Nucleophosmin (NPM1) exon-12 gene mutations are the hallmark of a large acute myelogenous leukemia (AML) subgroup with normal karyotype, but their prognostic value in this AML subset has not yet been determined. We screened 401 AML patients with normal karyotype treated within the German AML Cooperative Group Protocol 99 (AMLCG99) study for NPM1 mutations. Results were related with partial tandem duplications within the MLL gene (MLL-PTD), Fms-like tyrosine kinase 3–length mutations (FLT3-LM), the tyrosine kinase domain of FLT3 (FLT3-TKD), NRAS, KIT, and CEBPA mutations and with clinical characteristics and outcome. NPM1 mutations were detected in 212 (52.9%) of 401 patients. Fourteen mutations, including 8 new variants, were identified. NPM1-mutated cases associated frequently with FLT3 mutations but rarely with other mutations. The NPM1-mutated group had a higher complete remission (CR) rate (70.5% vs 54.7%, P = .003), a trend to a longer overall survival (OS; median 1012 vs 549 days, P = .076), and significantly longer event-free survival (EFS; median 428 vs 336 days; P = .012). The favorable impact of NPM1 mutations on OS and EFS clearly emerged in the large group (264 [66.8%] of 395 cases) of normal-karyotype AML without FLT3-LM. This positive effect was lost in the presence of a concomitant FLT3-LM, since survival of the NPM1+/FLT3-LM+ double positive was similar to NPM1–/FLT3-LM+ cases. In conclusion, this study demonstrates that NPM1+/FLT3-LM– mutations are an independent predictor for a favorable outcome in AML with normal karyotype.
Introduction
Acute myelogenous leukemia (AML) is a clinically and molecularly heterogeneous disease.1 The World Health Organization (WHO) classification2 subdivides AMLs predominantly according to karyotype since recurrent chromosomal abnormalities identify distinct leukemia entities and have a major impact on prognosis.3,4 Almost 45% of AMLs show a normal karyotype by conventional cytogenetics, and the clinical and biologic features of this large cytogenetic subgroup are still poorly understood. Accordingly, most of these cases are presently classified in the WHO scheme2 under the term “acute myeloid leukemia not otherwise characterized.”
Molecular analyses indicate that AML with normal karyotype are a heterogeneous subgroup, as a number of distinct mutations have been identified. They include mutations affecting genes encoding for transcription factors (AML1, CEBPA; 2%-3% and 15%-20% of cases, respectively),5-9 receptor tyrosine kinases (FLT3, KIT; 25%-30% and of 1% of cases, respectively),10-12 and the RAS genes (10% of cases),13,14 and a partial tandem duplication within the MLL gene (MLL-PTD; 5%-10% of cases).15-19
Mutations at the exon-12 of the NPM1 gene have been recently identified as the underlying genetic lesion of a distinct, large subgroup of adult AML20 characterized by normal karyotype, aberrant cytoplasmic expression of the mutated nucleophosmin (NPM1) proteins (NPM1-cytoplasmic–positive [NPM1c+] AML), wide morphologic spectrum, multilineage involvement, increased frequency of FLT3 mutations, and CD34– negativity. NPM1c+ AML also shows a distinctive gene expression profile.21
Mutational analysis for NPM1 was restricted to a small cohort of patients, and no information on the impact of NPM1 mutations on outcome was available. The present study analyzed the type, prevalence, association with other mutations, and prognostic impact of NPM1 exon-12 mutations in 401 AML patients with normal karyotype that were treated within protocol 99 of the German AML Cooperative Group (AMLCG99).
Patients, materials, and methods
Patient samples
The study was focused on 401 AML patients with normal karyotype (age, 16.8-81.9 years; median, 60.3 years) who entered the AMLCG99 between 1999 and 2004.
Patient characteristics are given in Table 1. All samples of bone marrow or peripheral blood (with at least 70% circulating blast cells) were obtained at diagnosis and were sent to the AMLCG reference laboratory (Munich, Germany).
Characteristics of patients included in this study
Patients, no. . | 401 . |
|---|---|
| Median age, y (range) | 60.3 (16.8-81.9) |
| Sex, no. M/F | 202/199 |
| FAB subtype | |
| M0, no. (%) | 3 (0.7) |
| M1, no. (%) | 96 (23.9) |
| M2, no. (%) | 133 (33.2) |
| M4, no. (%) | 93 (23.2) |
| M5a, no. (%) | 14 (3.5) |
| M5b, no. (%) | 29 (7.2) |
| M6, no. (%) | 25 (6.2) |
| Not further specified, no. (%) | 8 (2.0) |
| Etiology, no. (%) | |
| Primary AML | 373 (93.0) |
| s-AML following MDS | 20 (5.0) |
| t-AML | 8 (2.0) |
Patients, no. . | 401 . |
|---|---|
| Median age, y (range) | 60.3 (16.8-81.9) |
| Sex, no. M/F | 202/199 |
| FAB subtype | |
| M0, no. (%) | 3 (0.7) |
| M1, no. (%) | 96 (23.9) |
| M2, no. (%) | 133 (33.2) |
| M4, no. (%) | 93 (23.2) |
| M5a, no. (%) | 14 (3.5) |
| M5b, no. (%) | 29 (7.2) |
| M6, no. (%) | 25 (6.2) |
| Not further specified, no. (%) | 8 (2.0) |
| Etiology, no. (%) | |
| Primary AML | 373 (93.0) |
| s-AML following MDS | 20 (5.0) |
| t-AML | 8 (2.0) |
s-AML indicates secondary AML after myelodysplasia; MDS, myelodysplastic syndrome; and t-AML, therapy-related AML.
All patients gave informed consent before entering the AMLCG99 study. The study design adhered to the principles of the Helsinki Declaration and was approved by the ethics committees of the participating institutions.
Treatment protocol of the German AMLCG study
Treatment consisted of double induction with a randomized comparison of TAD9 (9 days thioguanine, cytosine arabinoside, daunorubicin)/HAM (high-dose cytosine arabinoside, mitoxantrone) versus HAM/HAM followed by TAD consolidation. Patients entering complete remission (CR) received randomly assigned monthly maintenance for 3 years or autologous stem cell transplantation (AMLCG99).22 Maintenance therapy was applied every 4 weeks and consisted of 100 mg/m2 AraC (1-beta-D-arabinofuranosylcytosine) every 12 hours subcutaneously on days 1 to 5 in combination with either 45 mg/m2 daunorubicin on days 2 and 3 (courses 1, 5, 9, etc), 100 mg/m2 thioguanine every 12 hours on days 1 to 5 (courses 2, 4, 6, etc), or 1 g/m2 cyclophosphamide on day 3 (courses 3, 7, 11, etc). Treatment was delayed and doses were reduced for hematologic toxicity according to predefined criteria. Upon achievement of a cumulative dose of daunorubicin of 540 mg/m2, daunorubicin was replaced by thioguanine.
Cytogenetics
Mutational analysis
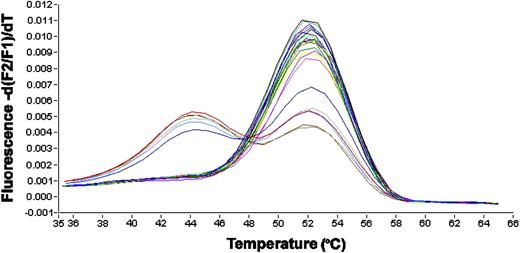
Mononucleated cells were isolated by standard Ficoll-Hypaque density gradient centrifugation. Nucleic acid isolation and cDNA synthesis were performed as previously described.10 Screening for NPM1 gene mutations was performed using a melting curve based LightCycler assay (Roche Diagnostics, Mannheim, Germany) with forward primer NPM1-F: TCCCAAAGTGGAAGCC; reverse primer NPM1-R: GGAAAGTTCTCACTCTGC; and hybridization probes NPM1–fluorescein (FL) sensor: CGGATGACTGACCAAGAGGCTATTCA-F and NPM1-anchor LC-Red-640-ATCTCTGGCCGTGGAGG-P, spanning the mutated region. The polymerase chain reaction (PCR) was carried out in a 20-μL reaction volume with 0.5 μM each of forward and reverse primer, 0.75 μM Hyb-Probes (Metabion, Munich, Germany), 4 mM MgCl2 and 2 μl LightCycler-FastStart DNA Master Hybridization Probes (Roche Diagnostics). LightCycler data were analyzed using LightCycler 3.0 software (Roche Diagnostics). Each 20 μL reaction contained 2 μL cDNA, an equivalent of about 3000 cells. Amplification was performed with 40 cycles using 55°C annealing temperature. Final melting-curve analysis was started at 28°C and continued to 70°C with slope of 0.2°C per second and continuous detection with channel F2/F1. A typical result is shown in Figure 1.
All cases that revealed an aberrant melting curve were subjected to nucleotide sequence analyses. Approximately 100 ng purified PCR products were directly sequenced with 3.3 pmol each of forward and reverse primer using the Big Dye Terminator Cycle Sequencing Kit (Applera, Darmstadt, Germany). After initial denaturation at 95°C for 5 minutes, 25 cycles at 94°C for 15 seconds and 60°C for 4 minutes were performed. Sequence analysis was performed on a 3100 Avant sequence detection system (Applied Biosystems, Foster City, CA).
Statistical analysis
Survival curves were calculated for overall survival (OS), event-free survival (EFS), and relapse-free survival (RFS) according to Kaplan-Meier and compared using the 2-sided log-rank test. OS was calculated from time of diagnosis to death and EFS was calculated from time of diagnosis to death, documentation of persistent leukemia, or relapse. Cox regression analysis related OS and EFS with analyzed parameters. Fisher exact test compared NPM1 mutation status and dichotomous variables and Student t test compared NPM1 mutation status and continuous variables. For all analyses, results were significant at a P level less than .05 at both sides. SPSS version 12.1.4 software (Chicago, IL) was used for statistical analysis.
Results
Frequency and type of NPM1 mutations
NPM1 gene mutations were detected in 212 (52.9%) of 401 AML patients. All cases were heterozygous for the mutation and retained a wild-type allele. Fourteen different mutations were characterized (Table 2). Type A, a tctg insertion between position nucleotide (nt) 960 and 961, was detected in 166 (78.3%) of 212 AML patients, followed by type D in 21 (9.9%) cases, and type B mutations in 13 (6.1%) cases. All other mutations were rare and were detected in only 1 to 3 individuals. One hundred sixty-two (97.6%) of 166 individuals had 4–base pair (bp) insertions of 10 different types between position nt 960 and 961. Two cases had 9-bp insertions between nt 965 and 966 (types E and F), 1 case had a 4-bp insertion between nt 964 and 965 (type M), and 1 case had 2 2-bp insertions between nt 960 and 961 and nt 962 and 963, respectively (type L; Table 2). All mutant proteins contained at their C-terminus mutations of at least 1 of the 2 tryptophans at position nt 288 and 290 and shared the last 5 amino acid residues VSLRK (Table 2). Moreover, all mutant proteins showed at the C-terminus a nuclear export signal (NES) motif (Table 2) that has been recently described in the original mutants A to F.27
NPM1 mutation variants in 401 AML patients with normal karyotype
Type . | n . | Wt . | Insertion . | Wt . | Insertion . | Wt . | Protein DLWQWRKSL (Wt) . |
|---|---|---|---|---|---|---|---|
| A* | 166 | gatctctg | tctg | gcagt | — | ggaggaagtctcttt | DLCLAVEEVSLRK |
| B* | 13 | gatctctg | catg | gcagt | — | ggaggaagtctcttt | DLCMAVEEVSLRK |
| C* | — | gatctctg | cgtg | gcagt | — | ggaggaagtctcttt | DLCVAVEEVSLRK |
| D* | 21 | gatctctg | cctg | gcagt | — | ggaggaagtctcttt | DLCLAVEEVSLRK |
| E* | — | gatctctg | — | gcagt | ctcttgccc | ggaggaagtctcttt | DLWQSLAQVSLRK |
| F* | — | gatctctg | — | gcagt | ccctggaga | ggaggaagtctcttt | DLWQSLEKVSLRK |
| GM | 1 | gatctctg | cagg | gcagt | — | ggaggaagtctcttt | DLCRAVEEVSLRK |
| KM | 3 | gatctctg | ccgg | gcagt | — | gaggaagtctcttt | DLCRAVEEVSLRK |
| LM | 1 | gatctctg | ccgcgg | agt | — | ggaggaagtctcttt | DLCRGVEEVSLRK |
| MM | 1 | gatctctg | — | gcag | agga | tggaggaagtcttcttt | DLWQRMEEVSLRK |
| NM | 2 | gatctctg | ccag | gcagt | — | ggaggaagtctcttt | DLCQAVEEVSLRK |
| OM | 1 | gatctctg | tttg | gcagt | — | ggaggaagtctcttt | DLCLAVEEVSLRK |
| PM | 2 | gatctctg | cttg | gcagt | — | ggaggaagtctcttt | DLCLAVEEVSLRK |
| QM | 1 | gatctctg | tcgg | gcagt | — | ggaggaagtctcttt | DLCRAVEEVSLRK |
Type . | n . | Wt . | Insertion . | Wt . | Insertion . | Wt . | Protein DLWQWRKSL (Wt) . |
|---|---|---|---|---|---|---|---|
| A* | 166 | gatctctg | tctg | gcagt | — | ggaggaagtctcttt | DLCLAVEEVSLRK |
| B* | 13 | gatctctg | catg | gcagt | — | ggaggaagtctcttt | DLCMAVEEVSLRK |
| C* | — | gatctctg | cgtg | gcagt | — | ggaggaagtctcttt | DLCVAVEEVSLRK |
| D* | 21 | gatctctg | cctg | gcagt | — | ggaggaagtctcttt | DLCLAVEEVSLRK |
| E* | — | gatctctg | — | gcagt | ctcttgccc | ggaggaagtctcttt | DLWQSLAQVSLRK |
| F* | — | gatctctg | — | gcagt | ccctggaga | ggaggaagtctcttt | DLWQSLEKVSLRK |
| GM | 1 | gatctctg | cagg | gcagt | — | ggaggaagtctcttt | DLCRAVEEVSLRK |
| KM | 3 | gatctctg | ccgg | gcagt | — | gaggaagtctcttt | DLCRAVEEVSLRK |
| LM | 1 | gatctctg | ccgcgg | agt | — | ggaggaagtctcttt | DLCRGVEEVSLRK |
| MM | 1 | gatctctg | — | gcag | agga | tggaggaagtcttcttt | DLWQRMEEVSLRK |
| NM | 2 | gatctctg | ccag | gcagt | — | ggaggaagtctcttt | DLCQAVEEVSLRK |
| OM | 1 | gatctctg | tttg | gcagt | — | ggaggaagtctcttt | DLCLAVEEVSLRK |
| PM | 2 | gatctctg | cttg | gcagt | — | ggaggaagtctcttt | DLCLAVEEVSLRK |
| QM | 1 | gatctctg | tcgg | gcagt | — | ggaggaagtctcttt | DLCRAVEEVSLRK |
— indicates no. cases detected.
LightCycler-based melting curve analysis. Depicted is the first derivation of the melting curve. The y-axis represents fluorescence intensity and the x-axis represents the temperature. Due to mismatches, mutations led to decreased melting temperatures of the hybridization probes from the amplification product. The peak at 52°C represents the wild-type allele, the peaks at 44°C represent mutated alleles. All mutated cases are heterozygous for the mutation and retain a wild-type allele, leading to double peaks in these cases.
LightCycler-based melting curve analysis. Depicted is the first derivation of the melting curve. The y-axis represents fluorescence intensity and the x-axis represents the temperature. Due to mismatches, mutations led to decreased melting temperatures of the hybridization probes from the amplification product. The peak at 52°C represents the wild-type allele, the peaks at 44°C represent mutated alleles. All mutated cases are heterozygous for the mutation and retain a wild-type allele, leading to double peaks in these cases.
NPM1 mutations and other gene mutations
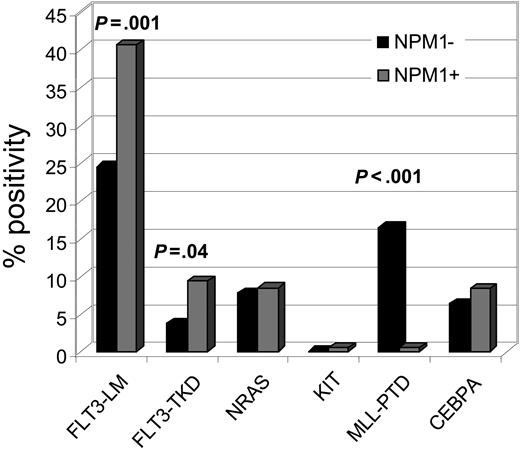
All NPM1-mutated cases were also analyzed for FLT3-LM, mutations in FLT3-TKD, NRAS- and KIT-mutations, as well as MLL-PTD. In addition, 83 cases were analyzed for CEBPA mutations in parallel. The results are shown in Figure 2. The NPM1-mutated group showed a significantly higher incidence of FLT3 mutations than the unmutated group (FLT3-LM: 40.6% vs 24.5%, P = .001; FLT3-TKD: 9.5% vs 3.8%, P = .040). MLL-PTD was detected in only 1 (0.5%) NPM1-mutated case compared with 16.4% MLL-PTD in the unmutated NPM1 group (P < .001). Thus NPM1 mutations and MLL-PTD are almost exclusive. Frequency of KIT, NRAS, and CEBPA mutations did not differ in mutated and unmutated NPM1 AML.
NPM1 mutations, morphology, and immunophenotype
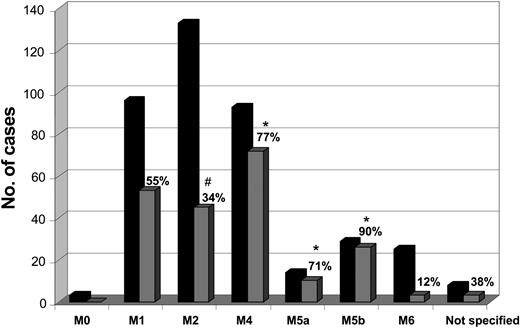
Frequencies of NPM1 gene mutations among French-American-British (FAB) subgroups were heterogenous. They were significantly lower in FAB M2 (34%) and higher in M4 (77%), M5a (71%), and especially in M5b (90%) compared with the total group (P < .001 for each comparison) (Figure 3).
The NPM1-mutated cases had higher expression of monocytic differentiation–associated antigens and a lower expression of CD34 (P < .001) and CD133 (P < .001) as well as of myeloperoxidase (P < .001) (Table 3).
Phenotypic profile of leukemic cells
. | Positive cells, % . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| Cell type, NPM1 mutation . | No. . | Mean . | SD . | P . | |||
| CD7 | NS | ||||||
| Yes | 133 | 15 | 15 | ||||
| No | 106 | 18 | 21 | ||||
| cCd3 | NS | ||||||
| Yes | 127 | 15 | 10 | ||||
| No | 99 | 16 | 12 | ||||
| CD2 | .023 | ||||||
| Yes | 136 | 7 | 8 | ||||
| No | 110 | 10 | 10 | ||||
| CD4 | < .001 | ||||||
| Yes | 131 | 36 | 25 | ||||
| No | 99 | 23 | 20 | ||||
| CD56 | .006 | ||||||
| Yes | 137 | 18 | 26 | ||||
| No | 109 | 10 | 20 | ||||
| CD19 | NS | ||||||
| Yes | 139 | 5 | 8 | ||||
| No | 114 | 5 | 11 | ||||
| Ccd22 | NS | ||||||
| Yes | 125 | 14 | 12 | ||||
| No | 97 | 12 | 8 | ||||
| CD13 | NS | ||||||
| Yes | 140 | 61 | 48 | ||||
| No | 117 | 56 | 24 | ||||
| CD33 | < .001 | ||||||
| Yes | 140 | 81 | 21 | ||||
| No | 119 | 60 | 31 | ||||
| CD65 | .043 | ||||||
| Yes | 132 | 37 | 26 | ||||
| No | 104 | 30 | 26 | ||||
| CD15 | .005 | ||||||
| Yes | 134 | 42 | 29 | ||||
| No | 108 | 32 | 26 | ||||
| CD14 | .015 | ||||||
| Yes | 131 | 25 | 57 | ||||
| No | 102 | 11 | 12 | ||||
| CD64 | < .001 | ||||||
| Yes | 131 | 36 | 30 | ||||
| No | 101 | 18 | 20 | ||||
| CD61 | .002 | ||||||
| Yes | 130 | 22 | 18 | ||||
| No | 99 | 15 | 15 | ||||
| CD235A | NS | ||||||
| Yes | 130 | 9 | 18 | ||||
| No | 102 | 13 | 19 | ||||
| Mpo | < .001 | ||||||
| Yes | 136 | 29 | 23 | ||||
| No | 113 | 41 | 25 | ||||
| Lactoferrin | < .001 | ||||||
| Yes | 135 | 8 | 10 | ||||
| No | 111 | 14 | 15 | ||||
| CD34 | < .001 | ||||||
| Yes | 139 | 8 | 13 | ||||
| No | 118 | 31 | 29 | ||||
| HLA-DR | NS | ||||||
| Yes | 140 | 51 | 31 | ||||
| No | 118 | 47 | 29 | ||||
| CD38 | .002 | ||||||
| Yes | 131 | 73 | 22 | ||||
| No | 101 | 63 | 26 | ||||
| CD117 | NS | ||||||
| Yes | 137 | 34 | 28 | ||||
| No | 111 | 39 | 25 | ||||
| TdT | NS | ||||||
| Yes | 127 | 29 | 24 | ||||
| No | 96 | 27 | 21 | ||||
| CD90 | .001 | ||||||
| Yes | 131 | 25 | 30 | ||||
| No | 106 | 13 | 21 | ||||
| CD11b | < .001 | ||||||
| Yes | 135 | 44 | 26 | ||||
| No | 107 | 32 | 23 | ||||
| CD36 | .004 | ||||||
| Yes | 132 | 38 | 27 | ||||
| No | 101 | 29 | 20 | ||||
| CD45 | < .001 | ||||||
| Yes | 133 | 95 | 11 | ||||
| No | 104 | 88 | 18 | ||||
| CD87 | < .001 | ||||||
| Yes | 129 | 28 | 25 | ||||
| No | 98 | 12 | 14 | ||||
| CD135 | < .001 | ||||||
| Yes | 137 | 53 | 28 | ||||
| No | 109 | 24 | 25 | ||||
| CD116 | < .001 | ||||||
| Yes | 131 | 47 | 30 | ||||
| No | 101 | 27 | 25 | ||||
| cCD79a | NS | ||||||
| Yes | 96 | 31 | 21 | ||||
| No | 76 | 30 | 22 | ||||
| NG2 | NS | ||||||
| Yes | 131 | 4 | 8 | ||||
| No | 101 | 3 | 4 | ||||
| CD133 | < .001 | ||||||
| Yes | 125 | 8 | 17 | ||||
| No | 96 | 18 | 22 | ||||
| CD9 | < .001 | ||||||
| Yes | 102 | 49 | 24 | ||||
| No | 76 | 34 | 25 | ||||
. | Positive cells, % . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| Cell type, NPM1 mutation . | No. . | Mean . | SD . | P . | |||
| CD7 | NS | ||||||
| Yes | 133 | 15 | 15 | ||||
| No | 106 | 18 | 21 | ||||
| cCd3 | NS | ||||||
| Yes | 127 | 15 | 10 | ||||
| No | 99 | 16 | 12 | ||||
| CD2 | .023 | ||||||
| Yes | 136 | 7 | 8 | ||||
| No | 110 | 10 | 10 | ||||
| CD4 | < .001 | ||||||
| Yes | 131 | 36 | 25 | ||||
| No | 99 | 23 | 20 | ||||
| CD56 | .006 | ||||||
| Yes | 137 | 18 | 26 | ||||
| No | 109 | 10 | 20 | ||||
| CD19 | NS | ||||||
| Yes | 139 | 5 | 8 | ||||
| No | 114 | 5 | 11 | ||||
| Ccd22 | NS | ||||||
| Yes | 125 | 14 | 12 | ||||
| No | 97 | 12 | 8 | ||||
| CD13 | NS | ||||||
| Yes | 140 | 61 | 48 | ||||
| No | 117 | 56 | 24 | ||||
| CD33 | < .001 | ||||||
| Yes | 140 | 81 | 21 | ||||
| No | 119 | 60 | 31 | ||||
| CD65 | .043 | ||||||
| Yes | 132 | 37 | 26 | ||||
| No | 104 | 30 | 26 | ||||
| CD15 | .005 | ||||||
| Yes | 134 | 42 | 29 | ||||
| No | 108 | 32 | 26 | ||||
| CD14 | .015 | ||||||
| Yes | 131 | 25 | 57 | ||||
| No | 102 | 11 | 12 | ||||
| CD64 | < .001 | ||||||
| Yes | 131 | 36 | 30 | ||||
| No | 101 | 18 | 20 | ||||
| CD61 | .002 | ||||||
| Yes | 130 | 22 | 18 | ||||
| No | 99 | 15 | 15 | ||||
| CD235A | NS | ||||||
| Yes | 130 | 9 | 18 | ||||
| No | 102 | 13 | 19 | ||||
| Mpo | < .001 | ||||||
| Yes | 136 | 29 | 23 | ||||
| No | 113 | 41 | 25 | ||||
| Lactoferrin | < .001 | ||||||
| Yes | 135 | 8 | 10 | ||||
| No | 111 | 14 | 15 | ||||
| CD34 | < .001 | ||||||
| Yes | 139 | 8 | 13 | ||||
| No | 118 | 31 | 29 | ||||
| HLA-DR | NS | ||||||
| Yes | 140 | 51 | 31 | ||||
| No | 118 | 47 | 29 | ||||
| CD38 | .002 | ||||||
| Yes | 131 | 73 | 22 | ||||
| No | 101 | 63 | 26 | ||||
| CD117 | NS | ||||||
| Yes | 137 | 34 | 28 | ||||
| No | 111 | 39 | 25 | ||||
| TdT | NS | ||||||
| Yes | 127 | 29 | 24 | ||||
| No | 96 | 27 | 21 | ||||
| CD90 | .001 | ||||||
| Yes | 131 | 25 | 30 | ||||
| No | 106 | 13 | 21 | ||||
| CD11b | < .001 | ||||||
| Yes | 135 | 44 | 26 | ||||
| No | 107 | 32 | 23 | ||||
| CD36 | .004 | ||||||
| Yes | 132 | 38 | 27 | ||||
| No | 101 | 29 | 20 | ||||
| CD45 | < .001 | ||||||
| Yes | 133 | 95 | 11 | ||||
| No | 104 | 88 | 18 | ||||
| CD87 | < .001 | ||||||
| Yes | 129 | 28 | 25 | ||||
| No | 98 | 12 | 14 | ||||
| CD135 | < .001 | ||||||
| Yes | 137 | 53 | 28 | ||||
| No | 109 | 24 | 25 | ||||
| CD116 | < .001 | ||||||
| Yes | 131 | 47 | 30 | ||||
| No | 101 | 27 | 25 | ||||
| cCD79a | NS | ||||||
| Yes | 96 | 31 | 21 | ||||
| No | 76 | 30 | 22 | ||||
| NG2 | NS | ||||||
| Yes | 131 | 4 | 8 | ||||
| No | 101 | 3 | 4 | ||||
| CD133 | < .001 | ||||||
| Yes | 125 | 8 | 17 | ||||
| No | 96 | 18 | 22 | ||||
| CD9 | < .001 | ||||||
| Yes | 102 | 49 | 24 | ||||
| No | 76 | 34 | 25 | ||||
NS indicates not significant.
Distribution of additional mutations in the NPM1-mutated group. Bars indicate the percentage of additional gene mutations in NPM1-mutated cases. FLT3-LM are frequent (40.6%) while MLL-PTD are extremely rare (0.5%).
Distribution of additional mutations in the NPM1-mutated group. Bars indicate the percentage of additional gene mutations in NPM1-mutated cases. FLT3-LM are frequent (40.6%) while MLL-PTD are extremely rare (0.5%).
NPM1 mutations and other biologic parameters
NPM1 mutations occurred at higher frequency in women (men, n = 89 [42%]; women, n = 123 [58%]; P = .04). There was no significant difference with regard to age (NPM1-mutated: median, 55.8 years; NPM1-unmutated: median, 58.1 years; P = .126). In the NPM1-mutated group the peripheral leukocyte count was significantly higher (mean, 61.1 × 109/L; median, 38.7 × 109/L; range, 0.2-486.0 × 109/L) than in the unmutated group (mean, 39.1 × 109/L; median, 10.0 × 109/L; range, × 109/L 0.1-361.0 × 109/L; P < .001).
Prognostic impact of NPM1 mutations
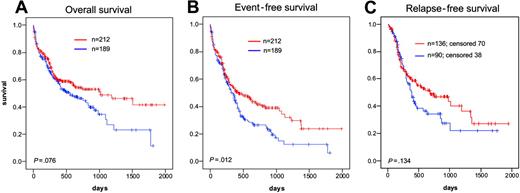
All 401 patients were evaluated for the prognostic impact of NPM1 gene mutations. The median follow-up time was 484 days. In NPM1-mutated cases, CR rates were significantly higher (70.5% vs 54.7%, P = .003); EFS was significantly longer (median, 428 vs 336 days; P = .012). Median OS showed a trend toward better prognosis (1012 days vs 549 days; P = .076) (Figure 4A). EFS was significantly longer (median, 428 vs 336 days; P = .012) (Figure 4B). RFS did not yet show significant difference (median, 473 vs 386 days; P = .134) (Figure 4C). However the curves split after 1 year and after longer follow-up a significant difference may come out. All of these differences were not related to different treatment modalities assigned by randomization.
Distribution of NPM1-mutated cases in cytomorphologic subtypes. The black columns give the total number of analyzed cases in this FAB group; gray columns give the number of positives within the respective FAB group. On top of the gray columns the percentages of positive cases within the respective FAB groups are indicated. *Significantly more than in the total. #Significantly less than in the total.
Distribution of NPM1-mutated cases in cytomorphologic subtypes. The black columns give the total number of analyzed cases in this FAB group; gray columns give the number of positives within the respective FAB group. On top of the gray columns the percentages of positive cases within the respective FAB groups are indicated. *Significantly more than in the total. #Significantly less than in the total.
Other factors impacting on EFS were leukocyte count (P < .001), platelets (P = .007), age (P < .001), and FLT3-LM (P = .006). Multivariate analysis showed that the prognostic impact of NPM1 mutations on EFS is independent of all these other factors (Table 4).
Summary of multivariate analysis on impact on EFS
. | P . | Relative risk . |
|---|---|---|
| NPM1+/FLT3-LM- | < .001 | 0.527 |
| WBC count | < .001 | 1.035* |
| Platelet count | NS | NA |
| Age | < .001 | 1.313† |
. | P . | Relative risk . |
|---|---|---|
| NPM1+/FLT3-LM- | < .001 | 0.527 |
| WBC count | < .001 | 1.035* |
| Platelet count | NS | NA |
| Age | < .001 | 1.313† |
WBC indicates white blood cell; NS, not significant; NA, not applicable.
Per 10 × 109/L.
Per 10 years.
Prognostic relevance of additional mutations
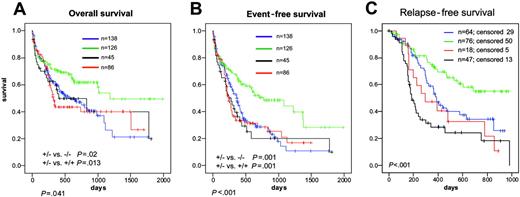
NPM1andFLT3-LMmutations. As there was a high coincidence of NPM1 mutations with FLT3-LM, survival data were evaluated for the 4 groups, NPM1–/FLT3-LM– (n = 138), NPM1–/FLT3-LM+ (n = 45), NPM1+/FLT3-LM– (n = 126), and NPM1+/FLT3-LM+ (n = 86). Median OS was 601, 405, 1183, and 321 days, respectively (P = .041). OS was significantly longer in NPM1+/FLT3-LM– than in both NPM1–/FLT3-LM– (P = .022) and NPM1+/FLT3-LM+ (P = .014) and especially in NPM1+/FLT3-LM– versus all others (P = .005) (Figure 5A). Thus, when FLT3-LM is associated with mutated NPM1 it shifts NPM1+ cases into the poor prognosis group. No difference in OS emerged between NPM1–/FLT3-LM+ and NPM1+/FLT3-LM+ cases (P = .644).
Kaplan-Meier analysis of AML with normal karyotype bearing mutated or mutated NPM1. (A) OS. (B) EFS. (C) RFS. Red lines indicate mutated NPM1; blue lines, unmutated NPM1. Results were significant at a level of P < .05 at both sides.
Kaplan-Meier analysis of AML with normal karyotype bearing mutated or mutated NPM1. (A) OS. (B) EFS. (C) RFS. Red lines indicate mutated NPM1; blue lines, unmutated NPM1. Results were significant at a level of P < .05 at both sides.
The same pattern was found for EFS. Median EFS was 773 days (NPM1+/FLT3-LM–), 365 days (NPM1–/FLT3-LM–), 279 days (NPM1–/FLT3-LM+), and 234 days (NPM1+/FLT3-LM+), respectively (P < .001). EFS was longer in NPM1+/FLT3-LM– compared with NPM1–/FLT3-LM– (P = .001) and NPM1+/FLT3-LM+ (P = .001) as well as to NPM1+/FLT3-LM– versus all others (P = .001). No difference in EFS emerged between NPM1–/FLT3-LM+ and NPM1+/FLT3-LM+ cases (P = .885) (Figure 5B). The RFS was significantly better for the NPM1+/FLT– group compared with all other 3 groups (P < .001).
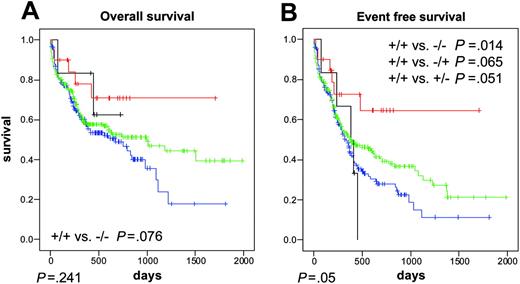
NPM1andFLT3-TKD mutations. We analyzed the combination of NPM1 and FLT3-TKD mutations: NPM1–/FLT3-TKD– (n = 153), NPM1–/FLT3-TKD+ (n = 6), NPM1+/FLT3-TKD– (n = 192), and NPM1+/FLT3-TKD+ (n = 20). Median OS was 676 days, not reached, 992 days, and not reached, respectively. There was a trend to a longer OS in the double-mutated NPM1+/FLT3-TKD+ than in the double-negative NPM1–/FLT3-TKD– (P = .076) (Figure 6A). Median EFS was 336 days, 385 days, 373 days, and median not reached, respectively (P = .051). EFS of the double-mutated NPM1+/FLT3-TKD+ was significantly longer than the double-negative NPM1–/FLT3-TKD– (P = .014) (Figure 6B).
NPM1and other mutations(NRAS, AML1, MLL-PTD, KIT,andCEBPA). Additional NRAS mutations (n = 18) did not have any prognostic impact in cases with mutated NPM1 (data not shown). AML1, MLL-PTD, KIT, or CEBP A mutations were rarely found in combination with NPM1, and thus prognostic relevance could not be analyzed.
Discussion
In this study, we determined the frequency and type of exon-12 NPM1 gene mutations in 401 AML patients with normal karyotype treated within study 99 of the AMLCG, determined their association with other gene mutations, and demonstrated the impact of NPM1 mutations on prognosis.
Kaplan-Meier analysis of AML with normal karyotype and different NPM1 and FLT3-LM status. The prognostically favorable impact of NPM1 mutations was overcome by an additional FLT3-LM. (A) OS. (B) EFS. (C) RFS. Blue lines indicate NPM1–/FLT3-LM–; green lines, NPM1+/FLT3-LM–; black lines, NPM1–/FLT3-LM+; and red lines, NPM1+/FLT3-LM+. Results were significant at a level of P < .05 at both sides.
Kaplan-Meier analysis of AML with normal karyotype and different NPM1 and FLT3-LM status. The prognostically favorable impact of NPM1 mutations was overcome by an additional FLT3-LM. (A) OS. (B) EFS. (C) RFS. Blue lines indicate NPM1–/FLT3-LM–; green lines, NPM1+/FLT3-LM–; black lines, NPM1–/FLT3-LM+; and red lines, NPM1+/FLT3-LM+. Results were significant at a level of P < .05 at both sides.
Point mutations at exon-12 of the NPM1 gene occurred more frequently than any other gene mutations (52.9% of all normal karyotype AMLs). Fourteen different NPM1 mutations were identified, including 8 previously unreported variants. All 14 mutations were heterozygous and resulted in frame shifts in the region encoding the C-terminus of the NPM1 protein that led to replacement of the last 7 amino acids with 11 different residues (the last 5 amino acids, VSLRK, being common to all variants) and substitution of at least one of 2 tryptophan residues at positions nt 288 and 290. Recently, Nakagawa et al27 reported a nuclear export signal (NES) motif in the first 6 described NPM1 mutants (A to F) and hypothesized that this motif might be responsible for the aberrant NPM1 cytoplasmic expression. This assumption is supported by the fact that all 8 newly detected NPM1 mutant proteins also all bear an NES motif at the protein C-terminus. Thus, despite genetic variations, all mutations result in distinct changes at the extreme C-terminus of the NPM1 mutant proteins.
Within the group of AML patients with normal karyotype, NPM1-mutated cases had a significantly higher rate of CR compared with wild-type NPM1 cases (70.5% vs 54.7%; P = .003). This finding confirms, at mutation level, previous findings mainly based on the aberrant cytoplasmic expression of the NPM1 protein.20
Most importantly, however, this study demonstrates a favorable impact of NPM1 mutations on outcome. In fact, NPM1-mutated AML patients with normal karyotype had a significantly longer EFS (median, 428 vs 336 days; P = .012) and a strong trend to a long OS (median, 1012 vs 549 days; P = .076) as compared with NPM1 wild-type cases. This finding is of major clinical importance since it strongly suggests that NPM1 mutations may allow dissection of the heterogenous group of AML with normal karyotype into prognostically different subgroups. This thesis is supported by multivariate analysis revealing NPM1 mutations as an independent prognostic factor.
Further analyses were performed to assess the impact of NPM1 mutations in the context of other molecular aberrations. The prognostic value of NPM1 mutations clearly emerged in the large group of AML patients with normal karyotype (about 70%) that lacked FLT3-LM. Analysis for NPM1 clearly identified patients with mutated NPM1 and FLT3-LM– as having the better prognosis. This subgroup accounted for 31.4% of all AML cases with normal karyotype. In the group of NPM1+/FLT3-LM– category, NPM1 mutations are presently the only identifiable genetic lesions that also promise to serve as a marker for monitoring minimal residual disease. The favorable impact of NPM1 mutations on prognosis was lost in the presence of a concomitant FLT3-LM, a recognized predictor of poor prognosis in AML with normal karyotype.10,11,14,25 Thus, prognosis of NPM1/FLT3-LM double-mutated cases was as unfavorable as the cases bearing NPM1 wild type and an FLT3-LM. The prognostically favorable effect of NPM1 mutations on RFS was especially strong when the FLT3-LM was taken into account. FLT3-LM has formerly been shown to be associated with a high relapse rate.10-12 In contrast, our data show that NPM1+/FLT3-LM– cases have a remarkably low relapse rate.
The overall prognostic significance of FLT3-TKD mutations is still unclear. However, in cases with mutated NPM1, FLT3-TKD mutations seem to be associated with further improvements in OS and EFS when compared with NPM1-mutated cases without FLT3-TKD mutations.
Partial tandem duplications within the MLL gene (MLL-PTD) and CEBPA mutations had a very low probability for coincidence with NPM1 mutations. In contrast, both FLT3-LM and mutations in the FLT3 tyrosine kinase domain (FLT3-TKD) occurred more frequently in cases with mutated genes than with unmutated NPM1 gene. This finding is in keeping with the hypothetical model that at least 2 hits of different mutation types are needed to induce AML28,29 (ie, type I mutations, such as in FLT3, KIT, or RAS genes that increase proliferation, and type II mutations such as AML1-ETO or PML-RARA fusion genes, which block differentiation).
So far, it is unclear why NPM1 mutations are associated with better response to induction therapy and outcome. Since the mutated NPM1 proteins maintain the dimerization domain and are therefore able to form heterodimers with the wild-type NPM1, it is likely that this may result in subcellular dislocation of the wild-type protein by the mutated NPM1. Altered distribution in cell compartments may possibly interfere with the normal functions of NPM1, a nucleocytoplasmic shuttling protein30 mainly found in the nucleolus,31,32 which plays a key role in the regulation of the adenosine diphosphate ribosylation factor–tumor protein 53 (ARF-p53) pathway33-35 and centrosome duplication.36 As daunorubicin was shown to induce nucleoplasmic dislocation of NPM1, which was associated with increased apoptosis,37 the subcellular relocation of wild-type NPM1 in NPM1c+ AML may possibly lead to increased sensitivity to chemotherapeutic agents.
Kaplan-Meier analysis of AML with normal karyotype and different NPM1 and FLT3-TKD status. The prognostically favorable impact of NPM1 mutations was overcome by an additional FLT3-LM. (A) OS. (B) EFS. Blue lines indicate NPM1–/FLT3TKD– (n=153); green lines, NPM1+/FLT3TKD– (n=192); black lines, NPM1–/FLT3TKD+ (n=6); and red lines, NPM1+/FLT3TKD+ (n=20). Results were significant at a level of P < .05 at both sides.
Kaplan-Meier analysis of AML with normal karyotype and different NPM1 and FLT3-TKD status. The prognostically favorable impact of NPM1 mutations was overcome by an additional FLT3-LM. (A) OS. (B) EFS. Blue lines indicate NPM1–/FLT3TKD– (n=153); green lines, NPM1+/FLT3TKD– (n=192); black lines, NPM1–/FLT3TKD+ (n=6); and red lines, NPM1+/FLT3TKD+ (n=20). Results were significant at a level of P < .05 at both sides.
The prevalence of NPM1 mutations in females (58% vs 42%; P = .04) detected in this study appears to be the first genetic aberration in AML with a sex prevalence. Sex-prevalent mutations have been found only for the 5q– syndrome in females,38 and hypereosinophilic syndrome (HES) in males.39
Gene expression profiling identified 2 prognostically relevant subgroups (named I and II) in AML with normal karyotype in a recent analysis.40 However, they clearly differ from our subgroups of mutated and unmutated NPM1 in terms of survival, FAB subtypes, frequency of FLT3-LM, sex, and white blood cell count. In the future, gene expression profiling and mutational analysis for NPM1 and FLT3 may be combined to build up a new prognostic model for AML with normal karyotype.
In conclusion, detection of NPM1 mutations by molecular techniques and/or looking at the aberrant cytoplasmic expression of the NPM1 protein by immunohistochemistry20 should be included in the diagnostic work-up of AML patients with normal karyotype, since it helps dissecting this heterogenous cytogenetic category into prognostically different subgroups.
Prepublished online as Blood First Edition Paper, August 2, 2005; DOI 10.1182/blood-2005-06-2248.
Supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC). An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors are indebted to all participating centers of the AMLCG. We are grateful to Madlen Fuchs, Theresa Förster, Nina Leopold, and Gudrun Mellert for excellent technical assistance. We thank Claudia Tibidò for helpful secretarial assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal