Abstract
The pathogenesis of acute myeloid leukemia (AML) involves the cooperation of mutations promoting proliferation/survival and those impairing differentiation. The RAS pathway has been implicated as a key component of the proliferative drive in AML. We have screened AML patients, predominantly younger than 60 years and treated within 2 clinical trials, for NRAS (n = 1106), KRAS (n = 739), and HRAS (n = 200) hot-spot mutations using denaturing high-performance liquid chromatography or restriction fragment length polymorphism (RFLP) analysis. NRAS mutations were confirmed in 11% of patients (126/1106) and KRAS mutations in 5% (39/739). No HRAS mutations were detected in 200 randomly selected samples. Codons most frequently mutated were N12 (43%), N13 (21%), and K12 (21%). KRAS mutations were relatively overrepresented in French-American-British (FAB) type M4 (P < .001). NRAS mutation was over-represented in the t(3;5)(q21∼25;q31∼q35) subgroup (P < .001) and underrepresented in t(15;17)(q22;q21) (P < .001). KRAS mutation was overrepresented in inv(16)(p13q22) (P = .004). Twenty-three percent of KRAS mutations were within the inv(16) subgroup. RAS mutation and FLT3 ITD were rarely coexistent (14/768; P < .001). Median percentage of RAS mutant allele assayed by quantitative RFLP analysis was 28% (N12), 19% (N13), 25% (N61), and 21% (K12). RAS mutation did not influence clinical outcome (overall/disease-free survival, complete remission, relapse rate) either for the entire cohort or within cytogenetic risk groups. (Blood. 2005;106:2113-2119)
Introduction
The cytogenetic characterization of subgroups of acute myeloid leukemia (AML) has promoted a risk-directed therapeutic approach to disease management. The recent identification of an internal tandem duplication (ITD) within the FLT3 gene provides a paradigm for molecular markers with additional prognostic power in AML, in this case predictive for reduced disease-free survival, event-free survival, and overall survival, and for increased relapse risk.1 Abnormalities of signal transduction pathways are common in AML, occurring in up to 50% of cases. These abnormalities comprise activating mutations in genes encoding receptor tyrosine kinases including FLT3 (ITD and D8352 mutation) and c-KIT,3 RAS protein activation (via mutation or loss of negative regulators such as nuclear factor-1 [NF-1]4 ), and phosphorylation of mitogen-activated protein (MAP) kinase.5
The RAS genes encode a family of membrane-associated proteins, which regulate signal transduction upon binding of ligand to a variety of membrane receptors. There are 3 functional RAS genes: N- (from a neuroblastoma cell line), K- (Kirsten), and H- (Harvey) RAS, each containing 4 exons. KRAS has an A and B protein encoded from alternative fourth exons. RAS gene mutations at codons 12, 13, and 61 confer constitutive activation of the RAS protein, which is held in the guanosine triphosphate (GTP)-bound state. RAS gene mutations were first reported in myeloid malignancies 17 years ago,6 and many groups have since attempted to study their frequency in small/medium-sized cohorts of AML patients.7-12 Several studies indicate that RAS gene mutation is associated with poor outcome in AML and myelodysplastic syndromes (MDSs),7,13,14 but historical data sets are of insufficient size to distinguish prognostic differences between subgroups.
We have assayed RAS mutational status by denaturing highperformance liquid chromatography (DHPLC) and restriction fragment length polymorphism (RFLP) in a large trial-based patient cohort. We have correlated this with presenting morphology, cytogenetics, and FLT3 ITD status where available. We demonstrate that RAS mutation frequency and spectrum differ between biologically distinct subtypes of AML but do not influence clinical outcome.
Patients, materials, and methods
Patients
The study cohort comprised 1106 patients at presentation with AML who were entered into the Medical Research Council (MRC) AML10 (n = 387) and AML12 (n = 719) clinical trials for patients younger than 56 and younger than 60 years, respectively. Informed consent for tissue collection and research studies was approved by the Research Ethics Committee for Wales (02/4560) and the Multi Research Ethics Committee for Wales (98/9/08). This study was approved by the MRC AML Trial Cell Bank Research Group, and samples were analyzed anonymously.
Cytogenetic definitions
G-banded karyotypes were collected centrally and described according to the International System for Human Cytogenetic Nomenclature (ISCN).15 Cytogenetic classification was performed as previously described.16 Briefly, a predefined list of specific clonal chromosomal abnormalities was used to classify patients into non-mutually exclusive subgroups. As all clonal aberrations were counted, individual patients may be counted more than once. Patients were then classified hierarchically into risk groups (favorable, intermediate, and adverse) according to the presence of specific primary aberrations and a complex karyotype, defined as one with at least 5 unrelated abnormalities.
Therapy
The AML10 clinical trial protocol has previously been published in detail.17 Patients in the AML12 trial were randomized to receive induction therapy with either ADE (cytarabine, daunorubicin, etoposide) 10 + 3 + 5orMAE (mitoxantrone, cytarabine, etoposide) 3 + 10 + 5 from 1994 to 1998. After 1998, induction therapy was modified to DAT (daunorubicin, cytarabine, thioguanine) 3 + 10 + 10 randomized to 2 different induction cytarabine doses, with an additional randomization to all-trans retinoic acid (ATRA) versus no ATRA. Bone marrow (BM) remission status was assessed and patients were randomized according to risk group (good, standard, or poor risk) defined on the basis of hierarchic cytogenetic classification16 and remission status after course 1.18 Consolidation chemotherapy for good/standard-risk groups comprised a second course of the induction regimen, followed by MACE (amsacrine, cytarabine, etoposide). Good-risk patients received a fourth course of either MidAC (mitoxantrone and cytarabine) or ICE (idarubicin, cytarabine, and etoposide) + MidAC as course 5; standard risk, either MidAC, ICE + MidAC, ICE + stem cell transplant (SCT; allogeneic or autologous depending on donor availability), or SCT alone. Poor-risk patients could be entered into the MRC Refractory/Relapse AML study as previously described.19
End points
Complete remission (CR) was defined as less than 5% bone marrow blasts. Full hematologic recovery was not required, though 97% of patients achieved neutrophil counts higher than 1 × 109/L and platelet counts higher than 100 × 109/L. Resistant disease (RD) was defined as more than 15% BM blasts and partial remission as 5% to 15% BM blasts after course 1. Induction death (ID) was defined as death within 30 days of entry; and deaths more than 30 days after entry were defined as RD. Overall survival (OS) was defined as the time from entry to death. Patients failing to achieve remission were considered to have an event on day 1. For patients achieving first CR, disease-free survival (DFS) was defined as the time from first CR to an event (death in CR or relapse). Relapse risk (RR) was the cumulative probability of relapse, censoring at death in CR; death in CR was the cumulative probability of dying in first CR, censoring at relapse.
NRAS mutation screen: denaturing high-performance liquid chromatography (DHPLC)
DNA was extracted predominantly from bone marrow cells (mononuclear cells or buffy coat) at AML presentation. The NRAS gene was screened for mutations at hot-spot codons 12, 13, and 61 as previously described.20 All samples with an abnormal DHPLC profile were reamplified from genomic DNA for confirmation on a second screen before sequencing. Our DHPLC assays cannot distinguish samples with 100% mutant DNA (biallelic mutation in all cells or loss of heterozygosity of the wild-type allele plus mutation of the retained allele) from 100% wild-type DNA. We therefore randomly selected 200 patient samples with known single DHPLC peaks (ie, apparently wild-type N12/13). Polymerase chain reaction (PCR) products from random pairs of these patients were mixed, before heteroduplex formation and reanalysis of these 100 profiles by DHPLC. Sensitivity of N12/13 mutation detection by DHPLC was assessed by spiking known wild-type NRAS DNA with different proportions of known cloned mutant NRAS DNA (MDS92 cell line, codon 12.2 G>C mutation).
KRAS codon 61 and HRAS codon 61 were assayed by DHPLC. Primers and conditions used are shown in Table 1.
Experimental conditions for RAS mutation analysis, screening by denaturing high-performance liquid chromatography (DHPLC), and restriction fragment length polymorphism (RFLP) analysis quantification of mutant allele percentage by PCR with an end-labeled radioactive primer
Method type/assay . | Primer sequence . | Annealing temperature, °C . | No. PCR cycles . | Amplicon size, bp . | RFLP enzyme . | RFLP digest product sizes, bp . |
|---|---|---|---|---|---|---|
| Mutation screening | ||||||
| DHPLC | ||||||
| K61F* | 5′-tgt gtt tct ccc ttc tca gga ttc-3′ | 60 | 35 | 158 | NA | NA |
| K61R | 5′-tgg caa ata cac aaa gaa agc c-3′ | 60 | 35 | 158 | NA | NA |
| K61F* | 5′-gat tcc tac cgg aag cag gt-3′ | 60 | 35 | 151 | NA | NA |
| K61R | 5′-tgg tgt tgt tga tgg caa ac-3′ | 60 | 35 | 151 | NA | NA |
| RFLP and sequencing | ||||||
| K12F (MM) | 5′-gac tga ata taa act tgt ggt agt tgg acc t-3′ | 56 | 35 | 121 | BstN1 (7.5 units) | Wt = 91 |
| K12R | 5′-aac aag att tac ctc tat tgt tgg atc a-3′ | 56 | 35 | 121 | (7.5 units) | Mut = 121 |
| H12F | 5′-agg aga ccc tgt agg agga-3′ | 60 | 35 | 170 | Msp1 (10 units) | Wt = 23, 56, 91 |
| H12R | 5′-gcg cta ggc tca cct cta t-3′ | 60 | 35 | 170 | (10 units) | Mut = 23, 147 |
| Mutant allele quantification: PCR with a end-labeled radioactive primer | ||||||
| N12F (MM) | 5′-act gag tac aaa ctg gtg gtg gtt gga cca-3′ | 63 | 25 | 172 | ScrF1 | Wt = 143 |
| N12R | 5′-tgg gta aag atg atc cga caa gtg a-3′ | 63 | 25 | 172 | ScrF1 | Mut = 172 |
| N13F | 5′-ctc cag aag tgt gag gcc gat-3′ | 63 | 25 | 250 | DpnII | Wt = 250 |
| N13R (MM) | 5′-ctg gat tgt cag tgc gct ttt ccc aag a-3′ | 63 | 25 | 250 | DpnII | Mut = 221 |
| N61F (MM) | 5′-tgt ttg ttg gac ata ctg gat aca gct gta-3′ | 63 | 25 | 225 | BsrG1 | Wt = 225 |
| N61R | 5′-tct tcc cta gtg tgg taa cct c-3′ | 63 | 25 | 225 | BsrG1 | Mut = 198 |
| K12F (MM) | 5′-gac tga ata taa act tgt ggt agt tgg acc t-3′ | 63 | 25 | 157 | ScrF1 | Wt = 127 |
| K12R | 5′-caa aga atg gtc ctg cac cag t-3′ | 63 | 25 | 157 | ScrF1 | Mut = 157 |
Method type/assay . | Primer sequence . | Annealing temperature, °C . | No. PCR cycles . | Amplicon size, bp . | RFLP enzyme . | RFLP digest product sizes, bp . |
|---|---|---|---|---|---|---|
| Mutation screening | ||||||
| DHPLC | ||||||
| K61F* | 5′-tgt gtt tct ccc ttc tca gga ttc-3′ | 60 | 35 | 158 | NA | NA |
| K61R | 5′-tgg caa ata cac aaa gaa agc c-3′ | 60 | 35 | 158 | NA | NA |
| K61F* | 5′-gat tcc tac cgg aag cag gt-3′ | 60 | 35 | 151 | NA | NA |
| K61R | 5′-tgg tgt tgt tga tgg caa ac-3′ | 60 | 35 | 151 | NA | NA |
| RFLP and sequencing | ||||||
| K12F (MM) | 5′-gac tga ata taa act tgt ggt agt tgg acc t-3′ | 56 | 35 | 121 | BstN1 (7.5 units) | Wt = 91 |
| K12R | 5′-aac aag att tac ctc tat tgt tgg atc a-3′ | 56 | 35 | 121 | (7.5 units) | Mut = 121 |
| H12F | 5′-agg aga ccc tgt agg agga-3′ | 60 | 35 | 170 | Msp1 (10 units) | Wt = 23, 56, 91 |
| H12R | 5′-gcg cta ggc tca cct cta t-3′ | 60 | 35 | 170 | (10 units) | Mut = 23, 147 |
| Mutant allele quantification: PCR with a end-labeled radioactive primer | ||||||
| N12F (MM) | 5′-act gag tac aaa ctg gtg gtg gtt gga cca-3′ | 63 | 25 | 172 | ScrF1 | Wt = 143 |
| N12R | 5′-tgg gta aag atg atc cga caa gtg a-3′ | 63 | 25 | 172 | ScrF1 | Mut = 172 |
| N13F | 5′-ctc cag aag tgt gag gcc gat-3′ | 63 | 25 | 250 | DpnII | Wt = 250 |
| N13R (MM) | 5′-ctg gat tgt cag tgc gct ttt ccc aag a-3′ | 63 | 25 | 250 | DpnII | Mut = 221 |
| N61F (MM) | 5′-tgt ttg ttg gac ata ctg gat aca gct gta-3′ | 63 | 25 | 225 | BsrG1 | Wt = 225 |
| N61R | 5′-tct tcc cta gtg tgg taa cct c-3′ | 63 | 25 | 225 | BsrG1 | Mut = 198 |
| K12F (MM) | 5′-gac tga ata taa act tgt ggt agt tgg acc t-3′ | 63 | 25 | 157 | ScrF1 | Wt = 127 |
| K12R | 5′-caa aga atg gtc ctg cac cag t-3′ | 63 | 25 | 157 | ScrF1 | Mut = 157 |
bp indicates base pair; F indicates forward; R, reverse; MM, mismatched; Wt, wild type; and Mut, mutant.
DHPLC temperature was 59°C for K61 and 64°C for H61.
KRAS and HRAS codons 12 and 13 mutation screening: restriction fragment length polymorphism (RFLP) analysis
RAS mutation confirmation/characterization
Samples with an abnormal RFLP or DHPLC profile were confirmed as mutant by DNA sequencing, using a fluorescent primer-adapted chain-termination method24 on an ABI 3100 sequencer (Applied Biosystems, Foster City, CA). When direct sequencing was negative, PCR products were cloned (Original TA Cloning Kit; Invitrogen, Groningen, the Netherlands) and sequenced.
Ratio of RAS mutant: wild-type alleles
Percent RAS mutant DNA was assayed for N12, N13 (G>A only), N61, and K12 by radioactive PCR-RFLP. PCR was performed as previously described using an end-labeled primer with 25 cycles of amplification at an annealing temperature of 63°C.1 Digestion products were separated on 6% denaturing polyacrylamide gel electrophoresis (PAGE), dried, autoradiographed, and quantitated by densitometry. Mutations could be detected at the following codons: N12.1 or N12.2; N13.2 G>A; N61.1 or N61.2; and K12.1 or K12.2. Mutant was expressed as percent of total for results.
FLT3 ITD mutation analysis
Statistical methods
Continuous data were compared using two sample t tests for data that were approximately normally distributed, and the nonparametric Wilcoxon rank-sum test for other types of distributions. Categoric data were compared using Fisher exact test for 2 × 2 tables, chi-squared tests for heterogeneity in larger tables, and Mantel-Haeszel tests for trend over ordered categories in 2 × 2 tables. Kaplan-Meier life tables were constructed for survival data and were compared using the log-rank test. Surviving patients for both AML10 and AML12 were censored on April 1, 2004. Median follow-up time for AML10 patients was 128 months (range, 52-189 months) and for AML12 patients was 72 months (range, 6-110 months), with a median follow-up time of 86 months for both trials when taken together. In order to build prognostic models associated with NRAS mutation or to adjust for multiple other factors, either logistic regression for categoric outcomes or proportional hazards regression (for time-to-event outcomes) was used, using forward selection techniques, with an entry probability of 0.01. All reported P values are 2-sided, and to allow for multiple testing, results are not considered statistically significant unless P is less than .01.
Results
Mutation frequency
NRAS mutations were confirmed in 11% of patients (126/1106) and KRAS mutations in 5% (39/739). KRAS mutation status was successfully analyzed in fewer patients because of poor efficiency of the K13 mutation assay and also insufficient DNA for all assays in some patients. Initial screening of randomly selected samples (n = 200) revealed no mutations at K61, H12, H13, or H61. FLT3 ITD was present in 26% of patients (287/1099) assayed for NRAS mutation and 24% of patients (180/736) assayed for KRAS mutation. FLT3 ITD status for 854 of these patients was reported previously.1
Combinations of NRAS and KRAS mutation and FLT3 ITD were coexistent in the same patient in only 14 of 739 patients, in whom all mutations were assayed (P < .001). Mutations that were coexistent broke down as follows: FLT3 ITD + NRAS, 11 (1%) of 1099; FLT3 ITD + KRAS, 3 (0.4%) of 736; and NRAS + KRAS 2 (0.3%) of 724.
We also directly sequenced DNA from 25 randomly selected AML patients from this cohort, with normal DHPLC profiles (N12/13 = 15, N61 = 10), and all were confirmed as wild-type NRAS. Dilution experiments showed that 10% or more of N12/13 RAS mutant DNA could be confidently detected within a background of wild-type sequence (data not shown). Mutations were confirmed by direct sequencing for 91 (80%) of 126 NRAS and 15 (38%) of 39 KRAS, while cloning and sequencing was required to confirm the remainder. One hundred DHPLC profiles from randomly paired samples with previously known single peaks remained as single peaks in the N12/13 assay (data not shown), confirming that biallelic mutation in all cells was likely to be a rare event.
RAS mutation spectrum
Codons most frequently mutated were N12 (43%), N13 (21%), and K12 (21%). Bases most frequently mutated were N12.2 (32%), K12.2 (20%), and N13.2 (16%). Of 39 KRAS mutations, 33 were at K12.2. G>A transition was the most common base substitution (62% NRAS, 54% KRAS). The next most frequent changes were G>C transversion, G>T transversion, then C>A transversion in NRAS, and G>T transversion then G>C transversion in KRAS. G>A transition predominated at base 2 of N12, N13, K12, and K13. At base 1, G>A transition predominated at N12 but was strikingly absent at N13, where G>C transversion predominated. All 9 substitutions at N61 base 1 were C>A transversion, while A>G transition predominated at base 2 and A>C transversion at base 3. While glycine → aspartate was the most common amino acid change at N12, N13, K12, and K13, glycine → serine and glycine → alanine were frequent at N12 but absent at N13. By contrast, glycine → arginine and glycine → valine were more common at N13 than N12. At N61 the most common amino acid change was glutamine → lysine followed by glutamine → arginine. Only one mutation was identified outside hot-spot codons 12, 13, and 61. This was at NRAS codon 22;1, C>A (glycine → lysine).
Ratio of RAS mutant: wild-type alleles
Percent RAS mutant DNA (clonal size) was assayed for N12, N13 (G>A only), N61, and K12 by radioactive PCR-RFLP. Median RAS mutant DNA percentage was 28% (N12), 19% (N13), 25% (N61), and 21% (K12).
RAS mutation and presenting clinical/morphologic patient characteristics
Central morphology review revealed that a small number of patients had acute lymphoblastic leukemia, but these cases were retained for completeness. RAS mutation frequency did not vary significantly with age, sex, presenting white cell count, WHO performance status, or de novo versus secondary AML (not shown). KRAS mutation frequency demonstrated significant heterogeneity among French-American-British (FAB) subgroups, more common in M4 (P < .001; Table 2).
RAS mutation status by French-American-British (FAB) subtype
. | NRAS . | . | KRAS . | . | ||
|---|---|---|---|---|---|---|
| FAB type . | Mutant, no. (%) . | Nonmutant, no. (%) . | Mutant, no. (%) . | Nonmutant, no. (%) . | ||
| AML M0 | 4 (3) | 20 (2) | 2 (5) | 12 (2) | ||
| AML M1 | 20 (16) | 159 (17) | 0 (0) | 108 (16) | ||
| AML M2 | 31 (25) | 254 (28) | 9 (24) | 187 (29) | ||
| AML M3 | 11 (9) | 170 (19) | 5 (13) | 129 (20) | ||
| AML M4 | 38 (30) | 173 (19) | 19 (50) | 131 (20) | ||
| AML M5 | 18 (14) | 87 (10) | 2 (5) | 59 (9) | ||
| AML M6 | 2 (2) | 19 (2) | 0 (0) | 13 (2) | ||
| AML M7 | 0 (0) | 11 (1) | 0 (0) | 7 (1) | ||
| RAEB | 2 (2) | 10 (1) | 1 (3) | 6 (1) | ||
| Bilineage | 0 (0) | 1 (< 0.5) | 0 (0) | 0 (0) | ||
| ALL | 0 (0) | 6 (1) | 0 (0) | 4 (1) | ||
| Other | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||
| Total with known FAB type | 126 (100) | 910 (100) | 38 (100) | 656 (100) | ||
| Unknown/unclassified | 0 (0) | 70 (8) | 1 (3) | 44 (7) | ||
. | NRAS . | . | KRAS . | . | ||
|---|---|---|---|---|---|---|
| FAB type . | Mutant, no. (%) . | Nonmutant, no. (%) . | Mutant, no. (%) . | Nonmutant, no. (%) . | ||
| AML M0 | 4 (3) | 20 (2) | 2 (5) | 12 (2) | ||
| AML M1 | 20 (16) | 159 (17) | 0 (0) | 108 (16) | ||
| AML M2 | 31 (25) | 254 (28) | 9 (24) | 187 (29) | ||
| AML M3 | 11 (9) | 170 (19) | 5 (13) | 129 (20) | ||
| AML M4 | 38 (30) | 173 (19) | 19 (50) | 131 (20) | ||
| AML M5 | 18 (14) | 87 (10) | 2 (5) | 59 (9) | ||
| AML M6 | 2 (2) | 19 (2) | 0 (0) | 13 (2) | ||
| AML M7 | 0 (0) | 11 (1) | 0 (0) | 7 (1) | ||
| RAEB | 2 (2) | 10 (1) | 1 (3) | 6 (1) | ||
| Bilineage | 0 (0) | 1 (< 0.5) | 0 (0) | 0 (0) | ||
| ALL | 0 (0) | 6 (1) | 0 (0) | 4 (1) | ||
| Other | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||
| Total with known FAB type | 126 (100) | 910 (100) | 38 (100) | 656 (100) | ||
| Unknown/unclassified | 0 (0) | 70 (8) | 1 (3) | 44 (7) | ||
Percentages are percentages of those with known FAB type. P = .04 for NRAS; P = .001 for KRAS. P values for those with known FAB type only. RAEB indicates refractory anemia with excess of blasts; ALL, acute lymphoblastic leukemia.
RAS/FLT3 mutation frequency varies between cytogenetic subgroups
Cytogenetic data were available for 922 patients (Table 3). RAS mutation frequency varied between karyotypic subtypes, with evidence for underrepresentation of NRAS mutation in t(15;17)(q22; q21) (P = .008) and strong evidence for overrepresentation of NRAS mutation in t(3;5)(q21∼25;q31∼q35) (P < .001) and KRAS mutation in inv(16)(p13q22) (P = .003). Twenty-three percent of KRAS mutations were within the inv(16) subgroup. No significant difference in NRAS or KRAS mutant frequency was found between cytogenetic risk groups (favorable/intermediate/adverse)(data not shown).
RAS mutation frequency within selected cytogenetic subgroups of AML patients for whom cytogenetic data were available (n = 922)
. | NRAS . | . | . | KRAS . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cytogenetic subgroup . | Mutant, no. (%) . | Nonmutant, no. (%) . | P . | Mutant, no. (%) . | Nonmutant, no. (%) . | P . | ||||
| No data | 17 | 167 | .3 | 8 | 131 | .8 | ||||
| Cytogenetics available | 109 | 813 | 31 | 569 | ||||||
| Favorable | ||||||||||
| t(15; 17) | 8 (7) | 138 (17) | .008* | 5 (16) | 109 (19) | .8 | ||||
| t(8; 21) | 5 (5) | 82 (10) | .07 | 3 (10) | 56 (10) | > .99 | ||||
| inv(16) | 11 (10) | 49 (6) | .14 | 7 (23) | 35 (6) | .004† | ||||
| Intermediate | ||||||||||
| Normal | 48 (44) | 292 (36) | .11 | 7 (23) | 191 (34) | .2 | ||||
| abn(11q) | 1 (1) | 11 (1) | > .99 | 1 (3) | 8 (1) | .3 | ||||
| abn(12p) | 1 (1) | 11 (1) | > .99 | 1 (3) | 7 (1) | .3 | ||||
| abn(15q) | 1 (1) | 5 (1) | .5 | 0 (0) | 5 (1) | > .99 | ||||
| abn(16q) | 1 (1) | 17 (2) | .7 | 2 (6) | 11 (2) | .14 | ||||
| abn(17p) | 1 (1) | 18 (2) | .7 | 0 (0) | 12 (2) | > .99 | ||||
| abn(1p) | 1 (1) | 14 (2) | > .99 | 1 (3) | 6 (1) | .3 | ||||
| abn(1q) | 2 (2) | 9 (1) | .6 | 1 (3) | 7 (1) | .3 | ||||
| abn(20q) | 0 (0) | 8 (1) | .6 | 0 (0) | 8 (1) | > .99 | ||||
| abn(21q) | 0 (0) | 8 (1) | .6 | 1 (3) | 3 (1) | .2 | ||||
| abn(6q) | 0 (0) | 9 (1) | .6 | 0 (0) | 5 (1) | > .99 | ||||
| abn(7p) | 1 (1) | 14 (2) | > .99 | 0 (0) | 10 (2) | > .99 | ||||
| add(5q) | 0 (0) | 5 (1) | > .99 | 0 (0) | 5 (1) | > .99 | ||||
| add(7q) | 0 (0) | 7 (1) | > .99 | 1 (3) | 6 (1) | .3 | ||||
| del(13q) | 0 (0) | 5 (1) | > .99 | 0 (0) | 2 (< 0.5) | > .99 | ||||
| del(7q) | 2 (2) | 23 (3) | .8 | 1 (3) | 19 (3) | > .99 | ||||
| del(9q) | 0 (0) | 18 (2) | .2 | 0 (0) | 10 (2) | > .99 | ||||
| i(8q) | 0 (0) | 3 (< 0.5) | > .99 | 0 (0) | 2 (< 0.5) | > .99 | ||||
| −3 | 0 (0) | 5 (1) | > .99 | 0 (0) | 5 (1) | > .99 | ||||
| −9 | 1 (1) | 2 (< 0.5) | .3 | 0 (0) | 3 (1) | > .99 | ||||
| −12 | 0 (0) | 9 (1) | .6 | 0 (0) | 8 (1) | > .99 | ||||
| −13 | 0 (0) | 8 (1) | .6 | 0 (0) | 6 (1) | > .99 | ||||
| −17 | 1 (1) | 14 (2) | > .99 | 0 (0) | 10 (2) | > .99 | ||||
| −18 | 0 (0) | 14 (2) | .4 | 0 (0) | 10 (2) | > .99 | ||||
| −20 | 0 (0) | 6 (1) | > .99 | 0 (0) | 2 (< 0.5) | > .99 | ||||
| −X | 0 (0) | 19 (2) | .15 | 0 (0) | 12 (2) | > .99 | ||||
| −Y | 2 (2) | 37 (6) | .3 | 1 (3) | 23 (4) | > .99 | ||||
| −X/Y | 2 (2) | 56 (7) | .04 | 1 (3) | 35 (6) | > .99 | ||||
| +4 | 2 (2) | 6 (1) | .2 | 0 (0) | 5 (1) | > .99 | ||||
| +6 | 1 (1) | 9 (1) | > .99 | 0 (0) | 5 (1) | > .99 | ||||
| +8 | 14 (13) | 69 (8) | .2 | 2 (6) | 63 (11) | .6 | ||||
| +11 | 1 (1) | 11 (1) | > .99 | 0 (0) | 4 (1) | > .99 | ||||
| +13 | 1 (1) | 9 (1) | > .99 | 1 (3) | 8 (1) | .4 | ||||
| +19 | 0 (0) | 6 (1) | > .99 | 0 (0) | 4 (1) | > .99 | ||||
| +21 | 2 (2) | 18 (2) | > .99 | 2 (6) | 13 (2) | .18 | ||||
| +22 | 3 (3) | 15 (2) | .5 | 2 (6) | 11 (2) | .14 | ||||
| t(6; 9) | 1 (1) | 4 (< 0.5) | .5 | 0 (0) | 4 (1) | > .99 | ||||
| t(9; 11) | 1 (1) | 1 (< 0.5) | .2 | 0 (0) | 1 (< 0.5) | > .99 | ||||
| t(9; 22) | 0 (0) | 9 (1) | .6 | 0 (0) | 5 (1) | > .99 | ||||
| t(10; 11) | 4 (4) | 9 (1) | .06 | 0 (0) | 9 (2) | > .99 | ||||
| t(11; 19) | 0 (0) | 4 (< 0.5) | > .99 | 1 (3) | 2 (< 0.5) | .15 | ||||
| t(11 q23) | 0 (0) | 11 (1) | .4 | 0 (0) | 6 (1) | > .99 | ||||
| abn(11p15) | 1 (1) | 5 (1) | .5 | 0 (0) | 1 (< 0.5) | > .99 | ||||
| abn(12p13) | 1 (1) | 8 (1) | > .99 | 0 (0) | 6 (1) | > .99 | ||||
| abn(8p 11) | 1 (1) | 2 (< 0.5) | .3 | 0 (0) | 2 (< 0.5) | > .99 | ||||
| Adverse | ||||||||||
| Complex | 2 (2) | 52 (6) | .08 | 1 (3) | 38 (7) | .7 | ||||
| −5 | 1 (1) | 18 (2) | .7 | 0 (0) | 13 (2) | > .99 | ||||
| del(5q) | 0 (0) | 21 (3) | .16 | 0 (0) | 16 (3) | > .99 | ||||
| −7 | 3 (3) | 33 (4) | .8 | 2 (6) | 24 (4) | .6 | ||||
| abn(3q) | 1 (1) | 12 (1) | .8 | 0 (0) | 10 (2) | > .99 | ||||
| inv(3) | 2 (2) | 9 (1) | .6 | 1 (3) | 9 (2) | .4 | ||||
| t(3; 5) | 5 (5) | 3 (< 0.5) | .001† | 0 (0) | 6 (1) | > .99 | ||||
. | NRAS . | . | . | KRAS . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cytogenetic subgroup . | Mutant, no. (%) . | Nonmutant, no. (%) . | P . | Mutant, no. (%) . | Nonmutant, no. (%) . | P . | ||||
| No data | 17 | 167 | .3 | 8 | 131 | .8 | ||||
| Cytogenetics available | 109 | 813 | 31 | 569 | ||||||
| Favorable | ||||||||||
| t(15; 17) | 8 (7) | 138 (17) | .008* | 5 (16) | 109 (19) | .8 | ||||
| t(8; 21) | 5 (5) | 82 (10) | .07 | 3 (10) | 56 (10) | > .99 | ||||
| inv(16) | 11 (10) | 49 (6) | .14 | 7 (23) | 35 (6) | .004† | ||||
| Intermediate | ||||||||||
| Normal | 48 (44) | 292 (36) | .11 | 7 (23) | 191 (34) | .2 | ||||
| abn(11q) | 1 (1) | 11 (1) | > .99 | 1 (3) | 8 (1) | .3 | ||||
| abn(12p) | 1 (1) | 11 (1) | > .99 | 1 (3) | 7 (1) | .3 | ||||
| abn(15q) | 1 (1) | 5 (1) | .5 | 0 (0) | 5 (1) | > .99 | ||||
| abn(16q) | 1 (1) | 17 (2) | .7 | 2 (6) | 11 (2) | .14 | ||||
| abn(17p) | 1 (1) | 18 (2) | .7 | 0 (0) | 12 (2) | > .99 | ||||
| abn(1p) | 1 (1) | 14 (2) | > .99 | 1 (3) | 6 (1) | .3 | ||||
| abn(1q) | 2 (2) | 9 (1) | .6 | 1 (3) | 7 (1) | .3 | ||||
| abn(20q) | 0 (0) | 8 (1) | .6 | 0 (0) | 8 (1) | > .99 | ||||
| abn(21q) | 0 (0) | 8 (1) | .6 | 1 (3) | 3 (1) | .2 | ||||
| abn(6q) | 0 (0) | 9 (1) | .6 | 0 (0) | 5 (1) | > .99 | ||||
| abn(7p) | 1 (1) | 14 (2) | > .99 | 0 (0) | 10 (2) | > .99 | ||||
| add(5q) | 0 (0) | 5 (1) | > .99 | 0 (0) | 5 (1) | > .99 | ||||
| add(7q) | 0 (0) | 7 (1) | > .99 | 1 (3) | 6 (1) | .3 | ||||
| del(13q) | 0 (0) | 5 (1) | > .99 | 0 (0) | 2 (< 0.5) | > .99 | ||||
| del(7q) | 2 (2) | 23 (3) | .8 | 1 (3) | 19 (3) | > .99 | ||||
| del(9q) | 0 (0) | 18 (2) | .2 | 0 (0) | 10 (2) | > .99 | ||||
| i(8q) | 0 (0) | 3 (< 0.5) | > .99 | 0 (0) | 2 (< 0.5) | > .99 | ||||
| −3 | 0 (0) | 5 (1) | > .99 | 0 (0) | 5 (1) | > .99 | ||||
| −9 | 1 (1) | 2 (< 0.5) | .3 | 0 (0) | 3 (1) | > .99 | ||||
| −12 | 0 (0) | 9 (1) | .6 | 0 (0) | 8 (1) | > .99 | ||||
| −13 | 0 (0) | 8 (1) | .6 | 0 (0) | 6 (1) | > .99 | ||||
| −17 | 1 (1) | 14 (2) | > .99 | 0 (0) | 10 (2) | > .99 | ||||
| −18 | 0 (0) | 14 (2) | .4 | 0 (0) | 10 (2) | > .99 | ||||
| −20 | 0 (0) | 6 (1) | > .99 | 0 (0) | 2 (< 0.5) | > .99 | ||||
| −X | 0 (0) | 19 (2) | .15 | 0 (0) | 12 (2) | > .99 | ||||
| −Y | 2 (2) | 37 (6) | .3 | 1 (3) | 23 (4) | > .99 | ||||
| −X/Y | 2 (2) | 56 (7) | .04 | 1 (3) | 35 (6) | > .99 | ||||
| +4 | 2 (2) | 6 (1) | .2 | 0 (0) | 5 (1) | > .99 | ||||
| +6 | 1 (1) | 9 (1) | > .99 | 0 (0) | 5 (1) | > .99 | ||||
| +8 | 14 (13) | 69 (8) | .2 | 2 (6) | 63 (11) | .6 | ||||
| +11 | 1 (1) | 11 (1) | > .99 | 0 (0) | 4 (1) | > .99 | ||||
| +13 | 1 (1) | 9 (1) | > .99 | 1 (3) | 8 (1) | .4 | ||||
| +19 | 0 (0) | 6 (1) | > .99 | 0 (0) | 4 (1) | > .99 | ||||
| +21 | 2 (2) | 18 (2) | > .99 | 2 (6) | 13 (2) | .18 | ||||
| +22 | 3 (3) | 15 (2) | .5 | 2 (6) | 11 (2) | .14 | ||||
| t(6; 9) | 1 (1) | 4 (< 0.5) | .5 | 0 (0) | 4 (1) | > .99 | ||||
| t(9; 11) | 1 (1) | 1 (< 0.5) | .2 | 0 (0) | 1 (< 0.5) | > .99 | ||||
| t(9; 22) | 0 (0) | 9 (1) | .6 | 0 (0) | 5 (1) | > .99 | ||||
| t(10; 11) | 4 (4) | 9 (1) | .06 | 0 (0) | 9 (2) | > .99 | ||||
| t(11; 19) | 0 (0) | 4 (< 0.5) | > .99 | 1 (3) | 2 (< 0.5) | .15 | ||||
| t(11 q23) | 0 (0) | 11 (1) | .4 | 0 (0) | 6 (1) | > .99 | ||||
| abn(11p15) | 1 (1) | 5 (1) | .5 | 0 (0) | 1 (< 0.5) | > .99 | ||||
| abn(12p13) | 1 (1) | 8 (1) | > .99 | 0 (0) | 6 (1) | > .99 | ||||
| abn(8p 11) | 1 (1) | 2 (< 0.5) | .3 | 0 (0) | 2 (< 0.5) | > .99 | ||||
| Adverse | ||||||||||
| Complex | 2 (2) | 52 (6) | .08 | 1 (3) | 38 (7) | .7 | ||||
| −5 | 1 (1) | 18 (2) | .7 | 0 (0) | 13 (2) | > .99 | ||||
| del(5q) | 0 (0) | 21 (3) | .16 | 0 (0) | 16 (3) | > .99 | ||||
| −7 | 3 (3) | 33 (4) | .8 | 2 (6) | 24 (4) | .6 | ||||
| abn(3q) | 1 (1) | 12 (1) | .8 | 0 (0) | 10 (2) | > .99 | ||||
| inv(3) | 2 (2) | 9 (1) | .6 | 1 (3) | 9 (2) | .4 | ||||
| t(3; 5) | 5 (5) | 3 (< 0.5) | .001† | 0 (0) | 6 (1) | > .99 | ||||
Patients with more than one cytogenetic abnormality will appear in more than one subgroup; therefore, total numbers in columns add up to more than the total number of patients. Percentages (in parentheses) are of total number of cases with available cytogenetic data. abn indicates abnormalities involving.
Significant underrepresentation of RAS mutation.
Significant overrepresentation of RAS mutation.
RAS mutational status: clinical outcome
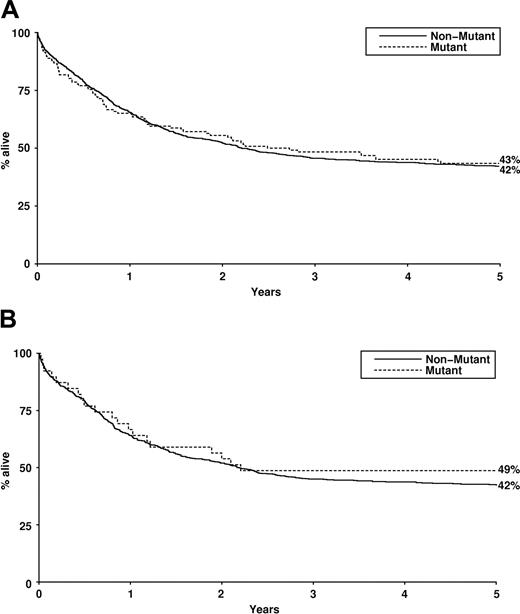
The presence of NRAS mutation did not significantly influence complete remission rate, induction death, resistant disease, relapse rate, disease-free survival, or overall survival either for the entire cohort (Table 4; Figure 1) or for cytogenetic risk groups (not shown), either in univariate analysis or in a proportional hazards model (including age, white cell count, cytogenetic risk group, de novo versus secondary, and FLT3 ITD).
Outcome data by NRAS and KRAS mutation status for patients treated within the MRC AML10 and AML12 clinical trials
. | NRAS . | . | . | KRAS . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | Mutant, % . | Nonmutant, % . | P . | Mutant, % . | Nonmutant, % . | P . | ||||
| Induction death | 9 | 8 | .7 | 8 | 8 | .9 | ||||
| Resistant disease | 10 | 9 | .8 | 3 | 10 | .16 | ||||
| Complete remission (CR) | 82 | 83 | .6 | 90 | 82 | .2 | ||||
| 5-y death in CR | 18 | 18 | .4 | 19 | 18 | .7 | ||||
| 5-y relapse rate | 48 | 48 | .6 | 44 | 47 | .8 | ||||
| 5-y disease-free survival | 42 | 42 | .9 | 45 | 43 | .9 | ||||
| 5-y overall survival | 43 | 42 | .9 | 49 | 43 | .5 | ||||
. | NRAS . | . | . | KRAS . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | Mutant, % . | Nonmutant, % . | P . | Mutant, % . | Nonmutant, % . | P . | ||||
| Induction death | 9 | 8 | .7 | 8 | 8 | .9 | ||||
| Resistant disease | 10 | 9 | .8 | 3 | 10 | .16 | ||||
| Complete remission (CR) | 82 | 83 | .6 | 90 | 82 | .2 | ||||
| 5-y death in CR | 18 | 18 | .4 | 19 | 18 | .7 | ||||
| 5-y relapse rate | 48 | 48 | .6 | 44 | 47 | .8 | ||||
| 5-y disease-free survival | 42 | 42 | .9 | 45 | 43 | .9 | ||||
| 5-y overall survival | 43 | 42 | .9 | 49 | 43 | .5 | ||||
Discussion
The frequency of NRAS mutation in our large cohort of AML patients is comparable with previous cohorts (12% to 44%6-8,11,25 ), the largest of which described data from 232 patients with 58 (28%) RAS mutations.26 Our analysis of a large cohort of AML patients was able to both strengthen previous observations from smaller cohorts (eg, a relative overrepresentation of RAS mutation in FAB type M47,8 ) and demonstrate new associations between RAS mutation frequency and biologically distinct subtypes of AML. No previous studies have meaningfully evaluated RAS mutation in association with karyotype. A high frequency of RAS mutation was found only in patients with t(3;5) (NRAS mutation) and inv(16) (KRAS mutation). Signal transduction pathway mutations are common in inv(16),27 although FLT3 ITD is relatively rare. Our sample size for t(3;5) is small, and independent confirmation of the high NRAS mutation frequency is required from larger cohorts. In contrast, NRAS mutation is relatively underrepresented in acute promyelocytic leukemia (FAB M3) with t(15;17), where FLT3 ITD is overrepresented.1 We confirmed that both RAS mutation and FLT3 ITD (and also 2 separate RAS mutations) are rarely present in the same tumor. Previous studies of clonogenic assays also confirm that 2 different NRAS mutations do not cosegregate within the same colony.28
Several different methods have previously been used for NRAS mutation detection, each with different sensitivities. Our data suggest that DHPLC is a more sensitive mutation screening method than direct sequencing. All abnormal DHPLC profiles were confirmed by cloning and sequencing, when direct sequencing failed to do so. DHPLC has been demonstrated to be more sensitive than both single-strand conformational polymorphism analysis and direct sequencing in the detection of germ-line and somatic mosaicism (reviewed in Xiao and Oefner29 ). DHPLC mutation screening also provides the opportunity to detect mutations that are outside “hot spots” at codons 12, 13, and 61 but that lie within the amplicon surrounding these codons. The hot spots are the most commonly mutated sites within RAS genes in human cancer, but other activating RAS mutations have been reported in AML patients (NRAS codon 60,11 KRAS codon 10/11 insertion30 ).
The most common RAS mutation base substitution reported in AML by us, and by others,8,10,31 is G>A transition. In hematopoietic tissue, this spectrum of RAS gene mutation (predominance of G>A transitions) is peculiar to the myeloid diseases AML and MDS. Using highly sensitive PCR mutation enrichment strategies, RAS mutations can be identified from peripheral blood of healthy hematologically normal individuals, but these are G>T transversions.32 NRAS G>T transversions are described in patients following cytotoxic chemotherapy for lymphoma,33 while patients with myeloma most commonly have N61 mutation.34 In solid tumors, the mutation spectrum of KRAS is consistent with exposure to different groups of carcinogens. The predominance of G>T transversions in lung cancer is consistent with the mutation spectrum induced by polycyclic aromatic hydrocarbons found in cigarette smoke, while the G>A transitions in gastrointestinal tumors are consistent with exposure to dietary carcinogens.35 In colorectal cancer KRAS G>A mutation may result from failure to repair the promutagenic 60-methylguanine DNA adduct. This adduct is produced as a consequence of exposure to selected carcinogens, including alkylators, and is read by DNA polymerase as adenine. In such patients, failure to repair this adduct is a consequence of promoter hypermethylation of the gene encoding the DNA repair enzyme 60-methylguanine-DNA methyltransferase.36 An alternative mechanism of G>A transition is spontaneous deamination of methylcytosine on the reverse DNA strand, but this is not likely as NRAS codons 12 and 13 are not at cytosine-phosphate-guanosine (CpG) islands.
It is clear that RAS gene mutation is a relatively frequent molecular event in AML and in MDS,37 occurring most commonly in NRAS, followed by KRAS, and least common in HRAS genes. This pattern is different from solid tumors such as gastrointestinal tumors in which KRAS is most commonly mutated. This may reflect a greater transforming capacity for NRAS mutation in hematopoietic cells,38 and/or the predominance of NRAS p21 protein in myeloid cells,39 leading to selective pressure for NRAS (compared with KRAS or HRAS) gene mutation.
RAS activation classically leads to proliferative signaling via Raf, MAP kinase, and activation of transcription factors such as activator protein 1 (AP-1). Although MAP kinase is constitutively activated in up to 50% of AML cases,5 this does not correlate with RAS mutational status.40 It is likely therefore that mutant RAS protein in AML blast cells signals via alternative signaling pathways downstream of RAS, which may include phosphatidylinositol 3 (PI3) kinase-Akt, Ral-guanine nucleotide exchange factors (GEFs), or Rac1.41 The precise role of RAS gene mutation in the pathogenesis of AML (or MDS) is yet to be defined. Clonogenic assays have identified a varying proportion of NRAS mutant and nonmutant colonies grown from AML bone marrow progenitors.28 That NRAS gene mutation was found in only more mature progenitors in some patients suggests that NRAS mutation is most likely a postinitiation event contributing to the progression/proliferation of subclones in AML. However, lethally irradiated mice that received transplants of bone marrow cells infected with mutated NRAS (N12) develop a myeloproliferative/AML-like disease.42 In vitro data also suggest that mutant RAS promotes a myeloid maturation defect, with relative sparing of the monocyte-macrophage lineage.43 This may be consistent with the overrepresentation of RAS mutation in M4/M5 FAB types.
The ratio of mutant to wild-type alleles showed considerable heterogeneity at presentation in patients with RAS mutant AML in our study. Assuming that the mutation was monoallelic, an average of 50% of cells harbored mutations. We cannot distinguish between variations in blast cell purity and clonal heterogeneity within the leukemic clone. While it is conceivable that NRAS mutation is never an essential component of the multistep pathogenesis in AML, this seems unlikely. We suggest that our data provide indirect evidence to implicate RAS mutation as an important functional pathologic event in selected cases of AML. Selection and expansion of RAS mutant clones may provide a differentiative stimulus toward the monocytic lineage, given that RAS mutation was overrepresented in FAB subtypes M4 and M5.
Although no previous study has been sufficiently large to definitively assess the influence of RAS mutation on clinical outcome in AML, in none of the 4 reported cohorts has a significant negative or positive effect been demonstrated.7,8,11,26 In contrast to previous studies in MDS,13,14 we found no influence of NRAS mutation on clinical outcome for our entire AML cohort or within cytogenetic risk groups. Mutation frequency was too low for meaningful assessment of clinical outcome within individual cytogenetic subgroups.
The rarity of the simultaneous presence of 2 different RAS mutations, or RAS mutations plus FLT3 ITD, is compatible with the notion that they all impart a proliferative/survival advantage through the same signaling pathway. Indeed FLT3 ITD is known to signal in part through the RAS pathway. It is of great interest therefore that whereas FLT3 ITD is associated with increased relapse rates, this is not the case for RAS mutation. This, in turn, suggests that the chemoresistance associated with FLT3 ITD is not due to the increased proliferative/survival signal per se that is common to both FLT3 ITD and RAS mutations. Either FLT3 ITD activates other pathways, not also activated by RAS mutation, or the mechanism by which ITDs are generated may activate different signaling pathways from those mechanisms that generate point mutations in the RAS gene. The clinical availability of therapeutic products with potential to target the RAS signaling pathway44 leads to the possibility that patients with RAS activation could respond well to these treatments. Our data may therefore not only identify novel associations between specific RAS mutations and biologic subtypes of AML but also have the potential to direct RAS-targeted therapy. A recent report of RAS-pathway “targeted” therapy (albeit relatively nonspecific) has demonstrated activity of a farnesyltransferase inhibitor, R115777, in patients with refractory AML.45 No NRAS or KRAS mutations were detected in responders, although 3 of 5 patients with chromosome 7 abnormalities (and presumed RAS activation) responded. Similarly, new therapeutic products aimed at common signaling pathways for FMS-like tyrosine kinase 3 (FLT3) and RAS (eg, MAP kinase) have the potential for response among other cytogenetic subgroups in which the frequency of either mutation is high. It is clear that the precise definition of downstream effector pathways of RAS signaling in myeloid leukemic cells will improve the understanding of mechanisms of RAS-induced leukemogenesis and may lead to further targets for the therapy of AML.
Overall survival by RAS mutant status. (A) Overall survival (Kaplan-Meier) of patients treated within AML10 and AML12 clinical trials by NRAS mutation status. (B) Overall survival (Kaplan-Meier) of patients treated within AML10 and AML12 clinical trials by KRAS mutation status.
Overall survival by RAS mutant status. (A) Overall survival (Kaplan-Meier) of patients treated within AML10 and AML12 clinical trials by NRAS mutation status. (B) Overall survival (Kaplan-Meier) of patients treated within AML10 and AML12 clinical trials by KRAS mutation status.
Prepublished online as Blood First Edition Paper, June 16, 2005; DOI 10.1182/blood-2005-03-0867.
Funded by the Leukaemia Research Fund (grant no. 98/35). The Acute Myeloid Leukemia (AML) DNA bank has been supported by the Medical Research Council, the Leukaemia Research Fund, and the Kay Kendall Leukaemia Fund. M.E.F. and P.D.K. were supported by the Leukaemia Research Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We wish to thank Ms Helen Walker for her assistance in characterizing the DNA bank samples. The authors also wish to thank David Baty and Dorothy Mechan for assistance with DHPLC assays.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal