We investigated the association between haplotypes of fibrinogen alpha (FGA), beta (FGB), and gamma (FGG), total fibrinogen levels, fibrinogen γ′ (γA/γ′ plus γ′/γ′) levels, and risk for deep venous thrombosis. In a population-based case-control study, the Leiden Thrombophilia Study, we typed 15 haplotype-tagging single nucleotide polymorphisms (htSNPs) in this gene cluster. None of these haplotypes was associated with total fibrinogen levels. In each gene, one haplotype increased the thrombosis risk approximately 2-fold. After adjustment for linkage disequilibrium between the genes, only FGG-H2 homozygosity remained associated with risk (odds ratio [OR], 2.4; 95% confidence interval [95% CI], 1.5-3.9). FGG-H2 was also associated with reduced fibrinogen γ′ levels and reduced ratios of fibrinogen γ′ to total fibrinogen. Multivariate analysis showed that reduced fibrinogen γ′ levels and elevated total fibrinogen levels were both associated with an increased risk for thrombosis, even after adjustment for FGG-H2. A reduced fibrinogen γ′ to total fibrinogen ratio (less than 0.69) also increased the risk (OR, 2.4; 95% CI, 1.7-3.5). We propose that FGG-H2 influences thrombosis risk through htSNP 10034C/T [rs2066865] by strengthening the consensus of a CstF site and thus favoring the formation of γA chain above that of γ′ chain. Fibrinogen γ′ contains a unique high-affinity, nonsubstrate binding site for thrombin, which seems critical for the expression of the antithrombin activity that develops during fibrin formation (antithrombin 1).
Introduction
Fibrinogen—the precursor of fibrin, the end-product of blood coagulation—is an essential component of the hemostatic system (for a review, see Mosesson et al1 ). Fibrinogen is converted to fibrin through limited proteolysis by thrombin, which exposes polymerization sites on the fibrin monomers. These monomers spontaneously associate to form insoluble fibrin. Activated factor XIII (subunit A) forms covalent bonds between adjacent fibrin monomers (for reviews, see Mosesson et al1 and Lorand2 ). These cross-links strengthen the fibrin clot and increase its resistance to degradation by the fibrinolytic system.3-5
Fibrinogen is a plasma glycoprotein with a molecular weight of 340 kDa and is primarily synthesized by hepatocytes. It circulates in plasma at a concentration of approximately 9 μM (3 g/L). Fibrinogen molecules are elongated, 45-nm long structures with 2 outer D domains that are connected by a coiled-coil segment to a central E-domain. They consist of 2 symmetric half molecules, each containing a set of 3 different polypeptide chains termed Aα, Bβ, and γ.1,6 The 3 chains are encoded by 3 separate genes, fibrinogen alpha (FGA), fibrinogen beta (FGB), and fibrinogen gamma (FGG), clustered in a region of approximately 50 kb on chromosome 4q31.3. The FGG gene contains 10 exons and is oriented in tandem with the FGA gene, which contains 6 exons. They are transcribed in the direction opposite the FGB gene, which is located downstream of the FGA gene and contains 8 exons.7 The most common haplotypes of each of the 3 fibrinogen genes have been reported by SeattleSNPs.8 These haplotypes are thought to represent all frequent gene and protein variants that exist in Americans of European descent.
Alternative splicing may occur in both the FGA and the FGG genes. The predominant Aα chain of circulating fibrinogen contains 610 amino acid residues, whereas the alternative Aα chain (1%-2% of Aα chains)9 contains 846 amino acid residues. The Bβ chain consists of 461 amino acids. The most abundant form of the γ chain, γA, consists of 411 amino acid residues, whereas the variant γ′ (γB) chain (7%-15% of γ chains)10 contains 427 amino acid residues.6
Abnormalities of fibrinogen have been reported to affect the risk for deep venous thrombosis. In a large case-control study, elevated levels of plasma fibrinogen were found to increase the risk for thrombosis, primarily in the elderly.11,12 The precise mechanism of this effect is unknown, though multiple mechanisms have been proposed.12,13 In addition, genetic variants of fibrinogen (dysfibrinogenemias) have been found in patients with thrombosis and prolonged thrombin time.14,15 Most of these patients have a mutation in the FGA or the FGG gene, though the precise relation between carriership of these mutations and venous thrombosis is poorly documented.16
We hypothesized that relatively common variations among the fibrinogen genes might exist and might influence the risk for deep venous thrombosis. Such variations would be part of the existing haplotypes and might affect fibrinogen levels, the formation of the fibrin network structure, or the sensitivity of the fibrin clot to the fibrinolytic system. To test our hypothesis, we selected haplotype-tagging single nucleotide polymorphisms (htSNPs) specific for each of the common haplotypes of the 3 fibrinogen genes and typed them in a large population-based case-control study on risk factors for venous thrombosis, the Leiden Thrombophilia Study (LETS). Subsequently, the efficiency of alternative splicing of the FGG gene was studied by measuring the levels of the alternatively spliced variant of the fibrinogen γ chain (fibrinogen γ′; γA/γ′ heterodimers plus γ′/γ′ homodimers) in all subjects.
Patients, materials, and methods
Study population
The design of LETS has been described in detail elsewhere.17,18 We included 474 consecutively diagnosed patients with objectively confirmed first episodes of deep vein thrombosis and 474 controls frequency matched for sex and age. Patients with active cancer were excluded. Control subjects were acquaintances or partners of the patients and had no cancer history. Mean age for both groups was 45 years (ranges, 15-69 years for patients; 15-72 years for controls). Each group consisted of 272 (57.4%) women and 202 (42.6%) men. Venous blood was collected into 0.1 vol of 0.106 M trisodium citrate. Plasma was prepared by centrifugation for 10 minutes at 2000g at room temperature and was stored at -70°C. High-molecular-weight DNA was isolated from leukocytes by standard methods and was stored at -20°C. DNA samples were available from 471 patients and 471 controls; plasma samples were available from 473 patients and 474 controls. Approval of this study was obtained from the Institutional Review Board of the Leiden University Medical Center. Informed consent was provided according to the Declaration of Helsinki.
Genetic analysis
All 3 fibrinogen genes were resequenced in 23 persons of European-American descent by SeattleSNPs.8 Sixty-five polymorphic sites were identified in these persons. For each gene, haplotypes for the 46 chromosomes were reconstructed from the unphased SNP genotype data using the software PHASE2.0.19 Five haplotypes in the FGG gene, 7 in the FGA gene, and 7 in the FGB gene were found.8,20 To tag these haplotypes, we identified 15 htSNPs (Figure 1A)21-23 —4 in FGG (129A>T [rs2066854], 5836G>A [rs2066860], 7874G>A [rs2066861], 9340T>C [rs1049636]), 5 in FGA (251G>A [rs2070006], 3655G>A [rs2070014], 3807T>C [rs2070016], 3845G>A [rs2070017], 6534A>G [rs6050]), and 6 in FGB (1038G>A [rs1800791], 1643C>T [rs1800788], 3471C>T [rs2227432], 9952A>C [rs2227421], 10149C>T [rs2227439], 11046C>T [rs209502]) (numbering according to SeattleSNPs8 ; GenBank accession numbers AF350254 for FGG, AF361104 for FGA, and AF388026 for FGB). For example, the FGG 9340T>C polymorphism is haplotype-tagging for FGG-H3 because the rare allele (9340C) is only present in FGG-H3. Genotyping of FGB was performed by polymerase chain reaction and restriction fragment length polymorphism analysis. Genotyping of FGA and FGG was performed using the 5′ nuclease/TaqMan assay.24 Polymerase chain reactions with fluorescent allele-specific oligonucleotide probes (Assay-by-Design/Assay-on-Demand; Applied Biosystems, Foster City, CA) were performed on a PTC-225 thermal cycler (Biozym, Hessisch Oldendorf, Germany), and fluorescence end point reading for allelic discrimination was performed on an ABI 7900 HT (Applied Biosystems). Primer sequences, probe sequences, and restriction enzymes used are available on request.
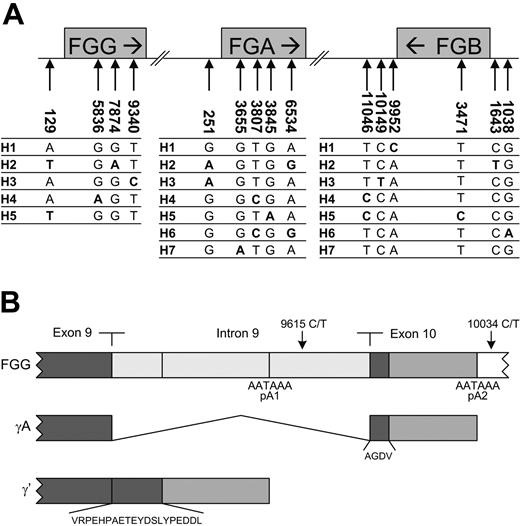
Haplotype structure of the fibrinogen gene cluster. (A) Haplotypes and typed htSNPs of the fibrinogen gene cluster. Numbering is according to Seattle-SNPs.8 The numbering of the haplotypes in the 3 genes is arbitrary. (B) Alternative mRNA processing of γ chain mRNA. The γA chain is translated from mRNA in which all 9 introns of the pre-mRNA were removed and polyadenylation occurred downstream of exon 10 at polyadenylation site 2 (pA2). In contrast, the γ′ chain arises from alternative processing of the FGG pre-mRNA. Intron 9 is not removed, and polyadenylation occurs at an alternative site located in this intron at polyadenylation site 1 (pA1). This leads to the translation of a polypeptide with a unique 20-amino acid extension encoded by intron 9 substituted for the carboxyl terminal 4 amino acids of the γ chain encoded by exon 10.21,22 This variant chain comprises approximately 7% to 15% of the fibrinogen γ chain found in plasma.10 Nearly all the γ′ protein occurs in vivo as a heterodimer with the γA variant in which one D region contains a γ′ carboxyl terminus and the other a γA carboxyl terminus (γA/γ′ fibrinogen).23 SNPs 9615C>T and 10034C>T are specific for haplotype 2 of FGG.
Haplotype structure of the fibrinogen gene cluster. (A) Haplotypes and typed htSNPs of the fibrinogen gene cluster. Numbering is according to Seattle-SNPs.8 The numbering of the haplotypes in the 3 genes is arbitrary. (B) Alternative mRNA processing of γ chain mRNA. The γA chain is translated from mRNA in which all 9 introns of the pre-mRNA were removed and polyadenylation occurred downstream of exon 10 at polyadenylation site 2 (pA2). In contrast, the γ′ chain arises from alternative processing of the FGG pre-mRNA. Intron 9 is not removed, and polyadenylation occurs at an alternative site located in this intron at polyadenylation site 1 (pA1). This leads to the translation of a polypeptide with a unique 20-amino acid extension encoded by intron 9 substituted for the carboxyl terminal 4 amino acids of the γ chain encoded by exon 10.21,22 This variant chain comprises approximately 7% to 15% of the fibrinogen γ chain found in plasma.10 Nearly all the γ′ protein occurs in vivo as a heterodimer with the γA variant in which one D region contains a γ′ carboxyl terminus and the other a γA carboxyl terminus (γA/γ′ fibrinogen).23 SNPs 9615C>T and 10034C>T are specific for haplotype 2 of FGG.
Sequencing
In selected participants (see “Results” for selection criteria), the promoter, 5′ untranslated region (UTR), exons, intron/exon boundaries, and 3′ UTR of the fibrinogen gamma gene were sequenced on an ABI PRISM 310 Genetic Analyzer (Perkin Elmer, Boston, MA). Reactions were performed using the ABI PRISM BigDye Terminator Cycle Sequencing kit (Perkin Elmer). Primer sequences are available on request.
Fibrinogen measurement
Total fibrinogen was determined previously11,12 according to the Clauss method using Dade thrombin reagent (Baxter, Miami, FL). The test was performed on an Electra 1000 (MLA; Pleasantville, NY). To simplify the calculation of the fibrinogen γ′ to total fibrinogen ratio (γ′/γ ratio), total fibrinogen levels were expressed in units per deciliter, where 100 U/dL corresponds to 2.79 g/L. Total fibrinogen as measured by the Clauss method corresponded well with total fibrinogen antigen as measured by enzyme-linked immunosorbent assay (ELISA) using commercial polyclonal rabbit antifibrinogen antibodies (DAKO A/S, Glostrup, Denmark) (R = 0.95; n = 60).
Fibrinogen γ′ antigen measurement
Fibrinogen γ′ (γA/γ′ plus γ′/γ′ fibrinogen) antigen levels were measured by ELISA using an antibody (2.G2.H9; Campro Scientific, Veenendaal, The Netherlands) raised against a peptide consisting of the carboxyterminal sequence (VRPEHPAETEYDSLYPEDDL) of the fibrinogen γ′ chain. This antibody recognizes an epitope including the high-affinity binding site for thrombin and is specific for the fibrinogen γ′ chain.25 We confirmed that its affinity for the carboxyterminal peptide was lost when the last 4 residues were removed26,27 (data not shown). Plastic 96-well microtiter plates (Greiner, Alphen a/d Rijn, The Netherlands) were coated (110 μL/well) with 2 μg/mL mouse anti-human γ′ fibrinogen during overnight incubation at 4°C. Plates were blocked with 110 μL 1% bovine serum albumin (BSA) in washing buffer (50 mM triethanolamine, 100 mM NaCl, 10 mM EDTA [ethylenediaminetetraacetic acid], 0.1% Tween-20, pH 7.5) for 1 hour at room temperature. One hundred microliters plasma sample diluted in dilution buffer (50 mM triethanolamine, 100 mM NaCl, 10 mM EDTA, 0.1% Tween-20, 10 mM benzamidine, pH 7.5) was added to the wells, and plates were incubated at room temperature for 1 hour. Sample dilutions were stable for at least 3 hours at room temperature. Bound fibrinogen γ′ was detected with 100 μL 1:20 000 diluted horseradish peroxidase (HRP)-conjugated rabbit anti-human fibrinogen (DAKO A/S). After a 1-hour incubation at room temperature, plates were incubated with 100 μL/well substrate buffer (0.1 M sodium acetate pH 5.0, 0.1 mg/mL tetramethylbenzidine, 0.01% H2O2). After 15 minutes, during the linear phase of the reaction, the reaction was stopped by the addition of 1 M H2SO4 (50 μL/well), and the absorbance at 450 nm was read spectrophotometrically. Between all incubation steps, wells were washed 3 times with washing buffer. Because a validated fibrinogen γA/γ′ standard with assigned potency was unavailable, the assay was calibrated using 1:2000 to 1:128 000 dilutions of pooled normal plasma that contained 100 U/dL fibrinogen γ′ (γA/γ′ plus γ′/γ′ fibrinogen) by definition. The calibration curve relating fibrinogen γ′ against A450 was linear from 0.0015 U/dL to 0.05 U/dL, corresponding with 1/64 000 and 1/2000 dilutions of pooled normal plasma, respectively. Fibrinogen γ′ antigen of a plasma sample was calculated as the mean result of the measurements of 2 different independent dilutions (1:8000 and 1:16 000). Results were expressed in units per deciliter. Variations in the results of the 2 independent dilutions were on average lower than 10%. Intra-assay variation was 4.5%. Interassay variation was 9.4%. Pooled normal plasma was prepared from the platelet-free plasma of 70 healthy volunteers (mean age, 38.7 years; 30 men and 40 women not using oral contraceptives).
Statistical analysis
In the healthy controls, Hardy-Weinberg equilibrium for each htSNP was tested by χ2 analysis. FGG, FGA, and FGB haplotypes could be assigned to 928 of 942 subjects directly from the genotypic data. No haplotypes could be assigned to 7 patients and 7 controls, because of genotyping failure or intragenic recombination. These samples were excluded from the analyses. For the other participants, all the identified haplotypes (R2h > 0.98)28,29 were in agreement with those reported by SeattleSNPs.8 Pearson χ2P values were calculated as a global test for differences in haplotype distribution between patients and controls. To investigate whether haplotypes of fibrinogen were associated with thrombosis, odds ratios (ORs) and 95% confidence intervals (95% CIs) according to Woolf30 were calculated as a measure of the relative risk, which indicated the risk for thrombosis in a category of exposure (eg, haplotype 2 carriers) relative to the reference category (eg, non-haplotype 2 carriers). To correct for multiple testing, we used the Bonferroni correction method.
The 3 fibrinogen genes are located on a single stretch of 50 kb DNA. This might result in a high degree of linkage disequilibrium (LD). The degree of LD between htSNPs in FGG, FGA, and FGB was estimated by calculating D′, which is a measure for LD using Haploview software version 2.05.31 Haplotypes across the 3 fibrinogen genes were constructed using Arlequin population genetics software (version 2).32 To account for LD between the FGG-H2, FGA-H2, and FGB-H2 haplotypes, we used logistic regression. To investigate the association between the various fibrinogen haplotypes, the plasma fibrinogen levels, the fibrinogen γ′ levels, and the γ′/γ ratio, mean levels with 95% confidence intervals were calculated. Quartiles of fibrinogen γ′ levels and quartiles of total fibrinogen levels, both as measured in the control subjects, were used as cutoff points to assess the association between fibrinogen γ′ level, total fibrinogen level and risk for venous thrombosis. The 10th percentile (P10) of the γ′/γ ratio, as measured in the control subjects, was used as the cutoff point to assess whether a low γ′/γ ratio was associated with the risk for venous thrombosis. Risk estimates were adjusted by residual analysis for all factors that influenced thrombosis risk in LETS and were associated with fibrinogen levels. These factors were age, sex, diabetes mellitus, body mass index (BMI),33 and high levels (higher than P90) of prothrombin,34 factor VIII,35 factor IX,36 factor XI,37 and C-reactive protein (CRP).38 Separate analyses were performed for patients with (n = 259) and without (n = 215) idiopathic thrombosis, in whom idiopathic was defined as an initial thrombotic event that occurred in the absence of pregnancy, puerperium, oral contraceptive use within 30 days, trauma, surgery, immobilization, or use of a plaster cast within 3 months before the event.39
Results
Fibrinogen haplotypes and risk for venous thrombosis
Genotyping of all 942 subjects showed that the 15 selected htSNPs identified all common haplotypes of FGG, FGA, and FGB (Figure 1A), except for FGG haplotype 5 (FGG-H5), FGA-H5, and FGA-H6 (R2h > 0.98).28,29 For all htSNPs, the distribution of the genotypes in the control subjects was in Hardy-Weinberg equilibrium. Haplotype frequencies in the control subjects (Table 1) were similar to those reported by SeattleSNPs.8 This shows that our population does not deviate importantly from the European-American descent population studied in Seattle. Inspection of the genotypic data by Haploview revealed, as expected,40,41 a high degree of linkage disequilibrium between htSNPs in the FGG, FGA, and FGB genes (data not shown). Arlequin analysis showed that haplotypes often extend over the 3 genes of the gene cluster (data not shown). Most recombination was found between the FGB and the FGA genes, as previously reported.42
Frequency distributions of haplotypes of the FGG, FGA, and FGB genes in patients and controls
Haplotype . | Patients . | Controls . | P . |
|---|---|---|---|
| FGG | .001 | ||
| H1 | 0.356 | 0.389 | |
| H2 | 0.336 | 0.270 | |
| H3 | 0.265 | 0.307 | |
| H4 | 0.043 | 0.034 | |
| FGA | .051 | ||
| H1 | 0.287 | 0.282 | |
| H2 | 0.344 | 0.287 | |
| H3 | 0.102 | 0.124 | |
| H4 | 0.106 | 0.129 | |
| H7 | 0.161 | 0.178 | |
| FGB | .138 | ||
| H1 | 0.351 | 0.334 | |
| H2 | 0.247 | 0.203 | |
| H3 | 0.094 | 0.113 | |
| H4 | 0.145 | 0.161 | |
| H5 | 0.012 | 0.019 | |
| H6 | 0.137 | 0.154 | |
| H7 | 0.014 | 0.015 |
Haplotype . | Patients . | Controls . | P . |
|---|---|---|---|
| FGG | .001 | ||
| H1 | 0.356 | 0.389 | |
| H2 | 0.336 | 0.270 | |
| H3 | 0.265 | 0.307 | |
| H4 | 0.043 | 0.034 | |
| FGA | .051 | ||
| H1 | 0.287 | 0.282 | |
| H2 | 0.344 | 0.287 | |
| H3 | 0.102 | 0.124 | |
| H4 | 0.106 | 0.129 | |
| H7 | 0.161 | 0.178 | |
| FGB | .138 | ||
| H1 | 0.351 | 0.334 | |
| H2 | 0.247 | 0.203 | |
| H3 | 0.094 | 0.113 | |
| H4 | 0.145 | 0.161 | |
| H5 | 0.012 | 0.019 | |
| H6 | 0.137 | 0.154 | |
| H7 | 0.014 | 0.015 |
FGG-H5 and FGA-H5 and -H6 were not found.
P values were determined by Pearson χ2.
Pearson χ2P values (Table 1) showed that a significant difference in the distribution of haplotype frequencies between patients and controls was observed only in FGG. In all 3 genes, subjects homozygous for one of the haplotypes—all accidentally designated as H2—were at significantly increased risk for thrombosis (Table 2). After Bonferroni correction for multiple testing (32 genotypes), only the risk associated with FGG-H2 homozygosity remained significant (P = .002; .05 divided by 32). Haploview analysis showed that haplotypes 2 of all 3 genes were linked to each other. D′ was 0.97 between FGG-H2 and FGA-H2 and 0.90 between FGA-H2 and FGB-H2. To identify the gene that harbors the risk-enhancing SNP, we calculated the odds ratios and 95% confidence intervals of the risk haplotype (H2) of each gene by means of logistic regression. By entering the 3 separate haplotypes in one model, we adjusted the effect of the haplotype of one gene for that of the haplotypes of the other 2 genes. After adjustment, the elevated risk associated with FGA-H2H2 and FGB-H2H2 largely disappeared, whereas the risk associated with FGG-H2H2 remained (OR, 3.5; 95% CI, 1.0-12.5), and the risk for heterozygous FGG-H2 carriers slightly increased (OR, 1.7; 95% CI, 0.8-3.7) (Table 3). Similar results were obtained when we restricted the analyses to patients with idiopathic thromboses (n = 259) or removed participants with FVLeiden, prothrombin 20210A, PC, PS, and AT3 deficiency (177 patients, 50 controls). We were then able to conclude that the risk-enhancing mutation was most likely located in the FGG gene.
Thrombosis risk for haplotypes of FGG, FGA, and FGB
. | FGG . | . | . | FGA . | . | . | FGB . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype . | No. patients (%) . | No. controls (%) . | OR (95% CI) . | No. patients (%) . | No. controls (%) . | OR (95% CI) . | No. patients (%) . | No. controls (%) . | OR (95% CI) . | ||||||
| Haplotype 1 | |||||||||||||||
| H1Hx | 222 (47.3) | 236 (50.1) | 0.8 (0.6-1.1) | 192 (41.0) | 200 (42.5) | 1.0 (0.7-1.3) | 212 (45.3) | 210 (45.1) | 1.1 (0.8-1.4) | ||||||
| H1H1 | 56 (11.9) | 65 (13.8) | 0.8 (0.5-1.2) | 39 (8.3) | 33 (7.0) | 1.2 (0.7-2.0) | 58 (12.4) | 50 (10.7) | 1.2 (0.8-1.9) | ||||||
| Haplotype 2 | |||||||||||||||
| H2Hx | 201 (42.8) | 198 (42.0) | 1.2 (0.9-1.5) | 200 (42.7) | 202 (42.9) | 1.1 (0.9-1.5) | 165 (35.3) | 151 (32.4) | 1.2 (0.9-1.6) | ||||||
| H2H2 | 57 (12.2) | 28 (6.0) | 2.4 (1.5-3.9) | 60 (12.8) | 34 (7.2) | 2.0 (1.3-3.2) | 33 (7.1) | 19 (4.1) | 1.9 (1.1-3.4) | ||||||
| Haplotype 3 | |||||||||||||||
| H3Hx | 185 (39.4) | 211 (44.8) | 0.8 (0.6-1.0) | 85 (18.2) | 103 (21.9) | 0.8 (0.6-1.1) | 78 (16.7) | 95 (20.4) | 0.8 (0.6-1.1) | ||||||
| H3H3 | 32 (6.8) | 39 (8.3) | 0.7 (0.4-1.2) | 5 (1.1) | 7 (1.5) | 0.7 (0.2-2.2) | 5 (1.1) | 5 (1.1) | 1.0 (0.3-3.3) | ||||||
| Haplotype 4 | |||||||||||||||
| H4Hx | 38 (8.1) | 32 (6.8) | 1.2 (0.7-2.0) | 94 (20.1) | 101 (21.4) | 0.9 (0.7-1.2) | 119 (25.4) | 123 (26.4) | 0.9 (0.7-1.3) | ||||||
| H4H4 | 1 (0.2) | 0 (0.0) | NA | 3 (0.6) | 10 (2.1) | 0.3 (0.1-1.1) | 8 (1.7) | 13 (2.8) | 0.6 (0.2-1.5) | ||||||
| Haplotype 5 | |||||||||||||||
| H5Hx | NA | NP | NA | NA | NP | NA | 11 (2.4) | 16 (3.4) | 0.7 (0.3-1.5) | ||||||
| H5H5 | NA | NP | NA | NA | NP | NA | 0 (0.0) | 1 (0.2) | NA | ||||||
| Haplotype 6 | |||||||||||||||
| H6Hx | NA | NA | NA | NA | NP | NA | 114 (24.4) | 131 (28.1) | 0.8 (0.6-1.1) | ||||||
| H6H6 | NA | NA | NA | NA | NP | NA | 7 (1.5) | 6 (1.3) | 1.1 (0.4-3.3) | ||||||
| Haplotype 7 | |||||||||||||||
| H7Hx | NA | NA | NA | 129 (27.6) | 142 (30.1) | 0.9 (0.6-1.2) | 13 (2.8) | 16 (3.4) | 0.8 (0.4-1.7) | ||||||
| H7H7 | NA | NA | NA | 11 (2.4) | 13 (2.8) | 0.8 (0.4-1.9) | 1 (0.2) | 1 (0.2) | 1.0 (0.1-15.9) | ||||||
. | FGG . | . | . | FGA . | . | . | FGB . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype . | No. patients (%) . | No. controls (%) . | OR (95% CI) . | No. patients (%) . | No. controls (%) . | OR (95% CI) . | No. patients (%) . | No. controls (%) . | OR (95% CI) . | ||||||
| Haplotype 1 | |||||||||||||||
| H1Hx | 222 (47.3) | 236 (50.1) | 0.8 (0.6-1.1) | 192 (41.0) | 200 (42.5) | 1.0 (0.7-1.3) | 212 (45.3) | 210 (45.1) | 1.1 (0.8-1.4) | ||||||
| H1H1 | 56 (11.9) | 65 (13.8) | 0.8 (0.5-1.2) | 39 (8.3) | 33 (7.0) | 1.2 (0.7-2.0) | 58 (12.4) | 50 (10.7) | 1.2 (0.8-1.9) | ||||||
| Haplotype 2 | |||||||||||||||
| H2Hx | 201 (42.8) | 198 (42.0) | 1.2 (0.9-1.5) | 200 (42.7) | 202 (42.9) | 1.1 (0.9-1.5) | 165 (35.3) | 151 (32.4) | 1.2 (0.9-1.6) | ||||||
| H2H2 | 57 (12.2) | 28 (6.0) | 2.4 (1.5-3.9) | 60 (12.8) | 34 (7.2) | 2.0 (1.3-3.2) | 33 (7.1) | 19 (4.1) | 1.9 (1.1-3.4) | ||||||
| Haplotype 3 | |||||||||||||||
| H3Hx | 185 (39.4) | 211 (44.8) | 0.8 (0.6-1.0) | 85 (18.2) | 103 (21.9) | 0.8 (0.6-1.1) | 78 (16.7) | 95 (20.4) | 0.8 (0.6-1.1) | ||||||
| H3H3 | 32 (6.8) | 39 (8.3) | 0.7 (0.4-1.2) | 5 (1.1) | 7 (1.5) | 0.7 (0.2-2.2) | 5 (1.1) | 5 (1.1) | 1.0 (0.3-3.3) | ||||||
| Haplotype 4 | |||||||||||||||
| H4Hx | 38 (8.1) | 32 (6.8) | 1.2 (0.7-2.0) | 94 (20.1) | 101 (21.4) | 0.9 (0.7-1.2) | 119 (25.4) | 123 (26.4) | 0.9 (0.7-1.3) | ||||||
| H4H4 | 1 (0.2) | 0 (0.0) | NA | 3 (0.6) | 10 (2.1) | 0.3 (0.1-1.1) | 8 (1.7) | 13 (2.8) | 0.6 (0.2-1.5) | ||||||
| Haplotype 5 | |||||||||||||||
| H5Hx | NA | NP | NA | NA | NP | NA | 11 (2.4) | 16 (3.4) | 0.7 (0.3-1.5) | ||||||
| H5H5 | NA | NP | NA | NA | NP | NA | 0 (0.0) | 1 (0.2) | NA | ||||||
| Haplotype 6 | |||||||||||||||
| H6Hx | NA | NA | NA | NA | NP | NA | 114 (24.4) | 131 (28.1) | 0.8 (0.6-1.1) | ||||||
| H6H6 | NA | NA | NA | NA | NP | NA | 7 (1.5) | 6 (1.3) | 1.1 (0.4-3.3) | ||||||
| Haplotype 7 | |||||||||||||||
| H7Hx | NA | NA | NA | 129 (27.6) | 142 (30.1) | 0.9 (0.6-1.2) | 13 (2.8) | 16 (3.4) | 0.8 (0.4-1.7) | ||||||
| H7H7 | NA | NA | NA | 11 (2.4) | 13 (2.8) | 0.8 (0.4-1.9) | 1 (0.2) | 1 (0.2) | 1.0 (0.1-15.9) | ||||||
All odds ratios were calculated with HxHx as the reference category (OR, 1).
Hx indicates all haplotypes but the one given; NA, not applicable; NP, not present in our study.
Thrombosis risk for haplotype H2 of a gene after adjustment for the effects of haplotype H2 of the other genes by multiple logistic regression analysis
. | OR (95% CI) . | . | . | ||
|---|---|---|---|---|---|
| Haplotype . | FGG . | FGA . | FGB . | ||
| Haplotype 2 | |||||
| H2Hx | 1.7 (0.8-3.7) | 0.6 (0.3-1.4) | 1.1 (0.8-1.6) | ||
| H2H2 | 3.5 (1.0-12.5) | 0.5 (0.1-1.9) | 1.3 (0.6-2.8) | ||
. | OR (95% CI) . | . | . | ||
|---|---|---|---|---|---|
| Haplotype . | FGG . | FGA . | FGB . | ||
| Haplotype 2 | |||||
| H2Hx | 1.7 (0.8-3.7) | 0.6 (0.3-1.4) | 1.1 (0.8-1.6) | ||
| H2H2 | 3.5 (1.0-12.5) | 0.5 (0.1-1.9) | 1.3 (0.6-2.8) | ||
All ORs were calculated with HxHx as a reference category (OR, 1).
Hx indicates all haplotypes but the one given.
Fibrinogen haplotypes and total fibrinogen levels
Since increased fibrinogen levels have been reported to be associated with risk for venous thrombosis,12 we investigated whether haplotypes of FGG, FGA, and FGB were associated with plasma levels of fibrinogen in the control subjects. Because haplotypes could be assigned to more than 98% of the subjects and because fibrinogen levels seemed normally distributed, we could confine the analysis to the calculation of means with 95% confidence intervals in homozygous carriers of the different haplotypes. However, none of the haplotypes was associated with plasma fibrinogen levels (Table 4).
Association of FGG, FGA, and FGB haplotypes with total fibrinogen levels in control subjects
Haplotype . | No. subjects . | Mean U/dL (95% CI) . |
|---|---|---|
| FGG | ||
| H1H1 | 65 | 115 (110-120) |
| H2H2 | 28 | 111 (105-118) |
| H3H3 | 39 | 114 (108-120) |
| FGA | ||
| H1H1 | 33 | 118 (109-127) |
| H2H2 | 34 | 112 (106-118) |
| H3H3 | 7 | 116 (101-132) |
| H4H4 | 10 | 120 (102-139) |
| H7H7 | 13 | 113 (100-125) |
| FGB | ||
| H1H1 | 50 | 115 (108-122) |
| H2H2 | 19 | 120 (111-129) |
| H3H3 | 5 | 112 (93-131) |
| H4H4 | 13 | 116 (104-129) |
| H5H5 | 1 | 101 |
| H6H6 | 6 | 101 (87-116) |
| H7H7 | 1 | 101 |
Haplotype . | No. subjects . | Mean U/dL (95% CI) . |
|---|---|---|
| FGG | ||
| H1H1 | 65 | 115 (110-120) |
| H2H2 | 28 | 111 (105-118) |
| H3H3 | 39 | 114 (108-120) |
| FGA | ||
| H1H1 | 33 | 118 (109-127) |
| H2H2 | 34 | 112 (106-118) |
| H3H3 | 7 | 116 (101-132) |
| H4H4 | 10 | 120 (102-139) |
| H7H7 | 13 | 113 (100-125) |
| FGB | ||
| H1H1 | 50 | 115 (108-122) |
| H2H2 | 19 | 120 (111-129) |
| H3H3 | 5 | 112 (93-131) |
| H4H4 | 13 | 116 (104-129) |
| H5H5 | 1 | 101 |
| H6H6 | 6 | 101 (87-116) |
| H7H7 | 1 | 101 |
Fibrinogen levels are measured in units per deciliter.
Haplotype 2 sequence variations
We did not observe a quantitative effect of the risk-enhancing haplotype FGG-H2 on plasma fibrinogen levels; hence, we reasoned that this haplotype should confer a qualitative defect. FGG-H2 contains the rare alleles of 3 htSNPs—one in intron 8 (7874G>A [rs2066861]), one in intron 9 (9615C>T [rs2066864]), and one downstream from the 3′ untranslated region (10034C>T [rs2066865]).8 In addition, we considered the rare allele of the 129A>T [rs2066854] polymorphism also as tagging for FGG-H2. It is present in both H2 and H5 (Figure 1A),8 but FGG-H5 was absent in our study population. Given that none of these 4 SNPs changed the amino acid sequence, the presence of an additional variation in the coding region of the FGG gene in a subset of the FGG-H2 haplotype was considered. Therefore, we sequenced the genes of 10 patients with venous thrombosis who were homozygous for FGG-H2 (20 FGG-H2 alleles), including the promoter, 5′ UTRs, exons, intron/exon boundaries, and 3′ UTR, but no novel sequence variations were found. This indicated that most likely one of the 4 FGG-H2 tagging SNPs is the risk-enhancing SNP. We hypothesized that the 9615C>T or the 10034C>T SNP influenced the efficiency of alternative splicing of the FGG pre-mRNA by its proximity to the polyadenylation sites of the fibrinogen γ′ and γA transcripts, respectively, and therefore could alter fibrinogen γ′ expression (Figure 1B). To test our hypothesis, we developed an ELISA for the measurement of fibrinogen γ′ levels in plasma and measured fibrinogen γ′ levels (ie, γA/γ′ plus γ′/γ′) in all subjects.
FGG haplotypes and fibrinogen γ′ level
Fibrinogen γ′ levels and total fibrinogen levels were measured in 473 patients and 474 controls. FGG-H2, which we identified as the risk haplotype, was strongly associated with reduced fibrinogen γ′ levels and even more strongly with the fibrinogen γ′ to total fibrinogen ratio (γ′/γ ratio) (Table 5). A clear allele-specific and dose-dependent effect of the FGG-H2 haplotype was observed on both parameters, with homozygous FGG-H2 carriers having the lower levels and intermediate values for carriers of one FGG-H2-allele. Figure 2 shows that for each FGG-H2 genotype (H2H2, H2Hx, HxHx), fibrinogen γ′ levels were strongly associated with total fibrinogen levels. This indicated that fibrinogen γ′ levels and total fibrinogen levels were strongly dependent and that, for each of the FGG-H2 genotypes, the percentage of FGG pre-mRNA spliced along the γ′ pathway was independent of the rate of transcription of the FGG gene.
Association of FGG haplotypes with fibrinogen γ′ levels and the γ′/γ ratio in control subjects
Haplotype FGG . | No. subjects . | Fibrinogen γ′ level, mean U/dL (95% CI) . | γ′/γ ratio, mean (95% CI) . |
|---|---|---|---|
| H2H2 | 28 | 67 (59-76) | 0.60 (0.55-0.65) |
| H2H1 | 106 | 96 (90-101) | 0.81 (0.79-0.84) |
| H2H3 | 83 | 103 (95-111) | 0.89 (0.86-0.92) |
| H2H4 | 8 | 85 (70-101) | 0.81 (0.71-0.90) |
| H1H1 | 65 | 113 (106-120) | 0.99 (0.95-1.02) |
| H1H3 | 117 | 131 (125-138) | 1.09 (1.05-1.12) |
| H1H4 | 13 | 123 (106-140) | 1.06 (0.96-1.16) |
| H3H3 | 39 | 131 (122-140) | 1.15 (1.11-1.19) |
| H3H4 | 11 | 140 (120-160) | 1.10 (1.00-1.21) |
Haplotype FGG . | No. subjects . | Fibrinogen γ′ level, mean U/dL (95% CI) . | γ′/γ ratio, mean (95% CI) . |
|---|---|---|---|
| H2H2 | 28 | 67 (59-76) | 0.60 (0.55-0.65) |
| H2H1 | 106 | 96 (90-101) | 0.81 (0.79-0.84) |
| H2H3 | 83 | 103 (95-111) | 0.89 (0.86-0.92) |
| H2H4 | 8 | 85 (70-101) | 0.81 (0.71-0.90) |
| H1H1 | 65 | 113 (106-120) | 0.99 (0.95-1.02) |
| H1H3 | 117 | 131 (125-138) | 1.09 (1.05-1.12) |
| H1H4 | 13 | 123 (106-140) | 1.06 (0.96-1.16) |
| H3H3 | 39 | 131 (122-140) | 1.15 (1.11-1.19) |
| H3H4 | 11 | 140 (120-160) | 1.10 (1.00-1.21) |
Fibrinogen levels are measured in units per deciliter.
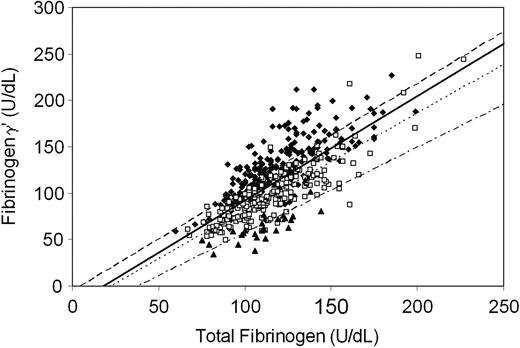
Association between total fibrinogen levels and fibrinogen γ′ levels. Levels were measured in units per deciliter. n = 471 control subjects. Regression coefficients were βHxHx (1.12; 95% CI, 1.01-1.23), βH2Hx (1.05; 95% CI, 0.96-1.14), βH2H2 (0.92; 95% CI, 0.56-1.27), and βall (1.13; 95% CI, 1.00-1.22). Correlation coefficients were RHxHx 0.78, RH2Hx 0.85, RH2H2 0.72, and Rall 0.74. ♦ (dashed line) indicates FGG HxHx; □ (dotted line) FGG H2Hx; ▴ (dash/dot line), FGG H2H2; (solid line), all.
Association between total fibrinogen levels and fibrinogen γ′ levels. Levels were measured in units per deciliter. n = 471 control subjects. Regression coefficients were βHxHx (1.12; 95% CI, 1.01-1.23), βH2Hx (1.05; 95% CI, 0.96-1.14), βH2H2 (0.92; 95% CI, 0.56-1.27), and βall (1.13; 95% CI, 1.00-1.22). Correlation coefficients were RHxHx 0.78, RH2Hx 0.85, RH2H2 0.72, and Rall 0.74. ♦ (dashed line) indicates FGG HxHx; □ (dotted line) FGG H2Hx; ▴ (dash/dot line), FGG H2H2; (solid line), all.
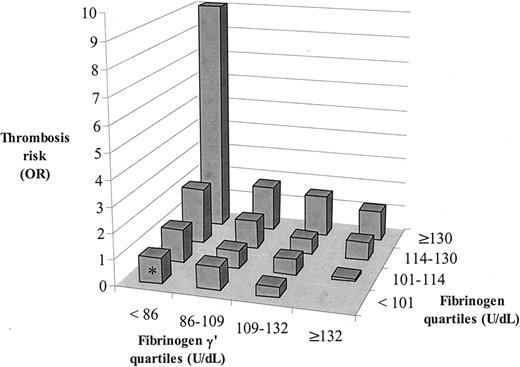
Venous thrombosis risk for quartiles of fibrinogen γ′ and quartiles of total fibrinogen. Values are measured in units per deciliter. For each of the 16 strata, the odds ratios were calculated. Because only 2 patients, but no controls, were present in the most logical reference category with fibrinogen γ′ 132 or greater and total fibrinogen less than 101, the stratum with fibrinogen γ′ less than 86 and total fibrinogen less than 101, which contained 68 patients and 69 controls, was set as the reference category (OR, 1) (marked by an asterisk in the figure).
Venous thrombosis risk for quartiles of fibrinogen γ′ and quartiles of total fibrinogen. Values are measured in units per deciliter. For each of the 16 strata, the odds ratios were calculated. Because only 2 patients, but no controls, were present in the most logical reference category with fibrinogen γ′ 132 or greater and total fibrinogen less than 101, the stratum with fibrinogen γ′ less than 86 and total fibrinogen less than 101, which contained 68 patients and 69 controls, was set as the reference category (OR, 1) (marked by an asterisk in the figure).
Fibrinogen γ′ level and risk for venous thrombosis
We have demonstrated that the FGG-H2 allele is associated with an increased risk for venous thrombosis (Table 2) and reduced fibrinogen γ′ levels and γ′/γ ratios (Table 5). The next question was whether reduced plasma fibrinogen γ′ or a reduced γ′/γ ratio would influence thrombosis risk. This analysis is complicated by the association between fibrinogen γ′ levels and total fibrinogen levels (Figure 2) and by the previous finding (also in LETS12 ) that elevated total fibrinogen levels were associated with increased risk for venous thrombosis. Therefore, we estimated in a preliminary analysis the risk for venous thrombosis (odds ratio) for fibrinogen γ′ levels and for total fibrinogen levels (both stratified into quartiles, as measured in control subjects) (Figure 3). In each total fibrinogen quartile, the risk for venous thrombosis increased when fibrinogen γ′ levels decreased, whereas in each fibrinogen γ′ quartile the thrombosis risk increased when total fibrinogen levels increased, indicating that reduced fibrinogen γ′ levels and elevated fibrinogen levels are separate risk factors for venous thrombosis. Subsequently, we calculated the risk for venous thrombosis for quartiles of fibrinogen γ′ levels and for quartiles of total fibrinogen levels adjusted for each other by means of logistic regression. This analysis showed that reduced fibrinogen γ′ levels and increased total fibrinogen levels were associated with increased risk for venous thrombosis (Table 6). This means that a reduced fibrinogen γ′ level is an independent risk factor for venous thrombosis but that the magnitude of the risk depends on the actual fibrinogen level (Figure 3). Because FGG-H2 was associated with reduced fibrinogen γ′ level and increased risk for thrombosis, we added FGG-H2 to the model (Table 6). The risks associated with reduced fibrinogen γ′ levels and elevated fibrinogen levels did not change, whereas the risk associated with FGG-H2 homozygosity almost completely disappeared (OR, 1.4; 95% CI, 0.8-2.5). Similar results were obtained when fibrinogen γ′ levels and total fibrinogen levels were entered into the logistic regression model as continuous variables. Adjustment for age, sex, diabetes mellitus, BMI and high levels (higher than P90) of prothrombin, factor VIII, factor IX, factor XI, and CRP slightly reduced the OR for high fibrinogen levels but slightly increased the OR for low fibrinogen γ′ levels (Table 6). When the same analyses were performed in the subgroup of patients with idiopathic thrombosis (n = 259) or after the removal of participants with FVLeiden, prothrombin 20210A, PC, PS, and AT3 deficiency (177 patients, 50 controls), essentially the same results were obtained. This indicated that the FGG-H2 haplotype increased the risk for venous thrombosis through its effect on fibrinogen γ′ levels and that there should be additional causes of low fibrinogen γ′ levels.
Thrombosis risk for fibrinogen γ′ levels and total fibrinogen levels
. | ORadj (95% CI) . | . | . | ||
|---|---|---|---|---|---|
. | A . | B . | C . | ||
| Fibrinogen γ′ levels | |||||
| More than 132 | 1* | 1* | 1* | ||
| 109-132 | 1.2 (0.8-1.8) | 1.2 (0.8-1.9) | 1.5 (1.0-2.4) | ||
| 86-109 | 1.9 (1.2-3.0) | 1.9 (1.1-3.0) | 2.3 (1.3-4.0) | ||
| Less than 86 | 3.0 (1.8-4.9) | 2.9 (1.6-5.4) | 3.6 (1.9-7.0) | ||
| Total fibrinogen levels | |||||
| Less than 101 | 1* | 1* | 1* | ||
| 101-114 | 1.1 (0.7-1.6) | 1.0 (0.7-1.5) | 0.9 (0.6-1.4) | ||
| 114-130 | 1.6 (1.1-2.5) | 1.6 (1.0-2.4) | 1.4 (0.9-2.2) | ||
| More than 130 | 3.2 (2.0-5.2) | 3.2 (1.8-5.4) | 2.2 (1.2-4.0) | ||
| FGG haplotype 2 | |||||
| HxHx | NA | 1* | 1* | ||
| H2Hx | NA | 0.9 (0.6-1.2) | 0.8 (0.6-1.2) | ||
| H2H2 | NA | 1.4 (0.8-2.5) | 1.3 (0.7-2.4) | ||
. | ORadj (95% CI) . | . | . | ||
|---|---|---|---|---|---|
. | A . | B . | C . | ||
| Fibrinogen γ′ levels | |||||
| More than 132 | 1* | 1* | 1* | ||
| 109-132 | 1.2 (0.8-1.8) | 1.2 (0.8-1.9) | 1.5 (1.0-2.4) | ||
| 86-109 | 1.9 (1.2-3.0) | 1.9 (1.1-3.0) | 2.3 (1.3-4.0) | ||
| Less than 86 | 3.0 (1.8-4.9) | 2.9 (1.6-5.4) | 3.6 (1.9-7.0) | ||
| Total fibrinogen levels | |||||
| Less than 101 | 1* | 1* | 1* | ||
| 101-114 | 1.1 (0.7-1.6) | 1.0 (0.7-1.5) | 0.9 (0.6-1.4) | ||
| 114-130 | 1.6 (1.1-2.5) | 1.6 (1.0-2.4) | 1.4 (0.9-2.2) | ||
| More than 130 | 3.2 (2.0-5.2) | 3.2 (1.8-5.4) | 2.2 (1.2-4.0) | ||
| FGG haplotype 2 | |||||
| HxHx | NA | 1* | 1* | ||
| H2Hx | NA | 0.9 (0.6-1.2) | 0.8 (0.6-1.2) | ||
| H2H2 | NA | 1.4 (0.8-2.5) | 1.3 (0.7-2.4) | ||
All levels are measured in units per deciliter.
A, logistic regression using quartiles of fibrinogen γ′ and total fibrinogen.
B, logistic regression using quartiles of fibrinogen γ′, total fibrinogen, and FGG-H2 genotypes.
C, logistic regression using quartiles of fibrinogen γ′, total fibrinogen, and FGG-H2 genotypes, adjusted for age, sex, diabetes mellitus, BMI, and high levels (higher than P90) of prothrombin, factor VIII, factor IX, factor XI, and CRP.
NA indicates not applicable.
Reference category
Fibrinogen γ′ to total fibrinogen ratio and risk for venous thrombosis
Because the FGG-H2 haplotype was strongly associated with reduced γ′/γ ratios (Table 5) and because the plasma concentrations of fibrinogen γ′ and of total fibrinogen influenced thrombosis risk, we also analyzed the effect of the γ′/γ ratio on the risk for venous thrombosis. We found that the γ′/γ ratio was lower in patients (mean, 0.89; 95% CI, 0.87-0.92) than in controls (mean, 0.95; 95% CI, 0.93-0.97). Individuals with γ′/γ ratios lower than 0.69, which represents the 10th percentile (P10) as measured in control subjects, were at increased risk for venous thrombosis (OR, 2.4; 95% CI, 1.7-3.5) compared with those with γ′/γ ratios of 0.69 or greater. When we entered the FGG-H2 genotypes together with the P10 of the γ′/γ ratio in the same logistic regression model, the risk associated with a reduced γ′/γ ratio (less than 0.69) remained (OR, 2.2; 95% CI, 1.3-3.5), whereas the risk associated with FGG-H2 homozygosity largely disappeared (OR, 1.2; 95% CI, 0.6-2.3). These results did not change after adjustment for age, sex, diabetes mellitus, BMI, and high levels (higher than P90) of prothrombin, factor VIII, factor IX, factor XI, and CRP. This indicated that the FGG-H2 haplotype acts on the risk for venous thrombosis through reduction of the γ′/γ ratio. Eighty-two percent of controls and 91% of patients with γ′/γ ratios less than 0.69 were homozygous carriers of the FGG-H2 haplotype.
Discussion
We investigated the effect of the most common haplotypes of the FGG, FGA, and FGB genes on the risk for venous thrombosis in a large population-based case-control study, LETS. Only the haplotype distribution of FGG differed between patients and controls. Three haplotypes— FGG-H2, FGA-H2, and FGB-H2—were found to increase the risk for thrombosis. After adjustment for linkage disequilibrium among the 3 genes, only the FGG-H2 haplotype remained associated with an increased risk for venous thrombosis. Homozygous carriers of the FGG-H2 haplotype (5.9% of the Dutch population) had a 2.4-fold (95% CI, 1.5-3.9) increased risk for first venous thrombosis. It is evident that the results obtained in this study must be confirmed in subsequent independent studies before we can evaluate the importance of these findings.
We found that the FGG-H2 haplotype was associated with reduced plasma fibrinogen γ′ levels (γA/γ′ plus γ′/γ′ fibrinogen) and with a reduced fibrinogen γ′ to total fibrinogen ratio (γ′/γ ratio) (Table 5). We further found that fibrinogen γ′ levels were strongly associated with total fibrinogen levels (Figure 2) and that the risk associated with fibrinogen γ′ levels was dependent on the actual fibrinogen concentration and vice versa (Figure 3), even after adjustment for the FGG-H2 haplotype (Table 6). We concluded that the FGG-H2 haplotype increased the risk for venous thrombosis by decreasing the plasma level of fibrinogen γ′ and the γ′/γ ratio. Individuals with γ′/γ ratios lower than P10 of the distribution, as measured in healthy subjects (less than 0.69), were at more than 2-fold increased risk for venous thrombosis, even after adjustment for the FGG-H2 haplotype. Most, but not all, of these individuals were homozygous for the FGG-H2 haplotype.
These findings suggested that a mutation present in most, if not all, FGG-H2 haplotypes is responsible for reduced efficiency in the formation of the alternatively spliced γ′ chain. Most likely, such a mutation is located in the FGG pre-mRNA itself. Although the possibility that this haplotype is linked to a functional SNP in a gene coding for an essential component of the splicing machinery cannot be excluded, we consider this to be less likely. The risk haplotype FGG-H2 is defined by 3 completely linked polymorphisms, of which the rare alleles are unique for this haplotype: 7874G>A (rs2066861), 9615C>T (rs2066864), and 10034C>T (rs2066865).8 In our study population of 940 patients and controls, we did not find any recombination between the 7874G>A and the 10034C>T polymorphisms (data not shown). Because FGG-H5 was absent in our study population, the 129A>T [rs2066854] polymorphism could also be considered an FGG-H2 tagging SNP (Figure 1A). No additional sequence variations were found in 10 homozygous carriers of the FGG-H2 haplotype. We propose that it is the 10034C>T change that results in reduced plasma fibrinogen γ′ levels and reduced γ′/γ ratios. The 10034C>T polymorphism (underlined in the sequence) is located in a cleavage stimulatory factor (CstF) consensus 2a43 sequence (YGTGTYTTYAYTGNNYGT at nt 10030-10047) just downstream of the polyadenylation site (nt 9997-10002) of the fibrinogen γA-specific exon 10. CstF is a multi-subunit protein complex required for efficient cleavage and polyadenylation of pre-mRNA, which initially extends several hundred nucleotides beyond the ultimate polyadenylation site.43 In FGG-H2, which contains T at nucleotide 10034, this consensus sequence is stronger than it is in the other FGG haplotypes (H1, H3, H4, and H5), which all have C at position 10034. Such an improvement of the CstF site may result in more frequent cleavage downstream of the second polyadenylation site (nt 9997-10002) in FGG-H2 pre-mRNA and increased splicing of intron 9. FGG-H2 is, therefore, expected to produce more γA transcripts and fewer γ′ transcripts. In this view, formation of the γ′-specific mRNA is more a matter of alternative polyadenylation than of alternative splicing.
In our study, we did not find support for an influence of fibrinogen haplotypes on total plasma fibrinogen levels in healthy control subjects. We could not confirm earlier studies that reported an effect on fibrinogen concentration of the FGB promoter polymorphisms Bcl I (11046C>T [rs209502]) and 1437G>A (also termed -455G>A [rs1800790]) (for a review, see Humphries44 ). These polymorphisms are present in both FGB-H4 and FGB-H5. In our study none of these haplotypes was associated with altered fibrinogen levels. Although it has been reported that genetic factors contribute to the variation in plasma fibrinogen levels (heritability [h2] = 0.2-0.5),45-47 no evidence was found for linkage between the fibrinogen locus and the regulation of plasma fibrinogen levels.48,49
Several studies have reported that fibrinogen γA/γ′ heterodimers behave differently from fibrinogen γA/γA homodimers during the early stages of polymerization, ultimately leading to an altered fibrin structure that is more extensively cross-linked by activated factor XIII than γA/γA fibrin and also more resistant to fibrinolysis.50-52 This has been explained by the presence of a unique binding site for factor XIII B in the carboxy terminus of the γ′ chain.53,54 These findings predict that an increase in fibrinogen γ′ level or in the γ′/γ ratio would result in more stable and lysis-resistant clots and, therefore, would represent a prothrombotic state. We found, however, that reduced fibrinogen γ′ levels and, more specifically, reduced γ′/γ ratios were associated with an increased risk for venous thrombosis. A possible explanation for this finding might be that the alternative carboxy terminus of the γ′ chain defines not only a binding site for factor XIII but also a high-affinity, nonsubstrate binding site for thrombin.51,52,55 Studies on γA/γ′ fibrin(ogen) indicated that this binding site functioned as a thrombin inhibitor56,57 and, importantly, contributed to the antithrombin activity that develops during fibrin formation (antithrombin 1).58,59 Thrombin interacts with the carboxy terminal sequence of the γ′ chain through its exosite 2,26,27,57 which prevents the participation of this exosite in the binding of thrombin to heparin60 and which recently was found to be responsible for the observation that thrombin-induced FPA generation was slower with γA/γ′ fibrinogen than with γA/γA fibrinogen.61 Experiments in which thrombin generation was measured in afibrinogenemia plasma and fibrinogen-depleted plasma supplemented with γA/γA fibrinogen or γA/γ′ fibrinogen showed that γA/γ′ fibrinogen had a more profound effect in down-regulating thrombin generation than γA/γA fibrinogen.62 Future studies should reveal whether and how a 2-fold reduction of the γ′/γ ratio would influence fibrin formation and degradation.
Two studies reported the effects of plasma fibrinogen γ′ levels on the risk for disease.25,63 In a preliminary report, Drouet et al63 investigated the hypothesis that the plasma γ′/γ ratio was a marker for arterial thrombosis activity, but they did not perform a formal association study. Lovely et al25 reported that the fibrinogen γ′ level, but not the γ′/γ ratio, was associated with coronary artery disease (CAD; defined as luminal narrowing lesion of 50% or greater in at least one major coronary artery or branch). However, this was a small study (91 CAD-positive and 42 CAD-negative patients), and, in our opinion, the authors did not properly exclude the possibility that the fibrinogen γ′ level acted as a substitute marker for total fibrinogen (Figure 2), which is an established risk factor for CAD. Interestingly, Mannila et al41 recently reported that the *216C>T polymorphism (which is identical to FGG-H2 tagging SNP 10034C>T) was not associated with risk for myocardial infarction. This is not surprising because none of the established genetic risk factors for venous thrombosis is associated with arterial thrombosis.64
It should be noted that the FGA H2-specific 6534A>G [rs6050] polymorphism is strongly, but not completely, linked to FGG-H2 (D′ in our study was 0.97). This polymorphism is located in exon 5 of FGA and codes for a threonine-to-alanine substitution at position 312 of the Aα chain, in a region important for FXIIIa-dependent cross-linking at position 328.65 It has been reported that the Ala312 allele is associated with more rigid and less porous fibrin gel structures.66 Ala312 fibrin clots had thicker fibers and more extensive α-chain cross-linking than Thr312 clots.66 These results suggested that Ala312 fibrin clots are more resistant to fibrinolysis, which might increase the risk for thrombosis.56 However, because this polymorphism is located on FGA-H2, which is strongly linked to FGG-H2, it is not unlikely that the risk reported to be associated with this polymorphism67 is in fact caused by linkage with FGG-H2. In our study, we were unable to identify the FGA Ala312Thr polymorphism as an independent risk factor for venous thrombosis. The FGG-H2 haplotype was the only haplotype associated with increased risk for venous thrombosis.
Prepublished online as Blood First Edition Paper, September 6, 2005; DOI 10.1182/blood-2005-05-2180.
Supported by grant 912-02-036 from the Netherlands Organization for Scientific Research (NWO). The Leiden Thrombophilia Study (LETS) study was supported by grant 89-063 from the Netherlands Heart Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal