Abstract
The usage of the immunoglobulin (Ig) VH3-21 gene is associated with poor prognosis in B-cell chronic lymphocytic leukemia (B-CLL) despite VH gene mutation status. Many VH3-21+ patients also display restricted heavy- and light-chain Ig gene rearrangements, implying a role of antigen selection in disease development. To explore the specific phenotypic/genotypic features among VH3-21+ B-CLLs, we compared gene expression patterns in 15 VH3-21+ and 24 non-VH3-21 patients (11 with unmutated and 13 with mutated VH genes) using Affymetrix microarray analysis (∼12 500 genes). A distinct expression profile was identified for VH3-21+ patients in contrast to the Ig-unmutated and -mutated groups. By applying different algorithms, the data enabled an efficient class discrimination of the VH3-21+ subset based on 27 or 57 genes. A set of genes was sorted out which, using different analytical methods, consistently gave a distinction between VH3-21+ and non-VH3-21 samples. Several of these genes are involved in regulation of DNA replication/cell-cycle control, transcription and protein kinase activity, which may render the VH3-21+ cells with a higher proliferative drive. However, no clear evidence of increased B-cell receptor signaling was found in the VH3-21+ group. Altogether, our identification of a specific VH3-21 profile may provide insights into the pathogenesis of the VH3-21+ subgroup. (Blood. 2005;106:681-689)
Introduction
The somatic hypermutation status of the immunoglobulin variable heavy chain (IgVH) genes has in the last few years been identified as one of the most important prognostic markers in B-cell chronic lymphocytic leukemia (B-CLL). Roughly half of patients with B-CLL display mutations within their VH genes (Ig-mutated), which correlates with favorable prognosis, whereas the remaining patients have unmutated VH genes and a worse outcome (Ig-unmutated).1-4 The selection of VH genes has been shown to be a nonrandom process in B-CLL with overrepresentation of the VH1-69, VH3-7, VH3-21, and VH4-34 genes.2,5-11 The implication of preferential VH gene usage has been hypothesized to be a result of antigen selection of B cells with certain B-cell receptors (BCRs), which then might render a higher probability to undergo malignant transformation. In Ig-unmutated B-CLL, biased usage of the VH1-69 gene has been shown by several groups in a significant fraction of patients.2,5-11 Lately, small subsets of VH1-69+ cases with more or less identical heavy- and light-chain Ig rearrangement features have also been reported.12,13 These findings have led to the speculation that unknown antigens could be involved in the pathogenesis of VH1-69+ B-CLL.
Recently, an overrepresentation of the VH3-21 gene was demonstrated in B-CLL with mutated VH genes.10 Interestingly, VH3-21-utilizing patients showed short overall survival, similar to patients with unmutated VH genes, despite the fact that 2-thirds of them had mutated VH genes.10,11,14,15 Furthermore, many of these VH3-21+ rearrangements (both mutated and unmutated) displayed unusually short and highly homologous complementarity-determining regions 3 (CDR3s), predominant λ light chain expression, and restricted use of 1 particular Vλ gene, Vλ2-14.11 These latter findings have implicated that an unknown antigen epitope could be recognized by the homologous VH3-21+/Vλ2-14+ BCRs expressed in the different VH3-21+ tumors.16 Thus, VH3-21-utilizing patients comprise a new separate B-CLL entity independent of mutation status and an effort is therefore warranted to identify distinguishing features at the molecular level in order to reveal the biology behind VH3-21+ B-CLL.
Gene expression patterns typical for different hematologic malignancies have been identified using microarray techniques.17-19 Expression profiling of B-CLL has shown a characteristic B-CLL “signature” compared with other malignant lymphomas, but has so far revealed relatively uniform gene expression patterns between the Ig-unmutated and Ig-mutated form of B-CLL.20,21 Interestingly, the expression profiles of both these subsets resembled the signature of memory B cells,20 which is in contrast with the initial speculation that Ig-unmutated B-CLL should derive from a naive B cell.22
In the present study, we have analyzed gene expression patterns in samples from 39 patients with B-CLL with the specific aim to identify genes that may reflect disturbed regulatory pathways, such as genes involved in BCR signaling, in the subset of patients with VH3-21 usage compared with Ig-mutated and Ig-unmutated cases using other VH genes.
Patients, materials, and methods
Patient material
Tumor samples were collected from 39 patients with B-CLL at the University Hospitals in Uppsala (n = 17), Linköping (n = 2), and Huddinge (n = 20), Sweden, between 1982 and 2002. Twenty-four samples were obtained from B-CLLs with non-VH3-21 usage, 13 with mutated and 11 with unmutated VH genes, and 15 samples with VH3-21 usage. Tumor material was mainly obtained from peripheral blood but also from bone marrow and lymph nodes. Morphologic classification was performed according to the World Health Organization (WHO) classification and the tumors typically expressed CD5 and CD23 and showed a weak Ig expression.23 The median age at diagnosis was 63 years (range, 45 to 83 years), 25 were men, and 14 women with a male-female ratio of 1.8:1. The Binet/Rai stage was as follows: 10 patients A/0, 10 patients A/I, 4 patients A/III, 3 patients B/I, 1 patient B/II, 3 patients B/III, 2 patients C/III, and 2 patients C/IV. In 4 cases either Rai or Binet stage was available: 2 cases were Rai III, 1 Rai I, and 1 Binet C. Informed consent was provided according to the Declaration of Helsinki, and the study was approved by the ethical committees of Karolinska Institutet, Huddinge University Hospital (Dnr 184/01), and Uppsala University (Ups 01-082).
Isolation of mononuclear cells
Mononuclear cells were obtained by separation on a Lymphoprep gradient (Nycomed Pharma, Roskilde, Denmark). The number of leukemic cells was assessed by immunophenotyping. The median proportion of B cells with CLL phenotype (CD5+/CD19+/CD23+) was 90% (range, 73%-98%).
VH gene analysis
High-molecular-weight DNA was prepared using standard protocols including proteinase K treatment. VH gene family-specific polymerase chain reaction (PCR) amplification was performed using consensus VH/JH primers and the clonal VH gene rearrangements were sequenced as previously described.11,24 The sequences were aligned to germline Ig sequences in the GenBank, V-BASE (MRC Centre for Protein Engineering, Cambridge, United Kingdom), and International ImMuno GeneTics (IMGT) databases. VH gene sequences deviating more than 2% from the corresponding germ-line gene were defined as mutated. The length of the heavy-chain CDR3 was calculated between codons 95 and 102.
RNA isolation, probe preparation, and hybridization
Lymphocytes (50-100 × 106) were collected for total RNA isolation from fresh or frozen samples using the Rneasy Mini Kit (Qiagen, Valencia, CA). The RNA was checked qualitatively on a 1% agarose gel, and quantitatively by spectrophotometry. Probes were prepared according to the recommendations from Affymetrix (Santa Clara, CA) from 8 μg total RNA. HG-U95Av2 arrays, complementary to more than 12 500 sequences, were hybridized, developed, and scanned to obtain quantitative gene expression levels, using standard protocols.
Microarray data analysis
Affymetrix Micro Array Suite Software (MAS) version 5.0 was applied to analyze the scanned images, convert intensities to a numerical format, and obtain a signal value for each probe on the array. Target intensity values from each array were scaled/normalized to a value of 100 to enable different arrays to be compared. A detailed description of the statistical algorithms has been presented.25
A 3-stage approach was followed in the subsequent microarray data analyses. First, a marker analysis was performed using the GeneCluster2 software26 (Whitehead Institute/MIT Center for Genome Research [WICGR], MA) by computing a ranking list where all individual probes are sorted in descending order according to the degree of correlation between their expression level and the sample group compared. For each sample group in the comparison, this analysis calculates a correlation score to identify the genes (probes) that have consistently high average expression in this group and low in the others (ie, “marker” genes). A measure of the statistical significance of each score can be obtained through a permutation test, where the sample group assignment is randomized iteratively and the probability of obtaining a given score by chance in the present material is calculated. The scores actually obtained in a given sample group comparison can thus be assigned to significance levels of, for example, 1% and 5%.
Second, a class discrimination study was conducted. In contrast to the gene-by-gene examination above, this type of analysis seeks to identify a group of genes whose expression levels collectively can discriminate between 2 sample groups. A common application of such an analysis is to enable classification of unknown samples based on the gene expression signature alone. However, since the basis for classification of samples in the present study is a known genotype (VH3-21 and VH mutation status), such a diagnostic tool is not relevant. The purpose of the class-discriminant analyses used here was instead to suggest groups of genes that may be involved in alternative or complementary molecular pathways that are related to the phenotypic differences between sample groups. Two different algorithms were used for class discrimination: weighted voting (GeneCluster226 ) and linear discriminant analysis (dCHIP27 ; Department of Biostatistics, Harvard School of Public Health, MA).
Third, in an effort to pinpoint a more limited number of genes that may be of particular interest from a functional point of view, the gene lists from the marker and discriminant analyses were combined. Genes that appeared in more than one of the various analyses for the same group (for example, VH3-21) were identified and listed.
Detailed descriptions of all data analyses conducted in this study are included as supplemental data on the Blood website; click the Supplemental Materials link at the top of the online article.
For gene annotation, we used the Gene Ontology Tree Machine (GOTM) software (Graduate School in Genome Science and Technology, University of Tennessee-Oak Ridge National Laboratory, Oak Ridge, TN), a web-based data-mining tool using Gene Ontology (GO) hierarchies to identify significant GO categories in a gene set. A statistical analysis in GOTM indicates GO terms, such as “cell cycle” and “metabolism,” and thereby suggests biologic areas that warrant further study (P < .01).28
Real-time quantitative PCR
For real-time quantitative PCR, TaqMan Assays-on-Demand (Applied Biosystems, Warrington, United Kingdom) including probes and primers for PPP5C, E2F4, SHARP, CDKN1B, and PPIA = (cyclophilin A) were carried out. First-strand cDNA was synthesized with 1 μg of total RNA using High-Capacity cDNA Archive Kit with random primers (Applied Biosystems). PCR was performed with TaqMan Universal PCR Master Mix and an ABI PRISM 7000 Sequence Detection System (Applied Biosystems) according to the manufacturer's instructions. The relative expression of PPP5C, E2F4, SHARP, and CDKN1B mRNA was normalized to the amount of PPIA in the same cDNA using the standard curve method described by the manufacturer.
Results
VH gene analysis
A total of 46 VH gene rearrangements were amplified and sequenced from 39 B-CLL cases, including 7 cases with 2 rearrangements. Of the 24 non-VH3-21 patients, 13 showed somatically mutated VH genes (mean mutation frequency 7%) while 11 cases were considered unmutated. Among the 15 VH3-21+ patients, 9 were mutated (mean mutation frequency 4%) and 6 unmutated. Eight VH3-21 cases displayed VH rearrangements with a short and homologous CDR3 (7 codons without known D gene usage), whereas the remaining 7 cases had rearrangements with varying CDR3 lengths (10-23 codons). The proportion of cases with a mutated VH3-21 gene and with a short/homologous CDR3 reflects the proportion of cases within the VH3-21+ subset as previously reported11 and as found in an extended study of more than 80 VH3-21+ CLLs (R.R., unpublished data, April 2005). The sequence results have been published previously for all cases included.11 The most frequently used VH genes among the non-VH3-21 gene rearrangements were VH1-69 (n = 6, 20%), VH3-23 (n = 3, 10%), VH3-9 (n = 3, 10%), VH3-11 (n = 3, 10%), and VH2-5 (n = 3, 10%).
Identification of genes distinguishing between VH3-21+ and non-VH3-21 B-CLL samples: marker gene analysis
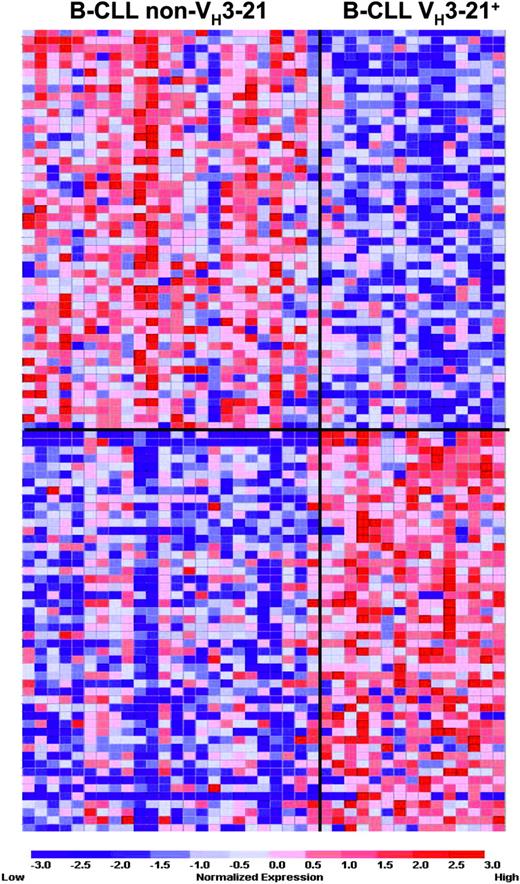
To identify genes correlating with VH3-21 and non-VH3-21 usage, a marker analysis was performed with the 4701 probes whose expression values passed the variation filter. A permutation testing on the 1000 best-correlated probes (see Supplemental Figure S1) showed that all of the 1000 probes were statistically significant on the 5% level for both VH3-21+ and non-VH3-21 samples, whereas at the 1% level 184 and 152 probes correlated with VH3-21 usage and non-VH3-21 usage, respectively. Figure 1 displays the 50 best-correlated probes for each group. In conclusion, the VH3-21 group was clearly and significantly distinguished by a large number of individual genes from B-CLLs with non-VH3-21 usage.
Results from a marker gene analysis, where the 50 genes that correlated best with either VH3-21 or non-VH3-21 usage were identified. The expression matrix depicts the gene expression values of the individual samples, with columns representing individual samples and rows representing individual genes. Genes are ranked in a “best-correlated” order. The color scale identifies relative gene expression changes normalized by the SD of 1, with 0 representing the mean expression level of a given gene across the panel. Gene lists are included as Table S1 in the Supplemental Materials.
Results from a marker gene analysis, where the 50 genes that correlated best with either VH3-21 or non-VH3-21 usage were identified. The expression matrix depicts the gene expression values of the individual samples, with columns representing individual samples and rows representing individual genes. Genes are ranked in a “best-correlated” order. The color scale identifies relative gene expression changes normalized by the SD of 1, with 0 representing the mean expression level of a given gene across the panel. Gene lists are included as Table S1 in the Supplemental Materials.
Sample class discriminator analysis
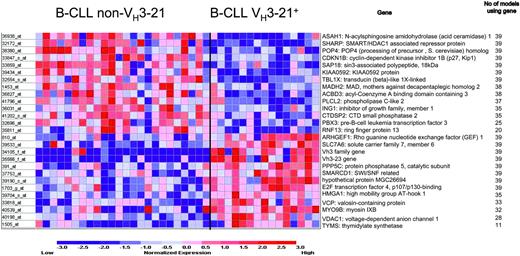
First, the weighted voting method for class discrimination was applied to the data, in which different numbers of individual probes are systematically tested for their ability to collectively discriminate between the sample groups. The range of 22 to 27 probes rendered the best statistically significant outcome in our data set; an evaluation with Fisher test yielded a P value of less than .001 for all class discriminators in the 22- to 27-probe range with a 90% correct classification. Figure 2 shows the outcome for the 27-probe model. With the “leave one out” cross validation, each of the 39 samples that were tested generated a new set of 27 probes. In the final model, 15 out of the 27 probes were common to all 39 samples.
Genes included in the weighted voting class discriminator model. Genes expressed at higher level in non-VH3-21 samples are listed on top and those that were more abundant in VH3-21+ B-CLL are shown on bottom. The highest accuracy was obtained using 27 probes: a total of 35 (90%) of 39 of samples were correctly predicted (P < .001).
Genes included in the weighted voting class discriminator model. Genes expressed at higher level in non-VH3-21 samples are listed on top and those that were more abundant in VH3-21+ B-CLL are shown on bottom. The highest accuracy was obtained using 27 probes: a total of 35 (90%) of 39 of samples were correctly predicted (P < .001).
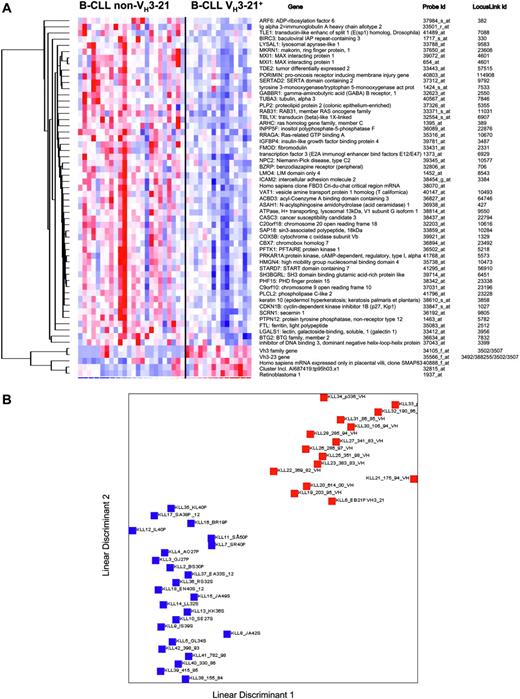
Second, another class discriminator algorithm, linear discriminant analysis (LDA), was also applied to ascertain if an even more accurate classification was possible. Fifty-seven probes were obtained after filtering (P =.01) in a comparison analysis and using an internal validation procedure, a 100% correct classification was achieved based on these 57 probes (Figure 3A). A graphical illustration of the separability of the VH3-21 and non-VH3-21 using groups is outlined in Figure 3B.
In summary, it was possible to construct class discriminators in the form of expression patterns based on approximately 30 to 60 probes that with 90% to 100% accuracy could distinguish cells with VH3-21 usage from those with non-VH3-21 usage.
Three-group marker analysis
In order to identify the probes that best correlate with the VH3-21 status when a mutated versus unmutated comparison is performed simultaneously, a 3-group marker analysis was also carried out. Figure 4 shows the 50 best-ranked probes for each group compared with both of the other groups. The permutation testing showed no significant association in either the Ig-unmutated or the Ig-mutated group (comparison A and B, Figure 4). However, 155 and at least 1000 up-regulated probes were associated to the VH3-21+ group at the 1% and 5% significance level, respectively (comparison C, Figure 4). Thus, the overall association of the marker genes for the VH3-21+ group was stronger than the marker genes for either the non-VH3-21-using Ig-unmutated or mutated group in this 3-group comparison.
LDA analysis in VH3-21+ and non-VH3-21 B-CLL. (A) Genes included in the LDA class discriminator. The expression matrix shows the 57 probes that are reliably differentially expressed between the 2 groups, VH3-21+ B-CLL and non-VH3-21 B-CLL. (B) LDA of VH3-21+ and non-VH3-21 B-CLL. Red squares represent VH3-21+ samples; blue squares, non-VH3-21 samples. LDA was applied to the data of the 39 samples by the 57-probe set. Linear discriminants were determined for each sample and plotted. The prediction rate for samples with known class was 100% (P = .01). The median false discovery rate for this predictor was 1.7%.
LDA analysis in VH3-21+ and non-VH3-21 B-CLL. (A) Genes included in the LDA class discriminator. The expression matrix shows the 57 probes that are reliably differentially expressed between the 2 groups, VH3-21+ B-CLL and non-VH3-21 B-CLL. (B) LDA of VH3-21+ and non-VH3-21 B-CLL. Red squares represent VH3-21+ samples; blue squares, non-VH3-21 samples. LDA was applied to the data of the 39 samples by the 57-probe set. Linear discriminants were determined for each sample and plotted. The prediction rate for samples with known class was 100% (P = .01). The median false discovery rate for this predictor was 1.7%.
Consistency of marker genes related to VH3-21 identified in different comparisons
A combination of the results from all the previous comparisons was performed to identify genes that most consistently could distinguish between VH3-21+ and non-VH3-21 samples, when using more than 1 comparison method. In order to obtain this list, the 20 best-ranked probes, up-regulated or down-regulated in VH3-21 B-CLLs, were listed from the 2 marker gene comparisons described previously. Out of 40 up-regulated and 40 down-regulated probes, 29 genes correlated with VH3-21 status and 38 with non-VH3-21 status. Eight genes were identified as up-regulated in VH3-21 in both marker gene analyses; 2 probes recognising VH3 family gene members, PPP5C: protein phosphatase 5, ARHGEF1: Rho guanine nucleotide exchange factor (GEF) 1, VCP: valosin-containing protein, VDAC1: voltage-dependent anion channel 1, TYMS: thymidylate synthetase, and SMARCD1: SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily d, member 1. Two genes were down-regulated in both marker gene analyses: PLCL2: phospholipase C-like 2 and SHARP: SMART/HDAC1-associated repressor protein.
Three-group marker analysis in Ig-mutated, unmutated, and VH3-21+B-CLL. Results from the 3-group marker gene analysis, where the 50 genes best-correlated with either (A) Ig-mutated, (B) Ig unmutated, or (C) VH3-21+ B-CLL were identified. Gene lists are included in the Supplementary Materials.
Three-group marker analysis in Ig-mutated, unmutated, and VH3-21+B-CLL. Results from the 3-group marker gene analysis, where the 50 genes best-correlated with either (A) Ig-mutated, (B) Ig unmutated, or (C) VH3-21+ B-CLL were identified. Gene lists are included in the Supplementary Materials.
The 27 probes that showed the best class discrimination in the weighted voting were all present and highly ranked among the 2 comparisons. Of the 57 probes that were obtained in the LDA analysis, 11 genes were in common with the genes identified in the 2 marker gene analysis; 2 out of 5 up-regulated in VH3-21+ samples and 9 out of 38 down-regulated. All comparisons are summarized in Table 1, and the genes are shown in a ranking order according to how many comparisons a given gene was found to be related to VH3-21 status (the VH3-21 vs non-VH3-21 comparison is designated comparison 1 and the 3-group marker analysis as comparison 2 in Table 1).
The highest-scored VH3-21+-related genes in different comparisons
Probe ID . | GenBank no. . | LocusLink no. . | Gene name . | Comparison* . |
|---|---|---|---|---|
| Up-regulated in VH3-21 + B-CLL (VH3-21-related) | ||||
| 35566_f_at | — | 3502, 3507 | VH3-23 gene† | 1,2,LDA,WV |
| 34105_f_at | — | 3492, 3500, 3502, 3507, 388255 | VH3 family gene (either the VH3-21, VH3-07 or VH3-74 gene)† | 1,2,LDA,WV |
| 391_at | X89416 | 5536 | PPP5C: protein phosphatase 5 | 1,2,WV |
| 810_at | U64105 | 9138 | ARHGEFI: Rho guanine nucleotide exchange factor (GEF) 1 | 1,2,WV |
| 33818_at | AC004472 | 7415 | VCP: valosin-containing protein | 1,2,WV |
| 40198_at | L06132 | 7416 | VDACI: voltage-dependent anion channel 1 | 1,2,WV |
| 1505_at | NM_001071 | 7298 | TYMS: thymidylate synthetase | 1,2,WV |
| 37753_at | U66617 | 6602 | SMARCDI: SW1/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 | 1,2,WV |
| 1703_g_at | NM_001950 | 1874 | E2FR: E2F transcription factor 4, p107/p130-binding | 1,WV |
| 39190_s_at | AC002126 | 284439 | MGC26694: hypothetical protein MGC26694 | 1,WV |
| 40539_at | U42391 | 4650 | MYO9B: myosin IXB | 1,WV |
| 39533_at | D87432 | 9057 | SLC7A6: solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 | 1,WV |
| 39704_s_at | L17131 | 3159 | HMGA1: high mobility group AT-hook 1 | 1,WV |
| 32844_at | AF104913 | 1981 | EIF4G1: eukaryotic translation initiation factor 4 gamma, 1 | 1,2 |
| 1306_at | D12686 | 1981 | 2 | |
| 33268_at | L25270 | 8242 | SMCX: Smcx homolog, X-linked (mouse) | 1,2 |
| 38259_at | AB002559 | 6813 | STXBP2: syntaxin binding protein 2 | 1 |
| 1760_s_at | D11327 | 5778 | PTPN7: protein tyrosine phosphatase, non-receptor type 7 | 1 |
| 41753_at | U48734 | 81 | ACTN4: actinin, alpha 4 | 1 |
| 34325_at | AJ005893 | 10084 | PQBP1: polyglutamine binding protein 1 | 2 |
| 34868_at | AB029012 | 23381 | EST1B: Est1p-like protein B | 1 |
| 591_s_at | M33684 | 5770 | PTPN1: protein tyrosine phosphatase, nonreceptor type 1 | 1 |
| 1553_r_at | U22028 | 1553 | CYP2A13: cytochrome P450, family 2, subfamily A, polypeptide 13 | 2 |
| 38522_s_at | X52785 | 933 | CD22: CD22 antigen | 2 |
| 2009_at | U33284 | 2185 | PTK2B: PTK2B protein tyrosine kinase 2 beta | 2 |
| 1994_f_at | AF001359 | 4292 | MLH1: mutL homolog 1, colon cancer, nonpolyposis type 2 (Escherichia coli) | 2 |
| 35620_at | AF043250 | 10452 | TOMM40: translocase of outer mitochondrial membrane 40 homolog (yeast) | 2 |
| 37095_r_at | M84562 | 2358 | FPRL1: formyl peptide receptor-like 1 | 2 |
| 40324_r_at | AF070585 | 8940 | TOP3B: topoisomerase (DNA) III beta | 2 |
| 36094_at | M21984 | 7140 | TNNT3: troponin T3, skeletal, fast | 2 |
| Down-regulated in VH3-21+ B-CLL (non-VH3-21-related) | ||||
| 41796_at | AB029015 | 23228 | PLCL2: phospholipase C-like 2 | 1,2‡,LDA,WV |
| 32172_at | AL096858 | 23013 | SHARP: SMART/HDAC1-assocjated repressor protein | 1,2§,WV |
| 33859_at | U96915 | 10284 | SAP18: sin3-associated polypeptide, 18kDa | 1,LDA,WV |
| 36938_at | U70063 | 427 | ASAH1: N-acylsphingosine amidohydrolase (acid ceramidase) 1 | 1,LDA,WV |
| 32554_s_at | Y12781 | 6907 | TBL1X: transducin (beta)-like 1X-linked | 1,LDA,WV |
| 33847_s_at | A1304854 | 1027 | CDKN1B: cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 1,LDA,WV |
| 38380_at | Y18863 | 10775 | POP4: POP4 (processing of precursor, Saccharomyces cerevisiae) homolog | 1,WV |
| 36827_at | AF020762 | 64746 | ACBD3: acyl-coenzyme A binding domain containing 3 | 1,WV |
| 1453_at | U68018 | 4087 | MADH2: MAD, mothers against decapentaplegic homolog 2 (Drosophila) | 1,WV |
| 41202_s_at | AF000152 | 10106 | CTDSP2: CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) | 1,WV |
| 36031_at | AB024401 | 3621 | ING1: inhibitor of growth family, member 1 | 1,WV |
| 32696_at | X59841 | 5090 | PBX3: pre-B-cell leukemia transcription factor 3 | 1,WV |
| 39434_at | AB011164 | 253725 | KIAA0592: KIAA0592 protein | 1,WV |
| 39345_at | AI525834 | 10577 | NPC2: Niemann-Pick disease, type C2 | 1,LDA |
| 654_at | L07648 | 4601 | MXI1: MAX interacting protein 1 | 1,LDA |
| 36894_at | AL031846 | 23492 | CBX7: chromobox homolog 7 | 2‡,LDA |
| 36502_at | AB020641 | 5218 | PFTK1: PFTAIRE protein kinase 1 | 2‡,LDA |
| 35811_at | AF037204 | 11342 | RNF13: ring finger protein 13 | 1 |
| 37035_at | AI557272 | 27230 | SERP1: stress-associated endoplasmic reticulum protein 1 | 1 |
| 1452_at | U24576 | 8543 | LMO4: LIM domain only 4 | 1 |
| 41379_at | AB011166 | 23137 | SMC5L1: SMC5 structural maintenance of chromosomes 5-like 1 (yeast) | 1 |
| 40063_at | U22897 | 10241 | NDP52: nuclear domain 10 protein | 1 |
| 37253_at | X92493 | 8395 | PIP5K1B: phosphatidylinositol-4-phosphate 5-kinase, type 1, beta | 2§ |
| 35145_at | X96401 | 4335 | MNT: MAX-binding protein | 2§ |
| 1797_at | U40343 | 1032 | CDKN2D: cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 2§ |
| 943_at | D43968 | 861 | RUNX1: runt-related transcription factor 1 (acute myeloid leukemia 1; aml 1 oncogene) | 2§ |
Probe ID . | GenBank no. . | LocusLink no. . | Gene name . | Comparison* . |
|---|---|---|---|---|
| Up-regulated in VH3-21 + B-CLL (VH3-21-related) | ||||
| 35566_f_at | — | 3502, 3507 | VH3-23 gene† | 1,2,LDA,WV |
| 34105_f_at | — | 3492, 3500, 3502, 3507, 388255 | VH3 family gene (either the VH3-21, VH3-07 or VH3-74 gene)† | 1,2,LDA,WV |
| 391_at | X89416 | 5536 | PPP5C: protein phosphatase 5 | 1,2,WV |
| 810_at | U64105 | 9138 | ARHGEFI: Rho guanine nucleotide exchange factor (GEF) 1 | 1,2,WV |
| 33818_at | AC004472 | 7415 | VCP: valosin-containing protein | 1,2,WV |
| 40198_at | L06132 | 7416 | VDACI: voltage-dependent anion channel 1 | 1,2,WV |
| 1505_at | NM_001071 | 7298 | TYMS: thymidylate synthetase | 1,2,WV |
| 37753_at | U66617 | 6602 | SMARCDI: SW1/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 | 1,2,WV |
| 1703_g_at | NM_001950 | 1874 | E2FR: E2F transcription factor 4, p107/p130-binding | 1,WV |
| 39190_s_at | AC002126 | 284439 | MGC26694: hypothetical protein MGC26694 | 1,WV |
| 40539_at | U42391 | 4650 | MYO9B: myosin IXB | 1,WV |
| 39533_at | D87432 | 9057 | SLC7A6: solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 | 1,WV |
| 39704_s_at | L17131 | 3159 | HMGA1: high mobility group AT-hook 1 | 1,WV |
| 32844_at | AF104913 | 1981 | EIF4G1: eukaryotic translation initiation factor 4 gamma, 1 | 1,2 |
| 1306_at | D12686 | 1981 | 2 | |
| 33268_at | L25270 | 8242 | SMCX: Smcx homolog, X-linked (mouse) | 1,2 |
| 38259_at | AB002559 | 6813 | STXBP2: syntaxin binding protein 2 | 1 |
| 1760_s_at | D11327 | 5778 | PTPN7: protein tyrosine phosphatase, non-receptor type 7 | 1 |
| 41753_at | U48734 | 81 | ACTN4: actinin, alpha 4 | 1 |
| 34325_at | AJ005893 | 10084 | PQBP1: polyglutamine binding protein 1 | 2 |
| 34868_at | AB029012 | 23381 | EST1B: Est1p-like protein B | 1 |
| 591_s_at | M33684 | 5770 | PTPN1: protein tyrosine phosphatase, nonreceptor type 1 | 1 |
| 1553_r_at | U22028 | 1553 | CYP2A13: cytochrome P450, family 2, subfamily A, polypeptide 13 | 2 |
| 38522_s_at | X52785 | 933 | CD22: CD22 antigen | 2 |
| 2009_at | U33284 | 2185 | PTK2B: PTK2B protein tyrosine kinase 2 beta | 2 |
| 1994_f_at | AF001359 | 4292 | MLH1: mutL homolog 1, colon cancer, nonpolyposis type 2 (Escherichia coli) | 2 |
| 35620_at | AF043250 | 10452 | TOMM40: translocase of outer mitochondrial membrane 40 homolog (yeast) | 2 |
| 37095_r_at | M84562 | 2358 | FPRL1: formyl peptide receptor-like 1 | 2 |
| 40324_r_at | AF070585 | 8940 | TOP3B: topoisomerase (DNA) III beta | 2 |
| 36094_at | M21984 | 7140 | TNNT3: troponin T3, skeletal, fast | 2 |
| Down-regulated in VH3-21+ B-CLL (non-VH3-21-related) | ||||
| 41796_at | AB029015 | 23228 | PLCL2: phospholipase C-like 2 | 1,2‡,LDA,WV |
| 32172_at | AL096858 | 23013 | SHARP: SMART/HDAC1-assocjated repressor protein | 1,2§,WV |
| 33859_at | U96915 | 10284 | SAP18: sin3-associated polypeptide, 18kDa | 1,LDA,WV |
| 36938_at | U70063 | 427 | ASAH1: N-acylsphingosine amidohydrolase (acid ceramidase) 1 | 1,LDA,WV |
| 32554_s_at | Y12781 | 6907 | TBL1X: transducin (beta)-like 1X-linked | 1,LDA,WV |
| 33847_s_at | A1304854 | 1027 | CDKN1B: cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 1,LDA,WV |
| 38380_at | Y18863 | 10775 | POP4: POP4 (processing of precursor, Saccharomyces cerevisiae) homolog | 1,WV |
| 36827_at | AF020762 | 64746 | ACBD3: acyl-coenzyme A binding domain containing 3 | 1,WV |
| 1453_at | U68018 | 4087 | MADH2: MAD, mothers against decapentaplegic homolog 2 (Drosophila) | 1,WV |
| 41202_s_at | AF000152 | 10106 | CTDSP2: CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) | 1,WV |
| 36031_at | AB024401 | 3621 | ING1: inhibitor of growth family, member 1 | 1,WV |
| 32696_at | X59841 | 5090 | PBX3: pre-B-cell leukemia transcription factor 3 | 1,WV |
| 39434_at | AB011164 | 253725 | KIAA0592: KIAA0592 protein | 1,WV |
| 39345_at | AI525834 | 10577 | NPC2: Niemann-Pick disease, type C2 | 1,LDA |
| 654_at | L07648 | 4601 | MXI1: MAX interacting protein 1 | 1,LDA |
| 36894_at | AL031846 | 23492 | CBX7: chromobox homolog 7 | 2‡,LDA |
| 36502_at | AB020641 | 5218 | PFTK1: PFTAIRE protein kinase 1 | 2‡,LDA |
| 35811_at | AF037204 | 11342 | RNF13: ring finger protein 13 | 1 |
| 37035_at | AI557272 | 27230 | SERP1: stress-associated endoplasmic reticulum protein 1 | 1 |
| 1452_at | U24576 | 8543 | LMO4: LIM domain only 4 | 1 |
| 41379_at | AB011166 | 23137 | SMC5L1: SMC5 structural maintenance of chromosomes 5-like 1 (yeast) | 1 |
| 40063_at | U22897 | 10241 | NDP52: nuclear domain 10 protein | 1 |
| 37253_at | X92493 | 8395 | PIP5K1B: phosphatidylinositol-4-phosphate 5-kinase, type 1, beta | 2§ |
| 35145_at | X96401 | 4335 | MNT: MAX-binding protein | 2§ |
| 1797_at | U40343 | 1032 | CDKN2D: cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 2§ |
| 943_at | D43968 | 861 | RUNX1: runt-related transcription factor 1 (acute myeloid leukemia 1; aml 1 oncogene) | 2§ |
Sample preparation control experiment
A methodologic issue regarding the collection of samples analyzed in this study relates to their variable sample collection procedure, and storage time. These considerations are normally addressed by including a large number of samples in a study, where these factors can be considered effectively randomized. In our material, however, there was a bias toward the VH3-21+ samples having been frozen before RNA extraction, whereas most of the non-VH3-21 samples had been harvested from fresh cells. We therefore performed a set of control experiments to assess the possible influence of freezing status on the results. Our conclusion is that such an effect is likely to be very minor, based on the following data. Five B-CLL samples were tested such that fresh and frozen samples from the same patient were compared in a marker analysis and probes related to freezing were identified. These probes were then compared with probes identified in the VH3-21+ versus non-VH3-21 comparison. A few probes (3 of 50) were found to be common in the comparisons. These genes were: EIF4G1: eukaryotic translation initiation factor 4 gamma, 1, probe id. 32 844_at, TOP3B: topoisomerase (DNA) III beta, probe id. 40 324_r_at and Cas-Br-M (murine) ectropic retroviral transforming sequence b, probe id. 514_at. It is therefore likely that these common probes related to freezing status and which were low ranked among VH3-21+-related probes, are of very limited relevance for the overall VH3-21+ versus non-VH3-21 comparison profile.
RQ-PCR
The differential expression of 4 genes (PPP5C, E2F4, SHARP, CDKN1B) was investigated by real-time quantitative (RQ)-PCR analysis of 4 VH3-21+ and 4 non-VH3-21 samples. RQ-PCR fold-change values for each gene were calculated and tabulated (Table 2), along with the fold-change analysis obtained from the microarray data. Three out of 4 genes showed changes in the same direction with both methods. However, SHARP, which was down-regulated in VH3-21+ B-CLLs, could not be verified with RQ-PCR. Mapping of the precise sequence coverage of the Affymetrix array probe target sequence and the RQ-PCR primers/probe in the ENSEMBL genome database (www.ensembl.org) showed a clear difference concerning which part of the SHARP mRNA that was detected with the 2 methods (data not shown). Thus, the discrepancy in the experimental results could be explained if SHARP is expressed as alternative mRNAs (eg, splice variants) and the differential regulation only concerns one of the mRNA variants. A specific investigation using multiple probes will be warranted to address this question.
Verification of differential expression by Taqman RQ-PCR
. | . | Fold change* . | . | |
|---|---|---|---|---|
| Gene . | Probe set . | Microarray . | RT-PCR . | |
| PPP5C | 391_at | 4.30 | 3.80 | |
| E2F-4 | 1703_g_at | 3.03 | 1.41 | |
| SHARP | 32172_at | −2.30 | 1.92 | |
| CDKN1B | 33847_s_at | −1.88 | −4.16 | |
. | . | Fold change* . | . | |
|---|---|---|---|---|
| Gene . | Probe set . | Microarray . | RT-PCR . | |
| PPP5C | 391_at | 4.30 | 3.80 | |
| E2F-4 | 1703_g_at | 3.03 | 1.41 | |
| SHARP | 32172_at | −2.30 | 1.92 | |
| CDKN1B | 33847_s_at | −1.88 | −4.16 | |
Fold change values from analysis of microarray and standard curve analysis of RQ-PCR data are given for each gene.
Biological annotation of identified genes
In order to obtain a broader, global description of the biologic patterns in the 2 groups correlating with VH3-21 and non-VH3-21 usage, the gene identifiers were linked to the Gene Ontology database. Pathways in biologic processes and molecular functions that showed significant enrichment for the genes that differed in expression between samples with VH3-21 and non-VH3-21 usage is shown in Table 3, which is based on the 200 best-ranked up- and down-regulated genes that were identified in the marker analysis. The table shows representative GO categories in “Biologic process” and “Molecular function” with the “parent” category and its corresponding “child” categories. For example, VH3-21 samples demonstrated statistical overrepresentation of genes in the functional categories “cell organization and biogenesis,” “cell cycle,” and “protein modification.”
Representative gene ontology categories enriched in VH3-21+ and non-VH3-21B-CLL
GO category . | O . | E . | R . | P . |
|---|---|---|---|---|
| Biologic process | ||||
| Up-regulated in VH3-21+ B-CLL | ||||
| Cell organization and biogenesis | 19 | 10 | 1.9 | .005 |
| Cell cycle | 23 | 10.77 | 2.14 | <.001 |
| Regulation of cell cycle | 15 | 6.49 | 2.31 | .002 |
| DNA replication and chromosome cycle | 8 | 2.78 | 2.88 | .006 |
| Protein modification | 30 | 17.42 | 1.72 | .002 |
| Protein amino acid dephosphorylation | 6 | 1.68 | 3.57 | .007 |
| Protein amino acid phosphorylation | 19 | 7.70 | 2.47 | <.001 |
| Transmembrane receptor protein tyrosine kinase signaling pathway* | ||||
| Signal complex formation | 2 | 0.12 | 16.67 | .006 |
| Down-regulated In VH3-21+ B-CLL | ||||
| Transport* | ||||
| Intracellular transport | 13 | 6.26 | 2.08 | .01 |
| Molecular function | ||||
| Up-regulated In VH3-21+ B-CLL | ||||
| Transferase activity, transferring phosphorus-containing groups | 25 | 11.7 | 2.14 | <.001 |
| Kinase activity | 24 | 10.39 | 2.31 | <.001 |
| Down-regulated In VH3-21+ B-CLL | ||||
| Protein kinase regulator activity | 4 | 0.76 | 5.26 | .007 |
| Protein kinase inhibitor activity | 3 | 0.38 | 7.89 | .006 |
| Hydrogen ion transport activity | 6 | 1.83 | 3.28 | .01 |
| Cytochrome-c oxidase activity | 3 | 0.34 | 8.82 | .004 |
GO category . | O . | E . | R . | P . |
|---|---|---|---|---|
| Biologic process | ||||
| Up-regulated in VH3-21+ B-CLL | ||||
| Cell organization and biogenesis | 19 | 10 | 1.9 | .005 |
| Cell cycle | 23 | 10.77 | 2.14 | <.001 |
| Regulation of cell cycle | 15 | 6.49 | 2.31 | .002 |
| DNA replication and chromosome cycle | 8 | 2.78 | 2.88 | .006 |
| Protein modification | 30 | 17.42 | 1.72 | .002 |
| Protein amino acid dephosphorylation | 6 | 1.68 | 3.57 | .007 |
| Protein amino acid phosphorylation | 19 | 7.70 | 2.47 | <.001 |
| Transmembrane receptor protein tyrosine kinase signaling pathway* | ||||
| Signal complex formation | 2 | 0.12 | 16.67 | .006 |
| Down-regulated In VH3-21+ B-CLL | ||||
| Transport* | ||||
| Intracellular transport | 13 | 6.26 | 2.08 | .01 |
| Molecular function | ||||
| Up-regulated In VH3-21+ B-CLL | ||||
| Transferase activity, transferring phosphorus-containing groups | 25 | 11.7 | 2.14 | <.001 |
| Kinase activity | 24 | 10.39 | 2.31 | <.001 |
| Down-regulated In VH3-21+ B-CLL | ||||
| Protein kinase regulator activity | 4 | 0.76 | 5.26 | .007 |
| Protein kinase inhibitor activity | 3 | 0.38 | 7.89 | .006 |
| Hydrogen ion transport activity | 6 | 1.83 | 3.28 | .01 |
| Cytochrome-c oxidase activity | 3 | 0.34 | 8.82 | .004 |
The table is based on the 200 best marker genes from the marker analysis of VH3-21+ versus non-VH3-21 B-CLL. O indicates observed gene number in the GO category; E, expected gene number in the GO category; R, ratio of enrichment for the GO category; and P, significance of enrichment for the GO category. Representative parent and child GO categories are shown. A detailed description of all GO categories is included as direct acyclic graphs (DAGs) in Supplemental Figure S2.
Nonsignificant GO category, but the “child” category showed significant enrichment.
BCR-signaling-related genes
Since the different comparisons in the previous paragraph did not reveal any obvious dysregulation of genes involved in BCR signaling, we also compared the expression levels of a selection of known BCR-related genes29 (BCR-signaling complex genes: SYK, LYN, FYN, BLNK, BLK, VAV1, VAV2, GRB2, ZAP-70; genes associated with BCR-signaling: CD19, CD22, CD23/FCER2; genes involved in downstream pathways affected by BCR signaling: PLCG2, PIK3CD, PIK3C2B, AKT, RAF1, ERK, PRKCB1, JNK, NFKBIE, NFKB2, NFKBIB, RELA, CCND2, NFATC1, NFATC3, GAB2), but only 3 of these genes were among the 200 highest up-regulated/down-regulated genes in the VH3-21 group: NFKB2, CD22, and NFATC1, none of which showed a high-ranking position (positions 133, 105, and 84, respectively).
Discussion
During recent years, the VH genes have been extensively studied in B-CLL and it has been observed, based on the mutation status, that Ig-unmutated B-CLLs have a more severe disease course than patients with B-CLL with mutated VH regions.1-4 Recently, Tobin et al11 showed that CLL patients who use the VH3-21 gene segment have a poor prognosis despite VH gene mutation status and that many of these cases also display peculiar molecular features with homologous heavy- and light-chain Ig rearrangements, which implies that antigen(s) could be involved in the disease pathogenesis. Based on these data, the VH3-21 subset has been suggested to comprise a third B-CLL entity.14 As a first attempt to investigate differences in gene expression in the VH3-21 group, we have analyzed the global gene expression profile in a group of 39 patients with B-CLL consisting of the 3 different subcategories (eg, Ig-unmutated, Ig-mutated, and VH3-21+ B-CLL).
VH3-21+ B-CLLs display a distinctive gene expression profile
First, a marker analysis revealed that VH3-21+ and non-VH3-21 B-CLLs were clearly separated and at least 1000 markers for each group were significant at the 5% level (Figure 1). Thus, VH3-21+ and non-VH3-21 B-CLLs are distinguished by pronounced differences in gene expression (Figure 1). Next, we attempted to build a model, which would correctly identify the VH3-21+ cases using the expression patterns of a number of genes. With the weighted voting algorithm and a 27-gene model, a 90% correct classification was achieved (Figure 2), whereas the LDA approach yielded an even more accurate classification based on 57 genes (Figure 3). Thus, it was possible to obtain a good discriminatory power for the distinction between VH3-21+ and non-VH3-21 B-CLLs using 2 different algorithms, which shows that these 2 sample groups are distinguishable based only on the expression patterns of a limited number of genes. In addition, the 3-group comparison (Figure 4), including all 3 subgroups in one marker analysis, demonstrated that the VH3-21 group was more clearly and significantly distinguished from the Ig-unmutated/mutated groups than either of these compared with each other. Taken together, our analyses underline the observation that the VH3-21+ group of B-CLL expresses a particular and identifiable gene expression profile, which may be searched for functional clues to the underlying pathology.
Based on the preceding analyses, a composite of the best-ranked identified genes from the 2 marker gene analyses was assembled in Table 1. This list also shows genes that were consistently identified as being related to VH3-21 status using different analytical approaches (eg, LDA and weighted voting). Two IgVH genes, 1 VH3-23 gene, and 1 VH3 family gene (which could represent either the VH3-21, VH3-07, or VH3-74 gene), were among the best reproduced VH3-21-related markers present in both marker gene analyses and the 2 class prediction models. The up-regulated VH3-23 serves as a positive marker and confirms the VH3-21 genotype, since VH3-23 is the product of the VH3-23 gene that is closely related to the VH3-21 gene.
Dysregulated transcription and cell-cycle control in the VH3-21 group
Among the highest ranked up-regulated genes in the VH3-21 group, several are involved in the regulation of transcription and cell-cycle control (eg, PPP5C, E2F4, SMARCD1, ARHGEF1, TYMS, HMGA1 and EIF4G1), which may provide cells with an increased global or specific transcription activity as well as a higher general proliferative drive. An increase in expression of protein phosphatase 5 has been shown to block p53-mediated p21 growth suppression,30 whereas overexpression of the E2F transcription factor 4 was reported to activate transcription.31 The SMARCD1/BAF60a have also been shown to be involved in regulation of transcription.32,33 The function of the up-regulated guanine nucleotide exchange factor (GEF) 1 is less well known. This protein belongs to the Rho guanosine triphosphatases (GTPases), which play a fundamental role in numerous cellular processes that are initiated by extracellular stimuli through G protein-coupled receptors. Furthermore, the up-regulated TYMS protein has been associated with drug resistance in colorectal cancer,34 and this protein as well as the HMGA1 and EIF4G1 proteins was shown to be overexpressed in recent microarray studies of different cancers.35-37
Among the down-regulated genes in the VH3-21 group, there are several examples of genes involved in repression of transcription and negative control of cell cycle progression (eg, SHARP, SAP18, TBL1X, MXI-1, CDKN1B, CDKN2D, and MADH2). The histone deacetylase (HDAC) complex is known to repress transcription and several proteins involved in this complex were found down-regulated in the VH3-21 group. For instance, reduced SHARP protein, SAP18, and TBL1X38 expression levels may result in reduced HDAC complex formation and thus decreased transcriptional repression. The down-regulated MXI-1 protein is another transcriptional repressor that has been shown to interact with MAX and result in MYC activation.39
The down-regulated cyclin-dependent kinase (CDK) inhibitor 1B (CDKN1B, p27, Kip1) and cyclin-dependent kinase inhibitor 2D (CDKN2D, p19, inhibits CDK4) are both proteins that bind to CDKs and prevents G1 progression.40 The ING1 tumor suppressor interacts with p53, thereby promoting cell-cycle arrest and apoptosis, and has been reported to show decreased expression in both lung cancer and acute lymphoblastic leukemia (ALL).41-43 MADH2/SMAD2 is involved in transforming growth factor (TGF)-β signaling,44 and mutations in MADH2/SMAD2 will provide resistance of cells to TGF-β induced growth inhibition.
The categorization of genes into functional groups also gives indications of the increased growth and cell proliferation among the VH3-21+ samples. Representative GO categories, such as “regulation of cell cycle,” “DNA replication and chromosome cycle,” and “kinase activity,” contain twice as many genes than expected that are involved in these processes (Table 3), and thus indicate that these enriched functional categories might be of importance for the more aggressive phenotype in VH3-21+ tumors.
No profound evidence of increased BCR signaling in the VH3-21+ group
Considering the speculated antigen involvement in the VH3-21+ group and the possibility of ongoing antigen activation through the VH3-21-encoded BCR, we were interested to study whether this subset displayed up-regulation of BCR-signaling pathways. We therefore compared the expression levels of a selection of BCR-signaling-related genes (“Results”), but no profound overexpression was evident in the VH3-21+ group. Furthermore, 1 signaling molecule that has caught much attention lately is the tyrosine kinase zeta-associated protein (ZAP)-70, which is overexpressed in unmutated B-CLLs and thought to be involved in increased BCR signaling within this subset.21,45-47 However, since we did not deplete the T cells, which also express ZAP-70, we cannot address the issue of ZAP-70 expression in our subgroups properly, in contrast to previous microarray studies.20,21
In conclusion, we have shown that the VH3-21+ group has a distinctive expression profile compared with Ig-unmutated and Ig-mutated CLLs, and that different class discriminator models can be applied to accurately classify a sample as being VH3-21+ solely based on gene expression data. Among the highest ranked up- and down-regulated genes, several were involved in regulation of DNA replication/cell cycling as well as transcriptional activation and kinase activity. It is thus tempting to speculate that these dysregulated genes could reflect a more proliferative phenotype in VH3-21+ tumors, which is ultimately expressed as a clinically more aggressive disease.
Prepublished online as Blood First Edition Paper, April 7, 2005; DOI 10.1182/blood-2004-10-4073.
Supported by grants from the Swedish Cancer Society and the Stockholm Cancer Society. The Wallenberg Consortium provided support to the Affymetrix core facility.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors are grateful to Emma Emanuelsson, Ann-Marie Forsblom, Monika Jansson, Ann Wallblom, Karin Karlsson, and Anna Laurell for skillful technical assistance and for assistance with clinical data, and to Ola Söderberg and Christer Sundström for providing CLL samples.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal