Abstract
Background: Cytogenetic risk categories based on conventional chromosome studies are widely used in clinical practice to make treatment decisions for AML. We evaluated the efficacy of interphase FISH to detect chromosome anomalies in the workup of young (<60 years) patients with AML.
Methods: Study subjects were enrolled in E1900, a front-line Eastern Cooperative Oncology Group (ECOG) clinical trial for AML. This is an on-going Phase III clinical trial with Daunorubicin dose-intensification ± Gemtuzumab-Ozogamicin consolidation therapy prior to stem cell transplant. This trial opened December 2002; as of February 2005, 223 patients were enrolled. The protocol was designed to collect bone marrow for both cytogenetic and FISH studies at study entry (diagnosis). Cytogenetic studies were done by local laboratories with results reviewed centrally by the ECOG Cytogenetics Committee. Each case was classified as acceptable or unacceptable based on predefined ECOG cytogenetic criteria. FISH for each patient was performed in the ECOG FISH laboratory at Mayo Clinic and utilized eight probe sets to detect t(8;21), t(9;22), t(11;var), t(15;17), inv(16), +8, −5/5q, and −7/7q (Vysis, Downer Grove, IL).
Results: 64 (29%) of 223 specimens had incomplete cytogenetic and/or FISH results. We analyzed the remaining 159 (71%) specimens with complete cytogenetic and FISH results. Results for each specimen were classified by probe set into one of the following categories:
Normal cytogenetics and normal FISH;
Abnormal cytogenetics and abnormal FISH for the anomaly the probe was designed to detect;
Abnormal cytogenetics and abnormal FISH for an anomaly the probe was not primarily designed to detect;
Normal cytogenetics and abnormal FISH;
Abnormal cytogenetics and normal FISH; or
Abnormal cytogenetics and abnormal FISH that further defined the karyotype.
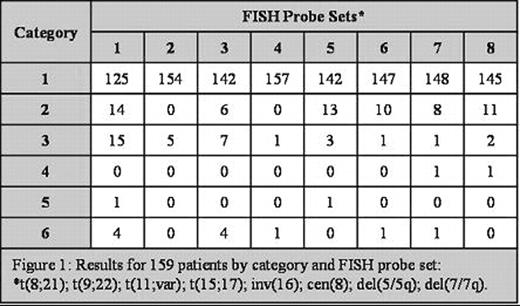
Results for 159 patients by category and FISH probe set: *t(8;21); t(9;22); t(11;var); t(15;17); inv(16); cen(8); del(5/5q); de(7/7q).
Results for 159 patients by category and FISH probe set: *t(8;21); t(9;22); t(11;var); t(15;17); inv(16); cen(8); del(5/5q); de(7/7q).
The concordance rate between cytogenetic and FISH results ranged from 97 to 100% for all probe sets and kappa analysis for concordance had a p value of <0.0001. Of the total 159 cases, discrepancies between FISH and cytogenetic results occurred in only 4 cases; two with normal cytogenetics and abnormal FISH and two with abnormal cytogenetics and normal FISH results.
Conclusions: The high level of agreement between cytogenetics and FISH demonstrates the accuracy of a panel of 8 FISH probe sets for the detection of significant abnormalities in AML. The data from this investigation support the use of FISH as an adjunct in cases of failed cytogenetic analyses to increase the yield of useful cytogenetic results in large cooperative trials. Furthermore, because of the strong correlation between cytogenetics and FISH, our results demonstrate the potential of FISH as a follow-up study of minimal residual disease in ECOG trials.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal