c-Rel serve distinct roles in conventional and plasmacytoid dendritic cells activated by engaging TLR-9. This diagram depicts the distinct roles for c-Rel and NF-κB1 in cDCs and pDCs prior to encountering microbial signals (steady state) and in response to CpG-ODN–mediated activation via TLR-9/MyD88. Survival-dependent and -independent mechanisms regulated by these transcription factors control splenic cDC and pDC numbers. For CpG-activated cDCs, c-Rel is necessary for IL-12p70 expression, whereas for CpG-stimulated pDCs, these transcription factors in addition to regulating IL-12p70 production, control IL-6 synthesis, morphologic transformation during differentiation, the expression of key activation markers, and survival.
The transcription factors NF-κB1 and c-Rel serve distinct roles in conventional and plasmacytoid dendritic cells activated by engaging TLR-9. This diagram depicts the distinct roles for c-Rel and NF-κB1 in cDCs and pDCs prior to encountering microbial signals (steady state) and in response to CpG-ODN–mediated activation via TLR-9/MyD88. Survival-dependent and -independent mechanisms regulated by these transcription factors control splenic cDC and pDC numbers. For CpG-activated cDCs, c-Rel is necessary for IL-12p70 expression, whereas for CpG-stimulated pDCs, these transcription factors in addition to regulating IL-12p70 production, control IL-6 synthesis, morphologic transformation during differentiation, the expression of key activation markers, and survival.
Despite the same Rel/NF-κB complexes being up-regulated in CpG1668-ODN–activated cDCs and pDCs, we establish that the defects associated with TLR-9 activation are remarkably different in Nfkb1–/–C-Rel–/– cDCs and pDCs (Figure 6). With the exception of reduced IL-12p70 expression, a finding consistent with p35 gene expression being regulated by c-Rel,8 the CpG1668-ODN–induced activation program in cDC appears to be mainly NF-κB1 and c-Rel independent. The finding that these transcription factors are largely dispensable for cDC activation by CpG1668-ODN is unlikely to reflect compensation by other Rel/NF-κB family members, the expression of which does not change in the absence of NF-κB1 and c-Rel (Ouaaz et al7 ; R.G., unpublished results, September 2004). While our findings for Nfkb1–/–C-Rel–/– cDC morphology and cell-surface–marker expression agree with published results for LPS activation of Nfkb1–/–C-Rel–/– cDC,7 stimulation of Nfkb1–/–C-Rel–/– cDCs with tumor necrosis factor (TNF)–receptor superfamily ligands CD40L or tumor necrosis factor activation–induced cytokine (TRANCE) leads to increased apoptosis.7 This indicates that a requirement for these transcription factors in the regulation of cDC survival depends on the nature of the activation signal.
In contrast, Nfkb1–/–C-Rel–/– pDCs display multifocal activation defects in the response to TLR-9 and TLR-7 ligands (Figure 6). These include a failure to up-regulate MHCII, CD40, and CD80, an absence of the cytoskeletal changes that typify DC morphology, plus diminished IL-12p70 and IL-6 expression. This was accompanied by high levels of cell death that coincided with a failure to induce A1 and Bcl-xL expression, BH3 prosurvival proteins encoded by Rel/NF-κB–regulated genes.29 Most of these NF-κB1/c-Rel–dependent TLR-7/9 activation defects were not simply an indirect consequence of apoptosis. Inhibition of TLR-mediated cell death by enforced Bcl-2 expression, while able to functionally substitute for the impaired expression of A1 and Bcl-xL, failed to rescue the defects associated with morphologic transformation, cell-surface marker, or cytokine expression.
Notably, the multitude and severity of the defects afflicting TLR-activated Nfkb1–/–C-Rel–/– pDCs resemble those displayed by TLR ligand–treated Nfkb1–/–C-Rel–/– B cells. In response to LPS,17,27 R848, or CpG1668-ODN (C. White, personal oral communication, June 2004), B cells lacking these transcription factors fail to up-regulate certain activation markers, blast formation and division is blocked, and apoptosis is elevated. With pDC exhibiting archetypal B-cell characteristics,34 our findings demonstrating the prominence of NF-κB1 and c-Rel as key transcriptional mediators of TLR-induced pDC activation indicate that the similarity between pDC and B cells also extends to the roles of the Rel/NF-κB signaling pathway.
The initial wave of CpG-ODN–induced gene expression is regulated through signaling pathways located downstream of TLR-9/MyD88, of which Rel/NF-κB, mitogen-activated protein kinase (MAPK), and interferon regulatory fact (IRF) feature prominently.14,35 Here we establish that of the cytokines known to be regulated by the NF-κB pathway that are expressed by TLR-activated pDCs, IL-6 and IL-12 are dependent on c-Rel and NF-κB1, whereas RANTES is regulated independently of these transcription factors. Although IFN-α genes do not appear to be direct transcriptional targets of the Rel/NF-κB pathway, a result consistent with previous findings,36 importantly in the case of pDCs, we establish that TLR-9–induced IFN-α expression is not dependent on the acquisition of a typical DC morphology or the up-regulation of costimulatory molecules. This compartmentalized regulation of activities may indicate that pDCs serve a multitude of immune functions, some of which are independent of IFN-α. Indeed, the inability of Nfkb1–/–C-Rel–/– pDC to promote the activation of naive T cells in culture, even though IFN-α expression is normal, shows that T-cell activation requires pDC functions other than or in addition to IFN-α production. With IFN-α secretion by pDCs normally preceding transformation associated with pDC activation, it is tempting to speculate that pDC function during an immune response occurs in distinct phases that come under the control of different signaling pathways. Whereas the primary or acute response might be largely dependent on high-level production of IFN-α, the subsequent evolving immune response may involve spatial or activity-dependent pDC functions akin to those typically associated with a fully activated cDC. The ability to separate the distinct events that occur during pDC activation offered by this model should in the future not only permit an in-depth genetic investigation of how TLR-mediated pDC differentiation is controlled, but may also allow an identification and dissection of distinct roles served by pDCs in an immune response.
Prepublished online as Blood First Edition Paper, July 21, 2005; DOI 10.1182/blood-2004-12-4965.
Supported by program grants from the National Health and Medical Research Council (NHMRC) (Australia) and the Leukemia and Lymphoma Society of America.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank D. Baltimore and J. Adams for mice, D. Huang for antibodies and advice, A. Strasser for critical reading of the manuscript, K. Brown and J. Merryfull for animal husbandry, M. Thomson for technical assistance, and the cell-sorting facility for expert help.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal