Abstract
The pericentric inversion of chromosome 16, inv(16)(p13q22), is associated with acute myeloid leukemia (AML) subtype M4Eo that is characterized by the presence of myelomonocytic blasts and atypical eosinophils. This rearrangement fuses the CBFB and MYH11 genes, with the latter encoding the smooth muscle myosin heavy chain (SMMHC). The myeloid transcription factor CCAAT/enhancer-binding protein α (CEBPA) is crucial for normal granulopoiesis. Alterations of structure and expression of CEBPA have been implicated in particular subtypes of AML. Here, we found that conditional expression of core-binding factor β (CBFB)-SMMHC in U937 cells suppresses CEBPA protein expression and binding activity. However, CEBPA mRNA levels remained unchanged. No differences were detected in CEBPA mRNA levels in patients with inv(16) AML-M4Eo (n = 12) compared to patients with AML with a normal karyotype and M4 subtype (n = 6), whereas CEBPA protein and binding activity were significantly reduced in patients with CBFB-SMMHC. Furthermore, calreticulin, an inhibitor of CEBPA translation, was induced on mRNA and protein level in CBFB-SMMHC patients with AML and after expression of CBFB-SMMHC in the U937-cell system. Inhibition of calreticulin by siRNA restored CEBPA levels. Our results suggest that modulation of CEBPA by calreticulin represents a novel mechanism involved in the differentiation block in CBFB-SMMHC AML. (Blood. 2005;106:1369-1375)
Introduction
A hallmark of acute myeloid leukemia (AML) is the association of distinct morphologic subtypes with particular chromosomal rearrangements.1,2 Whereas the AML1-ETO translocation or t(8;21) is usually observed in AML subtypes M1 or M2, the inversion of chromosome 16, inv(16)(p13q22), is a typical finding in AML-M4Eo.3 The latter is characterized by the presence of myelomonocytic blasts and atypical eosinophils. Inv(16) and the rarer t(16;16)(p13;q22) fuse the CBFB/PEBP2B and MYH11 genes, with MYH11 encoding the smooth muscle myosin heavy chain (SMMHC).4-8 The chimeric core-binding factor (CBF) protein CBFB-SMMHC contains the N-terminal 165 amino acids of CBFB fused to the C-terminal coiled-coil region of SMMHC. The phenotype of heterozygous cbfb-smmhc knock-in mice is embryonically lethal, with definitive hematopoiesis blocked at the stem-cell level.9 Expression of cbfb-smmhc in transgenic mice models predisposes to a disease morphologically similar to AML-M4Eo; however, by itself it is not sufficient to induce AML.9
The CBFB/PEBP2B gene forms together with its DNA-binding partner AML1/RUNX1 the heterodimeric transcription factor polyomavirus enhancer-binding protein 2 (PEBP2)/CBF.1-3,10,11 Both partner genes are frequently involved in chromosomal alterations in AML.10 In the t(8;21) rearrangement the N-terminal of AML1/RUNX1 is fused to almost the entire ETO gene.2,10 We previously demonstrated in AML patient samples that the AML1-ETO fusion suppresses the transcription factor CCAAT/enhancer-binding protein α (CEBPA) and that restoring CEBPA in these cells is sufficient to induce terminal neutrophil differentiation.12 Interestingly, we and others also found dominant-negative mutations of the CEBPA gene in a significant proportion of patients with myeloblastic subtypes (M1, M2) of AML.13-15 In addition, CEBPA expression or function can be abolished by the tyrosine kinase receptor FLT3 in AML.16,17 Posttranscriptional modulation of CEBPA can be mediated by the poly(rC)-binding protein hnRNP E2 in chronic myeloid leukemia (CML) blast crisis or by the leukemic fusion protein AML1-myelodysplasia syndrome-associated protein 1 (MDS1)-ecotropic viral integration site-1 (EVI1).18,19
Because we previously identified the CEBPA gene as an important downstream target of the AML1-ETO fusion,12 we here analyzed whether the leukemic fusion protein CBFB-SMMHC affects CEBPA similarly to AML1-ETO. Surprisingly, we found that CBFB-SMMHC suppresses CEBPA protein. In contrast to the AML1-ETO fusion, CBFB-SMMHC fails to affect CEBPA mRNA. As a novel mechanism involved in leukemia, we found that the translational inhibition of CEBPA is mediated by induction of calreticulin, a ubiquitous protein with calcium storage and chaperone function.
Patients, materials, and methods
Cells from patients with AML
Whole-cell lysates and nuclear extracts for Western blotting, Transam assays and UV cross-linking experiments were obtained from Ficoll-separated fresh mononucleated peripheral blood or bone marrow cells of patients with AML collected at the time of diagnosis before initiation of treatment. Conventional karyotype analysis was performed for each patient. Metaphase chromosomes were banded by conventional banding technique. A karyotype was considered normal if at least 20 metaphases remained without evidence of a clonal abnormality. Approval of these studies was obtained from the ethics committee of the University of Berne, Switzerland. Informed consent was provided according to the Declaration of Helsinki.
Conditional CBFB-SMMHC expression in myeloid cells
The U937 T-cell line was kindly provided by Gerard Grosveld (Memphis, TN). These cells are stably transfected with the tetracycline transactivator (tTA) under the control of a tetracycline (tet)-responsive element. To create a system for conditional expression of CBFB-SMMHC, a 4.0-kb Sca1/Xba1 fragment of the pcDNA3 vector, containing the neomycin resistance gene, was ligated with the 0.95-kb Sca1/Xba1 fragment of the tet-off response plasmid pTRE. A 2.0-kb EcoR1/Not1 fragment encoding for the entire CBFB-SMMHC cDNA was then introduced into the pTRE-neo plasmid. The plasmid was stably transfected into U937 T cells by electroporation. Twenty-four single-cell clones were expanded under neomycin and puromycin selection. To test the clones for inducible expression of CBFB-SMMHC, cells were washed 3 times in 50 mL phosphate-buffered saline (PBS) and seeded at a density of 2 × 105 cells/mL in the absence of tetracycline. The increase of CBFB-SMMHC mRNA transcripts was measured by quantitative real-time PCR (RT-PCR).
FACS analysis
Cells were incubated in PBS with 2% (wt/vol) bovine serum albumin (BSA) on ice for 1 hour with the antibody (50 ng CD11b antibody/106 cells; DakoCytomation, Glostrup, Denmark). Cells were washed and resuspended in PBS with 10% formaldehyde. We analyzed samples on a fluorescence-activated cell sorting (FACScan) flow cytometer (Becton Dickinson, San Jose, CA) using CellQuest software (Largo, FL).
Quantitative RT-PCR
RT-PCR was performed on the ABI PRISM 7700 Sequence Detection System using TaqMan Universal PCR Master Mix. For calreticulin and CEBPA mRNA quantitation Assays-on-Demand Gene Expression probes (Applied Biosystems, Foster City, CA) were used. For CBFB-SMMHC detection the primers were 5′-AAGACTGGATGGTATGGGCTGT-3′ and 5′-CAGGGCCCGCTTGGA-3′ and the probe was 5′-FAM-TGGAGTTTGATGAGGAGCGAGCCCT-TAMRA-3′. 7S was used as a reference gene for normalization, and the primers (Microsynth, Balgach, Switzerland) were 5′-ACCACCAGGTTGCCTAAGGA-3′ and 5′-CACGGGAGTTTTGACCTGCT-3′, the probe was 5′FAM-TGAACCGGCCCAGGTCGGAAAC-TAMRA-3′. The sensitivity was verified by stepwise diluting cDNA from U937 cells by 50%. Correct size of PCR products was verified by gel electrophoresis. Sequencing of the CEBPA gene was performed as previously described.13
EMSAs
A nuclear extraction kit (Active Motif, Carlsbad, CA) was used for extraction of nuclear extracts. The granulocyte colony-stimulating factor receptor (G-CSF R) promoter oligonucleotide (bp -57 to -38, with CEBP-binding site underlined) had the sequence 5′-AAGGTGTTGCAATCCCCAGC-3′. Electrophoretic mobility shift assays (EMSAs) were performed as described elsewhere.12,13,20,21 The CEBPA antibody (sc-61 X) was from Santa Cruz Biotechnology (Santa Cruz, CA) as well as the OCT-1 antibody (sc-232 X). Quantitative CEBPA- and CEBPB-binding activity was further assessed using an enzyme-linked immunosorbent assay (ELISA)-based assay (TransAM, Active Motif). Briefly, a 96-well plate was coated with the immobilized CEBP consensus-binding site oligo 5′-CTTGCGCAATCTATA-3′. Nuclear extracts were added together with a CEBPA antibody. Addition of a secondary antibody conjugated to horseradish peroxidase (HRP) provided sensitive colorimetric quantitation by conventional spectrophotometry. Specificity of the assay was further verified by the addition of excess oligo with a CEBP wild-type or mutated consensus-binding sequence.
UV cross-link assay for assessment of calreticulin mRNA-binding activity
A double-stranded RNA oligomer covering a calreticulin-binding site within the CEBPA mRNA was generated as follows. Oligomer A 5′-CCCCACGGGCGGCGGCGGCGGCGGCGACUU-3 containing CGG repeats, and oligomer B 5′-UAACCAGCCGCCGCCGCCGCCGCCGCCGCCGCCC-3′ containing CCG repeats were annealed. Double-stranded oligomers were separated from single-stranded oligomers by gel electrophoresis and subsequent extraction. The double-stranded oligomers were labeled using T4 kinase and p32-γ adenosine triphosphate (ATP). The UV cross-link essay was previously described in more detail.22 Results were analyzed using the software Quantity One 4.4.0 from Bio-Rad (Hercules, CA).
Assay for RNA interference
Calreticulin siRNA (Ambion, Austin, TX) had the sequences 5′-GGAGCAGUUUCUGGACGGATT-3′ and 5′-UCCGUCCAGAAACUGCUCCTT-3′. As control, the Silencer Negative Control no. 2 siRNA (Ambion; catalogue no. 4613) was used. U937 cells with inducible CBFB-SMMHC expression were set to a density of 1.4 × 106 in 100 μL Amaxa solution V (Nucleofactor Kit V, Amaxa, Cologne, Germany) and mixed with 800 ng siRNA. Cells were transfected by electroporation applying Nucleofector technology (software V2.1, Amaxa).
Western blot analysis
Whole-cell lysates were separated on 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane (Bio-Rad), blocked in 5% nonfat dry milk in Tris (tris(hydroxymethyl)aminomethane)-buffered saline with 0.1% Tween 20 (TBS-T) for 1 hour at room temperature, and then incubated with primary antibodies in TBS-T (with 2% nonfat dry milk) overnight at 4°C. CEBPA, CEBPB, CEBPE, G-CSF R, CBFB, and calreticulin were detected with rabbit polyclonal antibody against CEBPA(1:500; Santa Cruz Biotechnology, catalog no. sc-61), a rabbit polyclonal antibody against CEBPB (1:1000; Santa Cruz Biotechnology, catalog no. sc-150) a rabbit polyclonal antibody against CEBPE (1:1000; Santa Cruz Biotechnology, catalog no. sc-158), a rabbit polyclonal antibody against G-CSF R(1:500; Santa Cruz Biotechnology, catalog no. sc-694), a rabbit polyclonal antibody against CBFB (1:250;Active Motif, catalog no. 39501), and a rabbit polyclonal antibody against calreticulin (1:200 000; Sigma, St Louis, MO; catalog no. C4606), respectively, followed by an IgG-HRP-conjugated secondary antibody against rabbit (Amersham Biosciences, Freiburg, Germany; catalog no. NA934). CBFB proteins were detected with mouse polyclonal antibody against CBFB (1:1000, Sigma, catalog no. sc-17181). A monoclonal anti-mouse B-actin antibody served as a loading control (Sigma, catalog no. A2066).
Statistical analysis
Means and SDs were calculated. Statistical analysis was performed using the Mann-Whitney rank sum test (SigmaStat 3.0; Systat, Erkrath, Germany).
Results
Conditional expression of CBFB-SMMHC blocks myeloid differentiation
We expanded 24 single-cell clones of the myeloid leukemic cell line (U937) that conditionally expresses the CBFB-SMMHC protein following withdrawal of tetracycline. Quantitative RT-PCR with a probe specific against the CBFB-SMMHC transition sequence was performed. Seven clones showed a more than 10-fold increase in CBFB-SMMHC mRNA with a range from 12 to 1365-fold 2 days after withdrawal of tetracycline. Among these 7 clones, we arbitrarily selected no. 23 for the experiments presented in this study. Clone no. 23 reached a greater than 100-fold increase of CBFB-SMMHC mRNA on day 2 after withdrawal of tetracycline as compared with day 0.
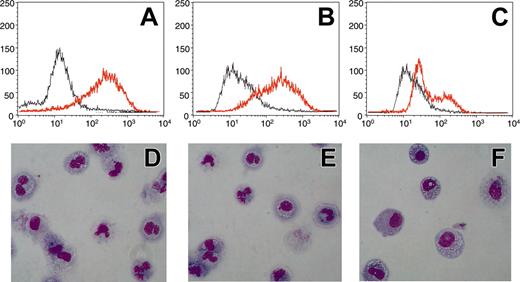
Conditional expression of CBFB-SMMHC in U937 leukemic cells blocks myeloid differentiation. U937 were treated with 0.5 μM ATRA for 5 days. CD11b expression was determined by FACS analysis (A-C) and morphology was examined by hematoxylin-eosin staining of cytospins at day 5 (D-F). Analysis was performed in U937T cells (A,D), in U937-tetoff-CBFB-SMMHC cells without induction of the CBFB-SMMHC fusion (B,E), and in U937-tetoff-CBFB-SMMHC cells following induction of CBFB-SMMHC protein (C,F). Gray lines indicate day 0; redlines, day 5 (A-C). Magnification (D-F) was × 40 with a Plan Fluor objective (Ph2 DLL; Nikon, Tokyo, Japan). Camera was a Nikon Digital Camera Dxm 12000. Acquisition software was from Lucia image (version 4; Prague, Czech Republic).
Conditional expression of CBFB-SMMHC in U937 leukemic cells blocks myeloid differentiation. U937 were treated with 0.5 μM ATRA for 5 days. CD11b expression was determined by FACS analysis (A-C) and morphology was examined by hematoxylin-eosin staining of cytospins at day 5 (D-F). Analysis was performed in U937T cells (A,D), in U937-tetoff-CBFB-SMMHC cells without induction of the CBFB-SMMHC fusion (B,E), and in U937-tetoff-CBFB-SMMHC cells following induction of CBFB-SMMHC protein (C,F). Gray lines indicate day 0; redlines, day 5 (A-C). Magnification (D-F) was × 40 with a Plan Fluor objective (Ph2 DLL; Nikon, Tokyo, Japan). Camera was a Nikon Digital Camera Dxm 12000. Acquisition software was from Lucia image (version 4; Prague, Czech Republic).
No morphologic changes were detectable over a period of 14 days after induction of CBFB-SMMHC (data not shown). To analyze whether CBFB-SMMHC affects myeloid differentiation in this system, we treated U937 cells with 0.5 μM all-trans-retinoic acid (ATRA) for 5 days. This treatment induces neutrophil differentiation in the parental U937 T cells (Figure 1D). U937-tetoff-CBFB-SMMHC cells, without induction of the fusion protein, also differentiate to neutrophils if treated with ATRA (Figure 1E). However, if the CBFB-SMMHC fusion is expressed following withdrawal of tetracycline, U937-tetoff-CBFB-SMMHC cells fail to differentiate (Figure 1F). Determination of CD11b expression as a marker for myeloid differentiation is shown in Figure 1A-C to further illustrate these observations. Whereas ATRA induces expression of CD11b in U937-T and in U937-tetoff-CBFB-SMMHC cells in the presence of tetracycline, this increase is not observed in U937 cells after CBFB-SMMHC induction. We thus conclude that forced expression of CBFB-SMMHC in U937 cells blocks ATRA-dependent neutrophil differentiation.
CBFB-SMMHC suppresses CEBPA protein in U937 leukemic cells
The transcription factor CEBPA is crucial for normal differentiation of granulocytes.1,21 In addition, an increasing number of reports indicate that CEBPA mutations as well as altered CEBPA expression are involved in the pathogenesis of particular AML subtypes.13-18 We therefore assessed whether the fusion protein CBFB-SMMHC affects CEBPA mRNA or protein or both.
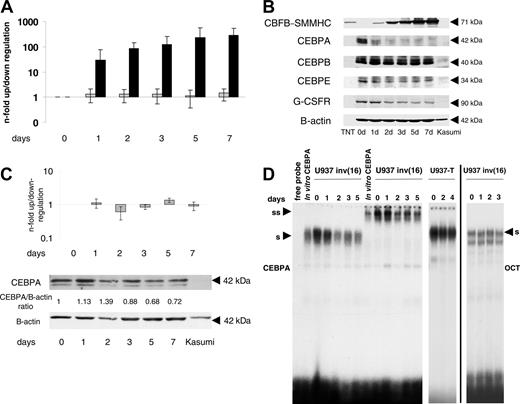
CEBPA mRNA levels were determined 48 hours after induction of CBFB-SMMHC in U937 cells by RT-PCR. CEBPA mRNA levels did not change in any of the 7 clones that show a more than 10-fold increase in CBFB-SMMHC mRNA (n-fold range, 0.66-1.32; mean 0.93; data not shown). Also, no CEBPA mRNA changes were observed in clone no. 23 depicted in Figure 2A representing pooled data from 5 independent experiments (n-fold range, 0.69-1.41). On day 2, the median absolute Ct value for CBFB-SMMHC was 31.78 (SD 0.58) and for CEBPA 17.86 (SD 0.61). These results suggest that CEBPA mRNA levels are not significantly affected by forced expression of CBFB-SMMHC protein.
Conditional expression of CBFB-SMMHC in U937 leukemic cells. U937 cells were analyzed before (day 0) and 1, 2, 3, 5, and 7 days after withdrawal of tetracycline. (A) Quantitative RT-PCR analysis for CBFB-SMMHC and CEBPAmRNAexpression. Gray bars indicate CEBPAmRNA levels, and black bars indicate induction of CBFB-SMMHC mRNA. Results of 5 independent experiments are expressed as n-fold up/down-regulation and compared to day 0. Mean values and SDs are shown. (B) Western blot analysis of whole-cell lysates harvested at the same time points noted in panel A. The CBFB-SMMHC fusion protein (71-kDa) was detected with a CBFB antibody. TNT is an in vitro-translated CBFB-SMMHC protein from a CBFB-SMMHC expression construct that served as positive control. The membrane was further incubated with antibodies against CEBPA, CEBPE, granulocyte-colony-stimulating factor receptor (G-CSFR) and B-actin. (C) Parental U937 cells with the tetracycline-inducible construct lacking the CBFB-SMMHC cDNA (U937-T) were analyzed by quantitative RT-PCR for CEBPA mRNA expression (top panel) and by Western blot analysis for CEBPA and β-actin protein expression (lower panels). Mean values and SDs are shown. (D) CEBPA-binding activity to a CEBP site present in the G-CSF R promoter was assessed by EMSA using nuclear extracts from time points as indicated after CBFB-SMMHC induction. In vitro-translated CEBPA protein served as a positive control. U937-T indicates the parental U937 cell line lacking the inducible CBFB-SMMHC construct; S, shifted CEBPA protein; SS, supershifted CEBPA complex. Binding of the extracts to an octamer (OCT) consensus site indicates equal loading and integrity of the samples.
Conditional expression of CBFB-SMMHC in U937 leukemic cells. U937 cells were analyzed before (day 0) and 1, 2, 3, 5, and 7 days after withdrawal of tetracycline. (A) Quantitative RT-PCR analysis for CBFB-SMMHC and CEBPAmRNAexpression. Gray bars indicate CEBPAmRNA levels, and black bars indicate induction of CBFB-SMMHC mRNA. Results of 5 independent experiments are expressed as n-fold up/down-regulation and compared to day 0. Mean values and SDs are shown. (B) Western blot analysis of whole-cell lysates harvested at the same time points noted in panel A. The CBFB-SMMHC fusion protein (71-kDa) was detected with a CBFB antibody. TNT is an in vitro-translated CBFB-SMMHC protein from a CBFB-SMMHC expression construct that served as positive control. The membrane was further incubated with antibodies against CEBPA, CEBPE, granulocyte-colony-stimulating factor receptor (G-CSFR) and B-actin. (C) Parental U937 cells with the tetracycline-inducible construct lacking the CBFB-SMMHC cDNA (U937-T) were analyzed by quantitative RT-PCR for CEBPA mRNA expression (top panel) and by Western blot analysis for CEBPA and β-actin protein expression (lower panels). Mean values and SDs are shown. (D) CEBPA-binding activity to a CEBP site present in the G-CSF R promoter was assessed by EMSA using nuclear extracts from time points as indicated after CBFB-SMMHC induction. In vitro-translated CEBPA protein served as a positive control. U937-T indicates the parental U937 cell line lacking the inducible CBFB-SMMHC construct; S, shifted CEBPA protein; SS, supershifted CEBPA complex. Binding of the extracts to an octamer (OCT) consensus site indicates equal loading and integrity of the samples.
Using protein extracts from the experiment described, Western blot analysis verified the strong induction of the 71-kDa CBFB-SMMHC fusion protein in accordance with the mRNA induction (Figure 2B). Interestingly and in contrast to CEBPA mRNA levels, CEBPA protein was gradually and strongly suppressed following withdrawal of tetracycline (Figure 2B). The same Western blot was subsequently incubated with antibodies directed against other CEBP family members and G-CSF R (Figure 2B). CEBPB protein remained unchanged after CBFB-SMMHC induction, consistent with our findings after conditional AML1-ETO induction in U937 cells.12 CEBPE represents a downstream target of CEBPA.12,13 Indeed, we observed a delayed but marked decrease in CEBPE protein following induction of CBFB-SMMHC. In addition, the G-CSF R protein as another direct target of CEBPA12,13,20,21 was similarly suppressed following CBFB-SMMHC induction.
To investigate whether the induction of the tetracycline-based system itself had any effect on CEBPA expression, parental U937 cells (U937-T) with the tTA constructs but lacking the CBFB-SMMHC cDNA were analyzed. No significant changes of CEBPA mRNA or protein levels were detectable after withdrawal of tetracycline (Figure 2C). We therefore conclude that conditional expression of the CBFB-SMMHC fusion gene suppresses CEBPA protein but not CEBPA mRNA levels.
DNA-binding activity of CEBPA was tested using nuclear extracts from the same time course experiment. In unstimulated U937 cells, almost the entire CEBP-binding activity to a downstream target such as the G-CSF R promoter is contributed by CEBPA.20,21 Starting 24 hours after CBFB-SMMHC induction we observed a consistent decrease of CEBPA binding to the G-CSF R promoter oligonucleotide (Figure 2D). Again, we observed no changes in DNA-binding activity in the parental U937 T cells after withdrawal of tetracycline (Figure 2D). In addition, equal binding of the extracts to an OCT consensus-binding site indicates equal loading and integrity of the samples. These experiments suggest that CBFB-SMMHC suppresses CEBPAprotein production and function.
In patients with AML-M4Eo, the CBFB-SMMHC fusion protein specifically suppresses CEBPA protein
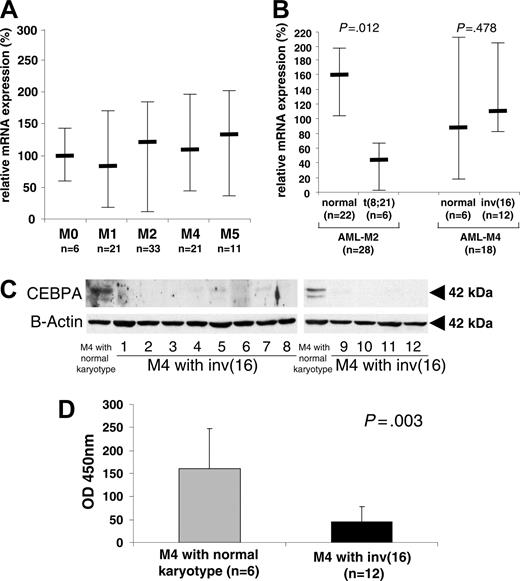
Little is known about CEBPA expression in patients with AML carrying the CBFB-SMMHC fusion gene. We previously reported that CEBPA mRNA is equally expressed in CBFB-SMMHC AML and in a group of normal-karyotype patients with AML.12 In contrast, patients with AML who had the AML1-ETO fusion expressed significantly less CEBPA mRNA than the other sub-groups mentioned.12 To extend these data, we screened 92 patients with AML of various FAB or cytogenetic subtypes for CEBPA mRNA expression. The results according to FAB subtypes are summarized in Figure 3A.
In particular, we compared 12 AML-M4Eo patients with CBFB-SMMHC to 6 AML-M4 patients with a normal karyotype as assessed by conventional cytogenetic analysis (Figure 3B). Clinical characteristics of these patients are presented in Table 1. Direct sequencing for CEBPA mutations was performed in all these patients, and none of the AML-M4 patients with a normal karyotype or with the CBFB-SMMHC fusion had any CEBPA mutations. Using quantitative RT-PCR, we found that similar CEBPA mRNA levels are observed in patients with AML with the CBFB-SMMHC translocation and in AML-M4 patients with a normal karyotype (Figure 3B). This analysis was repeated twice for each patient. In contrast, Figure 3B also demonstrates that, as reported previously by us in a different series of patients,12 CEBPA mRNA is suppressed in AML-M2 patients with the AML1-ETO fusion (t(8;21)) as compared to AML-M2 patients with a normal karyotype.
Clinical presentation of patients
No. . | Sex . | Age, y . | FAB . | Karyotype . | WBC count, × 109/L . | Blasts in PBLs, % . | LDH level, U/mL . |
|---|---|---|---|---|---|---|---|
| 1 | M | 58 | M4Eo | 47,XY,+8,inv(16)(p13q22) | 22.2 | 78 | 954 |
| 2 | M | 64 | M4Eo | 46,XY,inv(16)(p13q22) | 14.3 | 54 | 752 |
| 3 | F | 58 | M4Eo | 47,XX,inv(16)(p13q22),+22 | 13.2 | 58 | 674 |
| 4 | M | 46 | M4Eo | 46,XY,t(9;20;21),inv(16)(p13q22) | 38.4 | 95 | 1455 |
| 5 | F | 62 | M4Eo | 46,XX,inv(16)(p13q22) | 55.8 | 98 | 1025 |
| 6 | M | 54 | M4Eo | 46,XY,inv(16)(p13q22) | 18.2 | 45 | 770 |
| 7 | F | 62 | M4Eo | 46,XX,inv(16)(p13q22) | 8.2 | 25 | 482 |
| 8 | M | 49 | M4Eo | 47,XY,inv(16)(p13q22),+21 | 15.3 | 38 | 920 |
| 9 | F | 72 | M4Eo | 46,XX,inv(16)(p13q22) | 32.8 | 90 | 1285 |
| 10 | F | 48 | M4Eo | 46,XX,del(7q),inv(16)(p13q22) | 9.1 | 32 | 474 |
| 11 | M | 70 | M4Eo | 46,XY,inv(16)(p13q22) | 18.2 | 41 | 552 |
| 12 | M | 58 | M4Eo | 46,XY,inv(16)(p13q22) | 104.8 | 92 | 1485 |
| 13 | M | 44 | M4 | 46,XY | 6.6 | 30 | 380 |
| 14 | F | 61 | M4 | 46,XX | 22.8 | 68 | 884 |
| 15 | M | 58 | M4 | 46,XY | 14.4 | 55 | 710 |
| 16 | M | 58 | M4 | 46,XY | 98.8 | 98 | 1662 |
| 17 | F | 74 | M4 | 46,XX | 30.5 | 72 | 980 |
| 18 | M | 62 | M4 | 46,XY | 84.5 | 98 | 1585 |
No. . | Sex . | Age, y . | FAB . | Karyotype . | WBC count, × 109/L . | Blasts in PBLs, % . | LDH level, U/mL . |
|---|---|---|---|---|---|---|---|
| 1 | M | 58 | M4Eo | 47,XY,+8,inv(16)(p13q22) | 22.2 | 78 | 954 |
| 2 | M | 64 | M4Eo | 46,XY,inv(16)(p13q22) | 14.3 | 54 | 752 |
| 3 | F | 58 | M4Eo | 47,XX,inv(16)(p13q22),+22 | 13.2 | 58 | 674 |
| 4 | M | 46 | M4Eo | 46,XY,t(9;20;21),inv(16)(p13q22) | 38.4 | 95 | 1455 |
| 5 | F | 62 | M4Eo | 46,XX,inv(16)(p13q22) | 55.8 | 98 | 1025 |
| 6 | M | 54 | M4Eo | 46,XY,inv(16)(p13q22) | 18.2 | 45 | 770 |
| 7 | F | 62 | M4Eo | 46,XX,inv(16)(p13q22) | 8.2 | 25 | 482 |
| 8 | M | 49 | M4Eo | 47,XY,inv(16)(p13q22),+21 | 15.3 | 38 | 920 |
| 9 | F | 72 | M4Eo | 46,XX,inv(16)(p13q22) | 32.8 | 90 | 1285 |
| 10 | F | 48 | M4Eo | 46,XX,del(7q),inv(16)(p13q22) | 9.1 | 32 | 474 |
| 11 | M | 70 | M4Eo | 46,XY,inv(16)(p13q22) | 18.2 | 41 | 552 |
| 12 | M | 58 | M4Eo | 46,XY,inv(16)(p13q22) | 104.8 | 92 | 1485 |
| 13 | M | 44 | M4 | 46,XY | 6.6 | 30 | 380 |
| 14 | F | 61 | M4 | 46,XX | 22.8 | 68 | 884 |
| 15 | M | 58 | M4 | 46,XY | 14.4 | 55 | 710 |
| 16 | M | 58 | M4 | 46,XY | 98.8 | 98 | 1662 |
| 17 | F | 74 | M4 | 46,XX | 30.5 | 72 | 980 |
| 18 | M | 62 | M4 | 46,XY | 84.5 | 98 | 1585 |
FAB indicates French-American-British classification; WBC, white blood cell; PBLs, peripheral blood leukocytes; LDH, lactate dehydrogenase.
In accordance with the results obtained with conditional expression of the CBFB-SMMHC protein in the U937 cell line, no CEBPA protein was detectable by Western blot analysis in any of the patient samples with the CBFB-SMMHC rearrangement. However, significant amounts of CEBPA protein were observed in AML-M4 patients with a normal karyotype (Figure 3C). In 11 normal karyotype AML-M4 samples, we found high protein levels (as in lane 1 in both panels of Figure 3C) in 6 patients, 3 patients had weak CEBPA expression, and 2 patients had no detectable CEBPA protein.
CEBPA protein is specifically suppressed in AML-M4 patients with CBFB-SMMHC. (A) Ninety-two patients with AML of all subtypes were analyzed for CEBPA mRNA expression by quantitative RT-PCR. Differences among subtypes were not significant (P = .327). Mean values and standard deviation (error bars) are depicted. (B) Subgroup analysis using quantitative RT-PCR analysis of CEBPA mRNA levels from AML-M4Eo with CBFB-SMMHC (n = 12) and AML-M4 with a normal karyotype (n = 6). The n-fold expression levels were calculated. Mean values and SDs are shown. The difference between AML-M4Eo and normal karyotype AML-M4 was not significant (P = .478). In addition, AML-M2 patients with the AML1-ETO rearrangement were compared to AML-M2 patients with a normal karyotype. This difference was significant (P = .012). (C) Whole-cell lysates of 12 AML-M4Eo CBFB-SMMHC patient samples (lanes 1-8 and 9-12) and of 2 AML-M4 patients with a normal karyotype (first lane in left and right panels) were subjected to Western blotting with a CEBPA antibody (top blots). None of the samples from the AML-M4 patients with inv(16) had detectable CEBPA protein. Eleven patients with AML-M4 and normal karyotypes were tested, with 6 having high expression as shown in the first lane of the 2 panels, 3 having weak CEBPA expression, and 2 having no detectable CEBPA protein. The same membrane was subsequently incubated with an antibody against β-actin for control of loading and integrity (bottom blots). (D) CEBPA-binding activity was measured using the Transam assay. Mean values and SDs are shown. OD indicates spectophotometric result at 450 nm wavelength.
CEBPA protein is specifically suppressed in AML-M4 patients with CBFB-SMMHC. (A) Ninety-two patients with AML of all subtypes were analyzed for CEBPA mRNA expression by quantitative RT-PCR. Differences among subtypes were not significant (P = .327). Mean values and standard deviation (error bars) are depicted. (B) Subgroup analysis using quantitative RT-PCR analysis of CEBPA mRNA levels from AML-M4Eo with CBFB-SMMHC (n = 12) and AML-M4 with a normal karyotype (n = 6). The n-fold expression levels were calculated. Mean values and SDs are shown. The difference between AML-M4Eo and normal karyotype AML-M4 was not significant (P = .478). In addition, AML-M2 patients with the AML1-ETO rearrangement were compared to AML-M2 patients with a normal karyotype. This difference was significant (P = .012). (C) Whole-cell lysates of 12 AML-M4Eo CBFB-SMMHC patient samples (lanes 1-8 and 9-12) and of 2 AML-M4 patients with a normal karyotype (first lane in left and right panels) were subjected to Western blotting with a CEBPA antibody (top blots). None of the samples from the AML-M4 patients with inv(16) had detectable CEBPA protein. Eleven patients with AML-M4 and normal karyotypes were tested, with 6 having high expression as shown in the first lane of the 2 panels, 3 having weak CEBPA expression, and 2 having no detectable CEBPA protein. The same membrane was subsequently incubated with an antibody against β-actin for control of loading and integrity (bottom blots). (D) CEBPA-binding activity was measured using the Transam assay. Mean values and SDs are shown. OD indicates spectophotometric result at 450 nm wavelength.
Analysis of nuclear extracts from patient samples for functional assays such as EMSA is often hampered by limited quality and quantity of the material. The Transam assay provides significant advantages regarding these aspects and allows determination of DNA-binding activity of nuclear extracts from patient samples. Using this assay, we screened AML-M4 patient samples with and without CBFB-SMMHC for their binding activity to a CEBP consensus-binding site. We found a significantly decreased CEBPA-binding activity (71.6% reduction, P = .003) in the 12 samples with CBFB-SMMHC as compared with AML-M4 patients without CBFB-SMMHC (Figure 3D).
In conclusion, our results obtained from cell lines and AML patient samples with CBFB-SMMHC suggest that the CBFB-SMMHC fusion protein suppresses CEBPA protein and its DNA-binding activity. In contrast to our previous findings with the AML1-ETO fusion,12 we detected no changes of CEBPA mRNA levels following induction of CBFB-SMMHC protein. We therefore conclude that in AML with CBFB-SMMHC, an as-yet-unknown posttranscriptional mechanism, is involved in CEBPA regulation.
CBFB-SMMHC conditionally expressed in U937 cells or present in leukemic cells of AML-M4Eo patients induces calreticulin expression
Few reports exist on mechanisms involved in posttranscriptional regulation of CEBPA.18,19,22 It has been postulated that the poly(rC)-binding protein, hnRNP E2, inhibits CEBPA expression at the translational level in patients with chronic myeloid leukemia in blast crisis (CML-BC), but not in chronic phase.18 We observed no changes of hnRNP E2 mRNA and protein levels in U937 cells following induction of CBFB-SMMHC (data not shown). Apparently, CEBPA is not regulated by hnRNP E2 in CBFB-SMMHC cells.
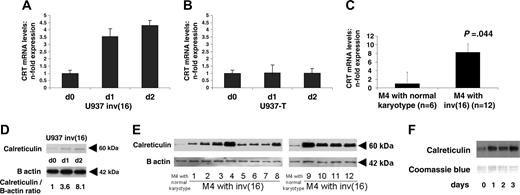
Calreticulin expression and activity are induced following conditional expression of CBFB-SMMHC in leukemic U937 cells and in CBFB-SMMHC AML-M4Eo patient samples. (A) Calreticulin (CRT) mRNA levels following induction of CBFB-SMMHC in U937 cells are determined by quantitative RT-PCR. Mean values and SDs (error bars) are shown. (B) Calreticulin mRNA levels are assessed by RT-PCR after withdrawal of tetracycline in the parental U937 T cells. (C) Calreticulin mRNA levels were determined in AML-M4 patients with CBFB-SMMHC compared to AML-M4 patients with a normal karyotype. (D) Calreticulin protein was analyzed by Western blot analysis in U937 cells following induction of CBFB-SMMHC by withdrawal of tetracycline. Quantification as calreticulin/β-actin ratio is indicated below. (E) Calreticulin protein was analyzed in AML-M4 patient samples with CBFB-SMMHC (lanes 1-8 and 9-12) and in representative AML-M4 patients with a normal karyotype (lanes 1 and 10). (F) Calreticulin activity of U937-CBFB-SMMHC cells following withdrawal of tetracycline was assessed by UV cross-linking to directly visualize the physical interaction between calreticulin protein and CEBPA mRNA.
Calreticulin expression and activity are induced following conditional expression of CBFB-SMMHC in leukemic U937 cells and in CBFB-SMMHC AML-M4Eo patient samples. (A) Calreticulin (CRT) mRNA levels following induction of CBFB-SMMHC in U937 cells are determined by quantitative RT-PCR. Mean values and SDs (error bars) are shown. (B) Calreticulin mRNA levels are assessed by RT-PCR after withdrawal of tetracycline in the parental U937 T cells. (C) Calreticulin mRNA levels were determined in AML-M4 patients with CBFB-SMMHC compared to AML-M4 patients with a normal karyotype. (D) Calreticulin protein was analyzed by Western blot analysis in U937 cells following induction of CBFB-SMMHC by withdrawal of tetracycline. Quantification as calreticulin/β-actin ratio is indicated below. (E) Calreticulin protein was analyzed in AML-M4 patient samples with CBFB-SMMHC (lanes 1-8 and 9-12) and in representative AML-M4 patients with a normal karyotype (lanes 1 and 10). (F) Calreticulin activity of U937-CBFB-SMMHC cells following withdrawal of tetracycline was assessed by UV cross-linking to directly visualize the physical interaction between calreticulin protein and CEBPA mRNA.
Calreticulin, a ubiquitous protein with calcium storage and chaperone function, has recently been reported in HeLa cells to interact with GC-rich sequences within the CEBPA mRNA and it would thereby repress translation of the 42- and 30-kDa isoforms of the CEBPA protein.22 We therefore hypothesized that the posttranscriptional down-regulation of CEBPA following conditional expression of CBFB-SMMHC might be caused by an increase of calreticulin expression or activity.
We thus investigated calreticulin mRNA and protein expression in the conditional CBFB-SMMHC cell line system as well as in AML-M4 samples with and without the CBFB-SMMHC rearrangement. Quantitative RT-PCR analysis revealed that calreticulin mRNA transcripts were induced within the first 24 hours after initiating CBFB-SMMHC expression (Figure 4A). A transcriptional mechanism of calreticulin induction thus has to be assumed. In contrast, removing tetracycline in the parental U937 T cells did not affect calreticulin mRNA levels (Figure 4B). In addition, we observed an 8.2-fold increase of calreticulin mRNA transcripts in 12 AML-M4 patients with CBFB-SMMHC as compared to 6 AML-M4 patients with a normal karyotype (Figure 4C). Furthermore, Western blot analysis demonstrated an 8-fold increase of calreticulin protein after 48 hours of conditional expression of CBFB-SMMHC (Figure 4D). Finally, AML-M4 patient samples with the CBFB-SMMHC rearrangement showed significantly higher calreticulin protein levels than AML-M4 patients with a normal karyotype (Figure 4E).
Calreticulin function can be assessed by a UV cross-linking experiment. Thereby, the direct interaction of calreticulin protein with GC-rich sequences within the CEBPA mRNA can be visualized and compared among different samples. Figure 4F gives evidence of a 12.5-fold increase in calreticulin binding to CEBPA mRNA starting early on day 1 following induction of CBFB-SMMHC. Early up-regulation of calreticulin activity appears to precede the suppression of CEBPA protein following CBFB-SMMHC induction. In conclusion, results obtained from patient samples and the cell lines with inducible CBFB-SMMHC indicate that CEBPA protein and binding activity appear to be regulated on a translational level by modulation of calreticulin mRNA and protein production as well as protein activity.
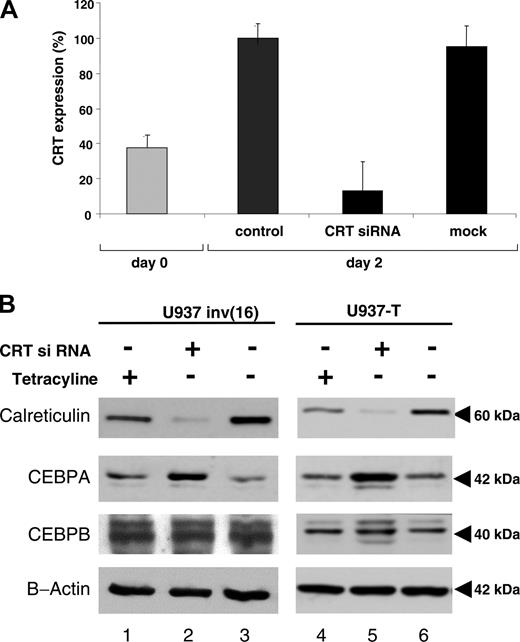
siRNA blocking calreticulin restores CBFB-SMMHC-mediated suppression of CEBPA protein
The experiments described suggest that down-regulation of CEBPA by CBFB-SMMHC is mediated by modulation of calreticulin. We therefore hypothesized that functional knock-down of calreticulin by siRNA might be able to restore efficient CEBPA translation. We thus induced the CBFB-SMMHC protein in U937 cells and transfected siRNA designed to target calreticulin. We found a 92% knock-down of calreticulin mRNA levels 48 hours after transfection (Figure 5A). Mock transfection did not interfere with calreticulin mRNA levels. Suppression of calreticulin protein became evident 48 hours after siRNA transfection (Figure 5B). The block of calreticulin protein expression was equally observed in U937 cells after CBFB-SMMHC induction as well as in the parental U937 T cells. Interestingly and in accordance with our hypothesis CEBPA mRNA levels were not induced by calreticulin siRNA. In fact, we even observed a slight reduction to 39.4% (mean value; 3 experiments) of CEBPA mRNA 48 hours after induction of CBFB-SMMHC and transduction with calreticulin siRNA. In the same series of experiments, induction of CBFB-SMMHC alone did not change CEBPA mRNA expression (mean value, 109%; 3 experiments) consistent with our experiments reported in Figure 2A.
Finally, transfection of calreticulin siRNA prevented suppression of CEBPA protein following CBFB-SMMHC induction. CEBPA protein levels even exceeded the baseline levels before CBFB-SMMHC induction and inhibition of calreticulin by siRNA (Figure 5B). In addition, the block of calreticulin in U937-T cells, thus in the absence of CBFB-SMMHC, also resulted in a significant increase of CEBPA protein. In contrast to CEBPA, no changes in CEBPB expression were observed. We therefore conclude that calreticulin in myeloid cells indeed is a potent inhibitor of CEBPA translation, whereas CEBPB is not affected.
Discussion
We report here that the CBFB-SMMHC fusion protein, the hallmark of leukemic cells from AML patients with subtype M4Eo, suppresses CEBPA protein expression and function. This suppression is mediated at a translational level by induced expression of calreticulin, an inhibitor of CEBPA translation.22 Block of calreticulin is sufficient to restore CEBPA protein in CBFB-SMMHC cells. This offers a novel model of how the CBFB-SMMHC gene fusion contributes to the leukemic phenotype in AML-M4Eo.
Inhibition of calreticulin expression by siRNA restores CEBPA protein expression. CBFB-SMMHC protein was induced in U937 cells by withdrawal of tetracycline. In addition, cells were either electroporated with siRNA designed to knock-down calreticulin or with mock siRNA as control, simultaneously with the onset of CBFB-SMMHC induction. (A) Quantitative RT-PCR of calreticulin mRNA at day 0 (▦) and 48 hours after induction of CBFB-SMMHC transfection with or without transduction with siRNA (▪). “Control” represents U937 cells after 48 hours of withdrawal of tetracycline and thus induction of CBFB-SMMHC, whereas “mock” represents U937 cells 48 hours after withdrawal of tetracycline and transduction with the Silencer Negative Control no. 2 siRNA (Ambion) to exclude unspecific effects of the transduction procedure. Mean values and SDs are shown. (B) Western blot analysis of calreticulin, CEBPA, CEBPB, and β-actin protein after withdrawal (-) of tetracycline or transduction (or both) with (+) calreticulin siRNA in the conditional U937 inv(16) cell line (left panels) and the parental U937-T cell line (right panels).
Inhibition of calreticulin expression by siRNA restores CEBPA protein expression. CBFB-SMMHC protein was induced in U937 cells by withdrawal of tetracycline. In addition, cells were either electroporated with siRNA designed to knock-down calreticulin or with mock siRNA as control, simultaneously with the onset of CBFB-SMMHC induction. (A) Quantitative RT-PCR of calreticulin mRNA at day 0 (▦) and 48 hours after induction of CBFB-SMMHC transfection with or without transduction with siRNA (▪). “Control” represents U937 cells after 48 hours of withdrawal of tetracycline and thus induction of CBFB-SMMHC, whereas “mock” represents U937 cells 48 hours after withdrawal of tetracycline and transduction with the Silencer Negative Control no. 2 siRNA (Ambion) to exclude unspecific effects of the transduction procedure. Mean values and SDs are shown. (B) Western blot analysis of calreticulin, CEBPA, CEBPB, and β-actin protein after withdrawal (-) of tetracycline or transduction (or both) with (+) calreticulin siRNA in the conditional U937 inv(16) cell line (left panels) and the parental U937-T cell line (right panels).
Our earlier studies indicated that CEBPA mRNA is similarly expressed in AML-M4Eo patient samples with CBFB-SMMHC compared to AML-M4 patients with a normal karyotype.12 These findings were verified in the present study in an independent panel of patients with AML. In addition, conditional induction of CBFB-SMMHC in U937 cells consistently failed to suppress CEBPA mRNA. This is in remarkable contrast to the closely related AML1-ETO fusion, which suppresses CEBPA mRNA both in a conditional myeloid cell line model as well as in t(8;21) AML patient samples.12
We observed increased expression of calreticulin mRNA and protein in inv(16) patients with AML and after conditional expression of inv(16) in myeloid cells. Thus, the mechanism of calreticulin induction is supposed to be transcriptional. However, a detailed analysis of the calreticulin promoter in myeloid cells has not been reported so far, and the precise mechanism of how calreticulin expression is modulated in the presence of CBFB-SMMHC remains to be elucidated. We have also observed that calreticulin mRNA is induced after conditional expression of the leukemic fusion protein AML1-MDS1-EVI1 in U937 cells,19 and this suggests an indirect link between CBFB-SMMHC and calreticulin. This might be further underlined by our observation that CBFB-SMMHC, like other leukemic fusion genes involving CBF family members such as AML1-MDS1-EVI1 and AML1-ETO, slows the cell cycle in U937 cells (data not shown). It raises the possibility that other factors involved in cell-cycle regulation may be involved in calreticulin modulation following expression of CBFB-SMMHC.
Interestingly, block of calreticulin by transient expression of calreticulin siRNA was sufficient to restore CEBPA protein. Even if calreticulin protein was not completely abolished 48 hours after calreticulin siRNA transfection, a strong increase in CEBPA protein expression was observed. Appropriate controls such as the silencer negative control excluded other possibilities of calreticulin inhibition such as nonspecific effects of electroporation per se. We did not observe any morphologic changes in U937 cells suggesting neutrophil differentiation after transient transfection of calreticulin siRNA. However, neutrophil differentiation after conditional expression of CEBPA in U937 cells requires about 2 weeks as previously demonstrated.13,21 This obviously is beyond the possibilities of a transient system.
A favorable course is generally observed in AML characterized by the presence of particular genetic abnormalities, notably the AML1-ETO fusion, the PML-RARA fusion, the CBFB-SMMHC fusion, or dominant-negative mutations in the CEBPA gene. AML1-ETO blocks CEBPA transcription,12 the PML-RARA fusion targets CEBPB expression,23 the CEBPA gene can be mutated per se,13 and we here show that the fourth member in this group CBFB-SMMHC blocks CEBPA translation. Targeting CEBP family members appears to be a common theme in this group of good-risk AML, which may perhaps be suitably relabeled as the so-called CEBP leukemias.
CEBP family members in general and CEBPA in particular are believed to suppress the leukemic phenotype through combined induction of direct transcriptional targets crucial for normal myeloid differentiation and inhibition of cell-cycle progression. We and others have shown in leukemic cell lines and transgenic mice models that restoring CEBPA expression is sufficient to induce terminal differentiation thereby pointing to potential therapeutic implications.12,13,17,21,24-26 Here, we report that block of calreticulin expression by siRNA relieves CEBPA suppression. Modulation of calreticulin expression might therefore be a novel potent therapeutic target for subsets of AML where CEBPA protein is suppressed.
Prepublished online as Blood First Edition Paper, April 26, 2005; DOI 10.1182/blood-2004-11-4392.
Supported by grants from the Swiss National Science Foundation SF 31-666899.01 (T.P.), SF 3100-67213 (M.F.F.), and SF 3100A0-100445 (B.U.M.).
D.H. and T.P. designed and performed research and wrote the paper; B.U.M. designed and performed research; N.A.T., J.S., and M.E. performed research; D.R.B., M.J., and S.M.-M. contributed vital material; and M.F.F. analyzed data.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank Daniel G. Tenen for helpful discussions and generously sharing reagents; Paul Liu for kindly providing the inv(16) cDNA; Barbara Huegli for technical support; Dominique Mühlematter, Valérie Parlier, and the entire staff of the Unit of Cancer Cytogenetics, Service of Medical Genetics, University Hospital, Lausanne, Switzerland, for assistance with cytogenetic analysis.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal