Abstract
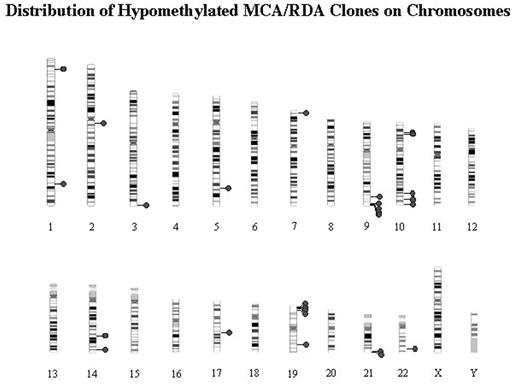
DNA methylation plays an important role in the development and aging of mammalian cells, and its dysregulation has been frequently observed in cancer cells. The purpose of this study is to investigate the involvement of aberrant DNA methylation in B chronic lymphocytic leukemia (B-CLL) cells. We compared methylation status of B-CLL cells isolated from patients with that of normal CD19+ cells isolated from health donors by methylated CpG island amplification/representative difference analysis method. 5 hypermethylated and 27 hypomethylated DNA regions were identified in B-CLL sample. Among the 27 hypomethylated regions, 5 located on chromosome 9q34, 3 on 10q25-26 and 4 on 19q13. Methylation status was confirmed by sequencing using sodium bisulfite-treated DNA samples. By comparing DNA samples from same patients at different clinical stages, we found that lower methylation density in these regions is linked with disease progression. Expression of 15 genes surrounding hypomethylated regions was studied by RT-PCR. Expression of laminin beta3 gene and melanotransferrin gene was found to be upregulated in all B-CLL cell lines as well as lymphoma cell lines comparing with normal CD19+ peripheral blood mononuclear cells. B-cell CLL/lymphoma 11b gene showed increased expression in only 2 B-CLL cell lines. For other genes, no transcriptional change was found regardless of changed DNA methylation. This study showed the predominance of DNA hypomethylation in B-CLL cells compared with hypermethylation. Hypomethylated regions clustered in a limited number of chromosomes and methylation density appeared to be inversely correlated with disease progress.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal