Abstract
The analysis of rare chromosomal translocations in myeloproliferative disorders has highlighted the importance of aberrant tyrosine kinase signaling in the pathogenesis of these diseases. Here we have investigated samples from 679 patients and controls for the nonreceptor tyrosine kinase JAK2 V617F mutation. Of the 480 myeloproliferative disorder (MPD) samples, the proportion of positive cases per disease subtype was 30 (20%) of 152 for atypical or unclassified MPD, 2 of 134 (2%) for idiopathic hypereosinophilic syndrome, 58 of 72 (81%) for polycythemia vera, 24 of 59 (41%) essential thrombocythemia (ET), and 15 of 35 (43%) for idiopathic myelofibrosis. V617F was not identified in patients with systemic mastocytosis (n = 28), chronic or acute myeloid leukemia (n = 35), secondary erythrocytosis (n = 4), or healthy controls (n = 160). Homozygosity for V617F was seen in 43% of mutant samples and was closely correlated with chromosome 9p uniparental disomy. Homozygosity was significantly less common in ET compared with other MPD subtypes. In 53 cases analyzed, the median level of PRV1 expression was significantly higher in V617F-positive cases compared with cases without the mutation. We conclude that V617F is widespread in MPDs. Detection of this acquired mutation is likely to have a major impact on the way patients with MPD are diagnosed, as well as serving as an obvious target for signal transduction therapy. (Blood. 2005;106:2162-2168)
Introduction
Chronic myeloproliferative diseases (CMPDs) are clonal hematopoietic stem cell disorders characterized by proliferation of one or more myeloid cell lineages in the bone marrow and increased numbers of mature and immature cells in the peripheral blood. CMPDs include polycythemia vera (PV), essential thrombocythemia (ET), idiopathic myelofibrosis (IMF) and chronic myeloid leukemia (CML), plus rarer subtypes such as chronic neutrophilic leukemia (CNL), hypereosinophilic syndrome (HES), and chronic eosinophilic leukemia (CEL). These diseases overlap with myelodysplastic/myeloproliferative diseases (MDS/MPDs) such as atypical CML (aCML) and chronic myelomonocytic leukemia (CMML), in which proliferation is accompanied by dysplastic features or ineffective hematopoiesis in other lineages.1 We refer here broadly to all these groups as myeloproliferative disorders (MPDs).
Although there are strict diagnostic criteria for MPD subtypes, precise categorization remains a subject of debate2 and furthermore, it can be difficult to differentiate some cases from reactive disorders. Only CML is characterized by a pathognomonic molecular marker, the BCR-ABL fusion, and the primary abnormalities driving excess proliferation in most other cases have been obscure. However, several lines of evidence have implicated aberrant tyrosine kinase signaling as the root cause of MPDs. Breakpoint cluster region-abelson (BCR-ABL) itself is a constitutively active tyrosine kinase that is believed to be the primary, and probably the only, driving force behind chronic-phase CML.3 Other gene fusions have been identified in rare cases of aCML, CMML, and HES/CEL that involve the tyrosine kinases PDGFRA, PDGFRB, FGFR1, and JAK2.4,5 In addition, the KIT receptor is activated by point mutation in the majority of cases of systemic mastocytosis, a disease that is classified separately by the World Health Organisation but which is clearly myeloproliferative in nature.1,6
Observations in other MPD subtypes have also implicated aberrant signaling in their pathogenesis, albeit indirectly. Myeloid cells from patients are hypersensitive to a number of growth factors and cytokines, with PV and IMF in particular being characterized by the presence of erythroid and megakaryocytic precursor cells, respectively, that grow spontaneously in culture.7-10 This factor-independent growth and/or terminal differentiation can be blocked by a range of signal transduction inhibitors.11,12 Finally, some patients show increased expression of the antiapoptotic factor B-cell leukemia-XL (Bcl-XL) and activation of signal transducer activator of transcription (STAT) 3 or STAT5,11,13,14 all of which are downstream elements of tyrosine kinase signaling. To investigate the molecular pathogenesis of MPDs we therefore initiated a mutation screen for genes encoding tyrosine kinases and downstream signaling components.
Patients, materials, and methods
Patients
We studied samples from a total of 679 individuals. Of these, 480 were patients with a known or suspected diagnosis of an MPD (268 males, 212 females) referred for analysis in Athens, Mannheim or Salisbury. Patients were referred for PV (n = 72, including one case who also had systemic mastocytosis), ET (n = 59), IMF (n = 35), idiopathic HES (n = 134), systemic mastocytosis (n = 28), CML-like diseases (aCML, CMML and related atypical MPDs, n = 99) or atypical, unclassified MPD (n = 53). All HES, systemic mastocytosis, CML-like diseases, and atypical, unclassified MPD cases were tested for BCR-ABL, FIP1L1-PDGFRA, KIT D816V (mastocytosis cases only), and other gene fusions as indicated by karyotype. None were BCR-ABL-positive, but 24 had rare tyrosine kinase fusion genes involving PDGFRA, PDGFRB, FGFR1, or JAK2, and 8 had the KIT D816V mutation. We also studied samples from patients with AML (n = 17), BCR-ABL-positive CML (n = 18) plus controls from healthy individuals (n = 160) or individuals with secondary erythrocytosis (n = 4). The study was approved by the Internal Review Boards from participating institutions and informed consent was provided according to the Declaration of Helsinki.
Mutation screening
Initially, we studied a set of 40 atypical, CML-like MPD patients by heteroduplex analysis (either denaturing high-performance liquid chromatography or conformation-sensitive capillary electrophoresis) to screen candidate genes encoding tyrosine kinase and downstream signaling components for mutations. We focused on known mutational hotspots seen in other malignancies or regions that were homologous to mutational hotspots in other genes. We screened KIT (exons 10, 11 and 17), PDGFRA (exons 12 and 18), PDGFRB (exons 12, 17 and 18), FMS (exons 7, 9, 12, 13, 18, 21 and 22), JAK2 (exon 20), CSK (exon 11), FES (exon 17), SYK (exon 11), STYK1 (exon 8), TIE1 (exons 18 and 19), PTPN11 (exons 3 and 13), and BRAF (exons 11-14), but no abnormalities were detected apart from known polymorphisms. All exons of HRAS, KRAS, and NRAS were also screened and 5 (12.5%) cases were found to have activating mutations of N-Ras (G12S, n = 2; G12D, n = 1; G12C, n = 1; and G13S, n = 1). Polymerase chain reaction (PCR) primer sequences and assay conditions are available on request. Following the recent report of the JAK2 V617F mutation (2343G > T; JAK2 exon 12; RefSeq ID NM_00497215 ) in patients with PV, ET, and IMF,16-19 we focused specifically on this abnormality.
V617F genotyping by amplification refractory mutation system (ARMS)
DNA was extracted by standard procedures after isolation of total leukocytes from peripheral blood following red cell lysis or mononuclear cells by density gradient centrifugation over Histopaque 1077 (Sigma-Aldrich, Ayrshire, United Kingdom). Patients and controls were genotyped initially by a DNA tetra-primer ARMS assay, a method that uses 2 primer pairs to specifically amplify the normal and mutant sequences plus a positive control band in a single reaction. Primers were designed using a ARMS design program20 and include mismatches to maximize discrimination of the 2 alleles (shown in lowercase) and mutant/wild-type-specific bases (underlined). PCR primers were: forward outer (FO), 5′-TCCTCAGAACGTTGATGGCAG-3′; reverse outer (RO), 5′-ATTGCTTTCCTTTTTCACAAGAT-3′; forward wild-type-specific (Fwt), 5′-GCATTTGGTTTTAAATTATGGAGTATaTG-3′; reverse-mutant-specific (Rmt), 5′-GTTTTACTTACTCTCGTCTCCACAaAA-3′. Amplifications were performed for 30 cycles with HotStar Taq polymerase (Qiagen, Crawley, United Kingdom), an annealing temperature of 60°C, 25 ng genomic DNA, and standard amplification conditions, except that the final concentrations of the outer primers and the mutant/wild-type-specific inner primers were 1 μM and 0.5 μM, respectively. Products were resolved on 3% agarose gels and visualized after staining with ethidium bromide. Control experiments (not shown) indicated that the assay gave identical results using 20 to 200 ng input DNA.
V617F genotyping and allele quantitation by Pyrosequencing
DNA samples were amplified using primers 5′-biotin-GAAGCAGCAAGTATGATGAGCA-3′ (forward; JAK2 exon 12) and 5′-TGCTCTGAGAAAGGCATTAGAA-3′ (reverse; JAK2 intron 12). Amplicons were generated in a 50 μL reaction volume with 15 pmol of forward and reverse PCR primers, 0.2mM dNTPs (deoxyribonucleoside triphosphates; Promega, Southampton, United Kingdom), 1.5 mM MgCl2, 1 ×Buffer II (Applied Biosystems, Warrington, Cheshire, United Kingdom), 1 unit AmpliTaq Gold (Applied Biosystems) using 10 ng genomic DNA. PCR conditions were 94°C for 7 minutes; 50 cycles with denaturation at 94°C for 30 seconds, annealing at 58°C for 30 seconds, and elongation at 72°C for 30 seconds; 1 cycle at 72°C for 7 minutes; and a final hold at 15°C. Single-stranded biotinylated PCR products were prepared for sequencing using the Pyrosequencing Vacuum Prep Tool (Biotage, Uppsala, Sweden). Three microliters Streptavidin Sepharose HP (Amersham Biosciences, Chalfont St Giles, United Kingdom) was added to 37 μL Binding buffer (10 mM Tris [tris(hydroxymethyl)aminomethane]-HCl, pH 7.6; 2 M NaCl; 1 mM EDTA [ethylenediaminetetraacetic acid]; 0.1% Tween 20) and mixed with 20 μL PCR product and 20 μL high-purity water for 10 minutes at room temperature using a Variomag Monoshaker (Camlab, Over, United Kingdom). The beads containing the immobilized templates were captured onto the filter probes after applying the vacuum and then washed with 70% ethanol for 5 seconds, denaturation solution (0.2 M NaOH) for 5 seconds, and washing buffer (10 mM Tris-acetate, pH 7.6) for 5 seconds. The vacuum was then released and the beads released into a PSQ 96 Plate Low (Biotage) containing 45 μL annealing buffer (20 mM Tris-acetate, 2 mM magnesium acetate, pH 7.6), 0.3 μM sequencing primer (5′-TCTCGTCTCCACAGA-3′; JAK2 exon 12, reverse orientation). The samples were heated to 80°C for 2 minutes and then allowed to cool to room temperature. Pyrosequencing reactions were performed according to the manufacturer's instructions using the PSQ 96 single nucleotide polymorphism (SNP) Reagent Kit (Biotage), which contained the enzyme and substrate mixture and nucleotides. The sample genotypes were determined using the Allele Frequency Quantification function in the SNP Software (Biotage, Uppsala, Sweden). Samples were scored as homozygous if the proportion of the mutant allele was greater than 50%, the maximum expected if a heterozygous mutant clone had expanded to include all cells in the sample. In practice this is likely to underestimate the number of homozygotes since our samples included lymphocytes (a proportion of which are probably not part of the mutant clone).
Quantification of PRV1 mRNA
Mononuclear cell RNA was extracted and reverse transcribed into cDNA with random hexamer primers using standard procedures. Real-time quantitative (RQ)-PCR was performed in 12.5 μL mastermix (TaqMan Universal PCR Master Mix, Applied Biosystems); using 0.25 μM PRV1-TM probe (5′-FAM[6-carboxy fluorescein]-TTGTCTGGTGTGGT*TCAACAAGAAGCT-3′, where * = 6-carboxy-tetramethylrhodamine [TAMRA]), 0.5 μM of each 3′ and 5′ oligonucleotide primers (PRV1-se 5′-CTCTCAGGAGGTGGGCTGT-3′ and PRV1-as 5′-GCGCAGAGAAGATCCCGA-3′ respectively), 5 μL cDNA in a final volume of 25 μL. Amplification was performed in a 2-step cycle (denaturation, 95°C for 15 seconds, annealing/extension at 62°C for 60 seconds) for 50 cycles. Amplification and postprocessing calculations were performed using the ABI PRISM 7000 Sequence Detection System (Applied Biosystems). The values obtained were normalized using the ABL gene as endogenous control21 using the deltadeltaCt method.22
Microsatellite analysis
DNA samples were amplified with a series of fluorescently labeled primer pairs flanking highly polymorphic microsatellite markers on chromosome 9p (D9S1779, D9S1858, D9S288, D9S1813, D9S286, D9S254, D9S157, D9S171). Standard conditions were used with 25 to 50 ng input genomic DNA, a final volume of 10 μL, 32 amplification cycles, and 55°C annealing temperature. Products were analyzed on an ABI 3100 genetic analyser using the Genotyper 2.0 program (Applied Biosystems, Foster City, CA). Peak heights were compared with controls and scored as heterozygous for each marker if there were 2 clear peaks of similar intensity, or the ratio of the 2 peaks were similar to healthy controls. Patients were scored as homozygous if only one peak was visible or, to allow for the presence of background normal cells, if one peak was one third or smaller than the expected size compared with controls. Uniparental disomy (UPD) was scored if 4 or more consecutive markers abutting or encompassing JAK2 were homozgygous (P < .05 of this occurring in the absence of UPD based on published rates of heterozygosity23 ).
Copy number determination by multiplex ligation probe amplification (MLPA)
MLPA primers were designed to nonpolymorphic exons of FES (15q26), JAK2 (9p24), SOAT1 (1q25), BTK (Xq22), TYK2 (19p13), MST1R/RON (3p21), and HCK (20q11), and obtained from Biomers (Ulm, Germany). Primers were between 43 and 69 bases in length and consisted of M13 tags, a random stuffer sequence of variable length in order to generate gene-specific products of different sizes and 20 bases homologous to the target of interest (primer sequences available on request). Primers were designed in pairs that would abut each other when hybridized to genomic DNA, and the right-hand primer in each pair was synthesised with a 5′phosphate group to enable ligation to the left oligonucleotide. Hybridization of probe pairs to genomic DNA, ligation, and amplification was performed as described,24 with the PCR products being separated on an ABI 3100 genetic analyzer. Peak areas were exported to an Excel spreadsheet (Microsoft, Redmond, WA), which was designed to assess the ratios of each test peak relative to all other peaks for that individual. Ratios of the JAK2 peak to other peaks in each patient sample were compared with the same ratios obtained for 2 healthy individuals, which were included in each run. For normal sequences a dosage quotient of 1.0 is expected; if a deletion or duplication is present the dosage quotient should be 0.5 and 1.5, respectively.25 BTK is on the X-chromosome and therefore provided an internal control that the assay was working (dosage quotient of 0.5 in males and 1.0 in females).
Statistical analysis
Observed and expected frequencies were compared by χ2 analysis. Mutation status was compared with PRV1 mRNA levels using the Mann-Whitney test.
Results
Incidence of V617F JAK2
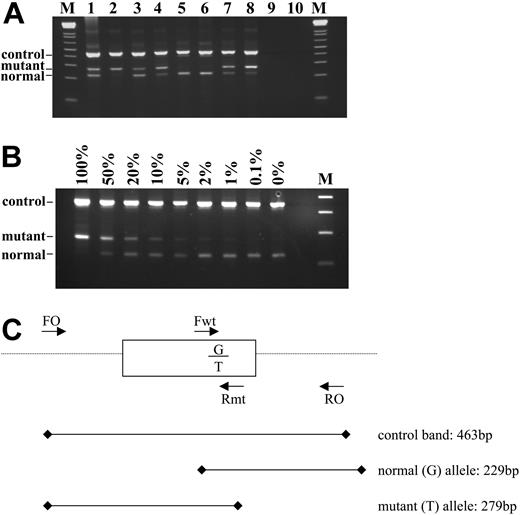
Of the 480 samples with a known or suspected diagnosis of an MPD, 129 (27%) were positive for the V617F JAK2 mutation by the ARMS assay and 351 (73%) were negative. Representative results are shown on Figure 1A. Sequence analysis of selected cases (n = 51) was fully concordant with the ARMS results (Figure 2A), although in some cases the proportion of the mutant allele was low and would have been difficult or impossible to detect by sequence analysis alone. Analysis of a dilution series indicated the sensitivity of the ARMS assay to be 1% to 2% (Figure 1B), whereas sequencing is generally accepted to have a sensitivity of only 20% to 30%, depending on the sequence context.
Using the ARMS assay, we found that the proportion of positive cases per disease subtype ranged from 58 (81%) of 72 for PV to 0 (0%) of 28 for mastocytosis (Table 1). Strikingly, V617F was detected in a substantial proportion of patients with CML-like diseases and atypical, unclassified MPD. V617F was not detected in any of the 480 MPD cases with rare tyrosine kinase fusion genes (n = 24), nor in any individual with CML (n = 18), AML (n = 17), or in healthy controls (n = 160). V617F and KIT D816V were found in one individual who had both PV and systemic mastocytosis, but not in any of the other 7 D816V-positive cases. Of the 14 V617F mutation-negative patients with PV, 13 were male (P = .005, χ2) but no other significant associations between sex and mutation status were identified. Mutation analysis of JAK2 exon 12 in V617F-negative cases did not reveal any additional sequence variants.
Patient details and summary of results
. | . | V617F-positive . | . | . | V617F-negative . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disease subtype . | No. patients . | No. patients (%) . | No. male/no. female . | Median age, y (range) . | No. patients (%) . | No. male/no. female . | Median age, y (range) . | V617F homozygotes no. (% of mutants) . | ||||
| PV | 72* | 58 (81) | 31/27 | 57 (26-76) | 14 (19) | 13/1 | 57 (33-78) | 24 (41) | ||||
| ET | 59 | 24 (41) | 15/9 | 65 (5-88) | 35 (59) | 23/12 | 56 (24-88) | 4 (17) | ||||
| IMF | 35 | 15 (43) | 10/5 | 65 (44-78) | 20 (57) | 15/5 | 62 (41-92) | 10 (67) | ||||
| Idiopathic HES | 134† | 2 (1.5) | 1/1 | 64 (63-65) | 132 (99) | 51/81 | 54 (3-89) | 2 (100) | ||||
| Mastocytosis | 28‡ | 0 (0) | NA | NA | 28 (100) | 16/12 | 53 (4-90) | NA | ||||
| CML-like MPDs | 99§ | 17 (17) | 13/4 | 62 (17-76) | 82 (83) | 60/22 | 66 (2-95) | 8 (47) | ||||
| Unclassified MPD | 53 | 13 (25) | 7/6 | 63 (17-77) | 40 (75) | 17/23 | 61 (21-87) | 7 (54) | ||||
| Total | 480 | 129 (27) | 77/52 | 58 (2-95) | 351 (73) | 195/156 | 62 (5-88) | 55 (43) | ||||
. | . | V617F-positive . | . | . | V617F-negative . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disease subtype . | No. patients . | No. patients (%) . | No. male/no. female . | Median age, y (range) . | No. patients (%) . | No. male/no. female . | Median age, y (range) . | V617F homozygotes no. (% of mutants) . | ||||
| PV | 72* | 58 (81) | 31/27 | 57 (26-76) | 14 (19) | 13/1 | 57 (33-78) | 24 (41) | ||||
| ET | 59 | 24 (41) | 15/9 | 65 (5-88) | 35 (59) | 23/12 | 56 (24-88) | 4 (17) | ||||
| IMF | 35 | 15 (43) | 10/5 | 65 (44-78) | 20 (57) | 15/5 | 62 (41-92) | 10 (67) | ||||
| Idiopathic HES | 134† | 2 (1.5) | 1/1 | 64 (63-65) | 132 (99) | 51/81 | 54 (3-89) | 2 (100) | ||||
| Mastocytosis | 28‡ | 0 (0) | NA | NA | 28 (100) | 16/12 | 53 (4-90) | NA | ||||
| CML-like MPDs | 99§ | 17 (17) | 13/4 | 62 (17-76) | 82 (83) | 60/22 | 66 (2-95) | 8 (47) | ||||
| Unclassified MPD | 53 | 13 (25) | 7/6 | 63 (17-77) | 40 (75) | 17/23 | 61 (21-87) | 7 (54) | ||||
| Total | 480 | 129 (27) | 77/52 | 58 (2-95) | 351 (73) | 195/156 | 62 (5-88) | 55 (43) | ||||
NA indicates not applicable.
Includes one case with PV plus systemic mastocytosis who was positive for V617F and KIT D816V.
Includes 7 FIP1L1-PDGFRA-positive CEL cases, all of whom were negative for V617F.
Includes 7 KITD816V-positive cases, all of whom were negative for V617F.
Includes 17 cases with rare tyrosine kinase fusions (ETV6-PDGFRB, n = 9; ZNF198-FGFR1, n = 2; BCR-FGFR1, n = 2; PCM1-JAK2, n = 2; BCR-PDGFRA, n = 1; TP53BP1-PDGFRB, n = 1), all of whom were negative for V617F.
ARMS assay to detect the JAK2 2343G > T in genomic DNA. (A) Representative results. Tracks 5 and 6 show a normal genotype; tracks 1 and 3 show a mutant band that is weaker or similar intensity to the normal band and were therefore scored as heterozygous for the 2343G > T mutation; tracks 2, 4, 7, and 8 show a mutant band that is stronger than the normal band and were therefore scored as homozygous for the mutation. Tracks 9 and 10 are negative controls and M is the 1-kilobase (kb)+ DNA ladder (Invitrogen, Paisley, United Kingdom). (B) Sensitivity of the ARMS assay. DNA from a homozygous patient with minimal residual wild-type allele was diluted with normal DNA and amplified. The ARMS assay is routinely capable of detecting V617F at a dilution of 1% to 2%. (C) Schematic outline of the assay. Primers FO and RO flank JAK2 exon 12 and should generate a control 463-bp band in all cases. Primers Fwt and RO generate a 229-bp wild-type (2343G)-specific product and primers FO and Rmt generate a 279-bp mutant (2343T)-specific product.
ARMS assay to detect the JAK2 2343G > T in genomic DNA. (A) Representative results. Tracks 5 and 6 show a normal genotype; tracks 1 and 3 show a mutant band that is weaker or similar intensity to the normal band and were therefore scored as heterozygous for the 2343G > T mutation; tracks 2, 4, 7, and 8 show a mutant band that is stronger than the normal band and were therefore scored as homozygous for the mutation. Tracks 9 and 10 are negative controls and M is the 1-kilobase (kb)+ DNA ladder (Invitrogen, Paisley, United Kingdom). (B) Sensitivity of the ARMS assay. DNA from a homozygous patient with minimal residual wild-type allele was diluted with normal DNA and amplified. The ARMS assay is routinely capable of detecting V617F at a dilution of 1% to 2%. (C) Schematic outline of the assay. Primers FO and RO flank JAK2 exon 12 and should generate a control 463-bp band in all cases. Primers Fwt and RO generate a 229-bp wild-type (2343G)-specific product and primers FO and Rmt generate a 279-bp mutant (2343T)-specific product.
Relative ratios of V617F and normal JAK2 alleles
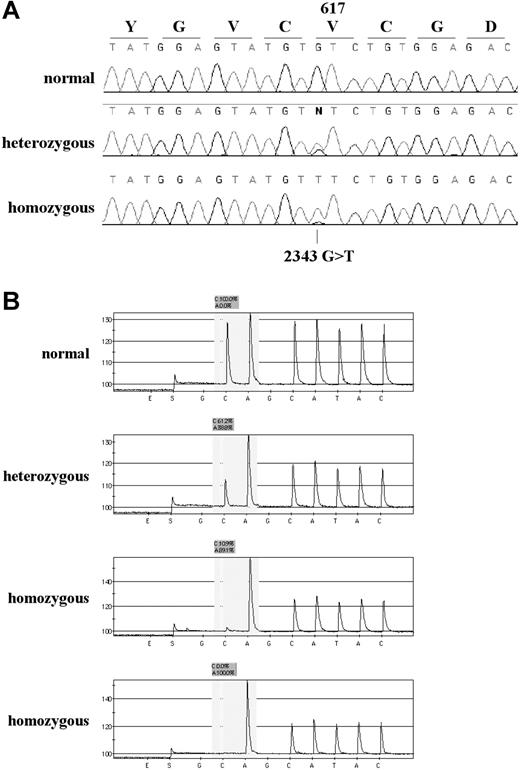
The ARMS and sequencing results for several patients showed mutant bands much stronger than the wild-type bands (Figures 1A and 2A) and in some cases the wild-type band was not visible at all, suggesting that the mutation may be homozygous in the mutant clone. Since ARMS and sequencing results are not always quantitative, we designed a Pyrosequencing assay for the V617F mutation, as this method is capable of providing robust allele ratios.26 Pyrosequencing was performed on the 90 mutant samples for which DNA was available, and confirmed homozygosity in most of the samples with a stronger mutant band by ARMS analysis (Figure 2B). Samples analyzed by ARMS only were therefore scored as homozygous if the ratio of mutant to wild-type bands was at least as strong as that seen in the 50% dilution control (Figure 1B, 50% track), as determined by visual inspection. Overall, homozygosity was seen in 55 (43%) of 129 mutant samples and results for each patient subgroup are summarized on Table 1. The frequency of homozygotes relative to heterozygotes was not significantly different from the average of all cases in any of the subgroups apart from ET, in which the proportion of homozygous mutants was significantly lower than average (P = .009, χ2). We found evidence of a residual wild-type signal in all but 2 homozygous cases by Pyrosequencing, with dilution experiments (not shown) indicating that the sensitivity of the assay was approximately 5%. Pyrosequence analysis of 23 cases that were normal genotype by the ARMS test yielded normal (100% wild-type allele) in all cases.
V617F and chromosome 9p uniparental disomy
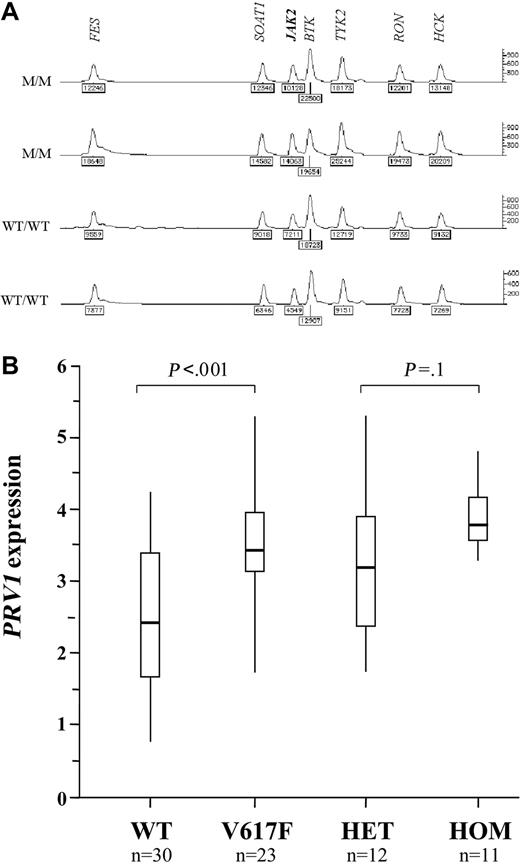
A homozygous V617F clone was seen in 24 (33%) of 72 cases of PV, 4 (7%) of 59 cases of ET, and 10 (29%) of 35 cases of IMF. This is strikingly similar to the frequencies of acquired chromosome 9p UPD (autosomal regions for which both copies are derived from a single parent) that has been described in these diseases. Furthermore, the minimal disomic region contains JAK2.27,28 To determine if UPD9 was associated with homozygosity for V617F, we initially performed microsatellite analysis on 57 MPD cases (wild type, n = 30; V617F homozygous, n = 27). Microsatellites were scored as homozygous or heterozygous and the results are summarized on Figure 3. In wild type individuals, most microsatellites are expected to be heterozygous. Consecutive tracts of homozygous markers usually indicate loss of heterozygosity (LOH) due to complete or partial chromosome loss, or UPD. We found significant tracts of homozygosity in the vicinity of JAK2 in 4 (13%) of the wild type cases and 25 (93%) of the V617F homozygotes (P < .001). To determine whether this homozygosity arose through LOH or UPD, we measured the number of copies of the JAK2 gene relative to control genes by MLPA. One copy of JAK2 would indicate LOH, whereas 2 copies would indicate UPD. All 7 cases analyzed that were homozygous for both V617F and at least 3 consecutive 9p markers had 2 copies of JAK2, consistent with UPD9 (Figure 4A).
Detection of the JAK2 by sequence analysis, and Pyrosequencing. (A) Sequence traces of 3 individuals found to be wild-type, heterozygous, and homozygous by the ARMS assay. (B) Representative pyrograms for one wild-type, one heterozygous, and 2 homozygous individuals. The sequencing primer is in reverse orientation (immediately abutting the site of the mutation) and the dispensation order GCAGCATAC was used (the 2 Gs are internal controls that should give no peak; E and S indicate enzyme and substrate, respectively). In a wild-type individual (sequence CACATAC) the peak heights of the first C and A are similar. In a heterozygous individual (sequence (C/A)ACATAC) the C peak is reduced in height and the A peak increased above all other peaks as both the mutant A and following A are being read in the same reaction. In a homozygous individual (sequence AACATAC) the first C peak is either much lower than the A peak (third trace) or, occasionally, absent (fourth trace). The ratio of the healthy (C) and mutant (A) alleles is calculated by the Pyrosequencing SNP analysis software. Values are log10 delta-delta CtPRV1/ABL.
Detection of the JAK2 by sequence analysis, and Pyrosequencing. (A) Sequence traces of 3 individuals found to be wild-type, heterozygous, and homozygous by the ARMS assay. (B) Representative pyrograms for one wild-type, one heterozygous, and 2 homozygous individuals. The sequencing primer is in reverse orientation (immediately abutting the site of the mutation) and the dispensation order GCAGCATAC was used (the 2 Gs are internal controls that should give no peak; E and S indicate enzyme and substrate, respectively). In a wild-type individual (sequence CACATAC) the peak heights of the first C and A are similar. In a heterozygous individual (sequence (C/A)ACATAC) the C peak is reduced in height and the A peak increased above all other peaks as both the mutant A and following A are being read in the same reaction. In a homozygous individual (sequence AACATAC) the first C peak is either much lower than the A peak (third trace) or, occasionally, absent (fourth trace). The ratio of the healthy (C) and mutant (A) alleles is calculated by the Pyrosequencing SNP analysis software. Values are log10 delta-delta CtPRV1/ABL.
Correlation between the presence of V617F and overexpression of PRV1
Overexpression of the PRV1 gene has been reported in the great majority of patients with PV and a subset of patients with ET and IMF.29-32 Although the mechanism of overexpression is not known, many groups consider quantification of PRV1 mRNA to be a useful supplementary test in the diagnosis of MPDs. To determine if PRV1 expression is associated with abnormalities of JAK2, we compared mRNA levels to mutational status in 53 cases (PV, n = 15; ET, n = 32; IMF, n = 6). Although there was considerable overlap between the range of PRV1 levels in normal JAK2 (n = 30, median = 293, range = 6-17 500) and V617F cases (n = 23, median = 2700, range = 59-208 000), the difference between the 2 groups was significant (P < .001, Mann-Whitney test). The median PRV1 level for homozygous V617F cases (median = 4940, n = 11) was greater than the median for heterozygous cases (median = 1650, n = 12), but the difference was not significant (P = .1, Mann-Whitney). Results are summarized in Figure 4B.
Discussion
We have identified the JAK2 V617F mutation in approximately one-fifth of patients with an atypical MPD, as well as confirming the presence of this mutation in the great majority of patients with PV and nearly half of cases with ET or IMF. The finding of an identical mutation in such clinically diverse (although clearly related) cases is quite remarkable. There is nothing in the sequence context to suggest that position 2343G might be particularly mutable and, since no other mutations were found in JAK2 exon 12, it seems likely that the V617F substitution must have very specific functional consequences. The mutation occurs in a highly conserved region of the pseudokinase (JH2) domain, a region that is homologous to the true tyrosine kinase domain but lacks key catalytic residues, and has been shown to constitutively activate JAK2.16,17,19 The pseudokinase domain is believed to negatively regulate JAK2 signaling by direct interaction with the kinase domain.33 Structural modeling has suggested that residues V617 to E621 form a loop connecting 2β-strands of the N-terminal lobe of the pseudokinase domain, with C618 contacting the kinase activation loop. It has been postulated that V617, C618, and other local residues inhibit movement of the activation loop from its inactive to its active conformation (ie, the V617 region plays a direct role in negatively regulating JAK2 signaling).34 The substitution of V617 by the large aromatic amino acid phenylalanine is likely to disrupt this negative regulation, although this remains to be proven biochemically. However, evidence that amino acid substitutions in the pseudokinase domain can activate JAK2 comes from the Drosophilia mutation hopscotch, in which a mutation corresponding to E695K in the human protein results in hematopoietic hyperplasia.35 In addition, JAK2 has been directly implicated in human malignancy, first by fusion to TEL/ETV6 and PCM1 as a consequence of chromosome translocations in leukemia36-38 and, second, by aberrant signaling in a wide range of solid tumors.39
Microsatellite analysis to identify regions of homozygosity. The V617F genotype of the patients is indicated as either homozygous normal (WT/WT; patients 1-30) or homozygous mutant (M/M; patients 31-57). Microsatellite markers on chromosome 9p are shown in order from the most telomeric (D9S1779) to the most centromeric (D9S171) and are scored as homozygous (black), heterozygous (white), or not done (gray). Based on published rates of heterozygosity,23 significant tracts of chromosome 9p homozygosity (LOH/UPD) were scored if 4 consecutive markers above or below JAK2 were homozygous.
Microsatellite analysis to identify regions of homozygosity. The V617F genotype of the patients is indicated as either homozygous normal (WT/WT; patients 1-30) or homozygous mutant (M/M; patients 31-57). Microsatellite markers on chromosome 9p are shown in order from the most telomeric (D9S1779) to the most centromeric (D9S171) and are scored as homozygous (black), heterozygous (white), or not done (gray). Based on published rates of heterozygosity,23 significant tracts of chromosome 9p homozygosity (LOH/UPD) were scored if 4 consecutive markers above or below JAK2 were homozygous.
JAK2 copy number and correlation of status with PRV1 expression. (A) Representative results of MLPA analysis to measure JAK2 copy number. The relative peak areas for JAK2 are the same for 2 homozygous V617F cases with 9p LOH (M/M) and 2 healthy controls (WT/WT), indicating the presence of 2 copies of JAK2. The second case is male and shows a reduced peak height for the X-linked gene BTK compared with the 3 other cases, all of which were female. (B) PRV1 mRNA levels (log10deltadeltaCtPRV1/ABL) determined by real-time PCR in 30 MPD patients with normal JAK2 (WT) and 23 MPD patients with the V617F mutation. These 23 patients are also shown split into the 12 heterozygous (HET) and 11 homozygous (HOM) cases. Vertical lines indicate the range of results, open boxes indicate the interquartile range, and thick horizontal lines indicate median values.
JAK2 copy number and correlation of status with PRV1 expression. (A) Representative results of MLPA analysis to measure JAK2 copy number. The relative peak areas for JAK2 are the same for 2 homozygous V617F cases with 9p LOH (M/M) and 2 healthy controls (WT/WT), indicating the presence of 2 copies of JAK2. The second case is male and shows a reduced peak height for the X-linked gene BTK compared with the 3 other cases, all of which were female. (B) PRV1 mRNA levels (log10deltadeltaCtPRV1/ABL) determined by real-time PCR in 30 MPD patients with normal JAK2 (WT) and 23 MPD patients with the V617F mutation. These 23 patients are also shown split into the 12 heterozygous (HET) and 11 homozygous (HOM) cases. Vertical lines indicate the range of results, open boxes indicate the interquartile range, and thick horizontal lines indicate median values.
JAK2 is a nonreceptor tyrosine kinase that plays a major role in myeloid development by transducing signals from diverse cytokines and growth factor receptors, including those for interleukin (IL)-3, IL-5, erythropoietin, granulocyte-macrophage-colony-stimulating factor (GM-CSF), G-CSF, and thrombopoietin.39,40 It seems likely that a mutation which interferes with the negative regulation of JAK2 could account for the observed hypersensitivity of myeloid cells from MPD patients to growth factors. However it is not clear why different individuals with V617F show preferential expansion of erythroid, granulocyte, megakaryocyte, monocyte, or eosinophil lineages. Potentially, this could be due to the identity of the cell in which the mutation arises, the constitutional genetic background of the individual, or to other, secondary acquired changes.
In addition to secondary or constitutional differences, V617F itself may also play a role in specifying disease phenotype since we found homozygosity to be significantly less common in ET compared with other MPD subtypes. However, definitive identification of V617F zygosity in individual cases is complicated by the fact that it is not possible to distinguish between a relatively small homozygous clone and a larger heterozygous clone when mixed populations of cells are analyzed. Nevertheless, we found a wide range (5%-100%) in the relative proportion of mutant alleles in V617F-positive cases by Pyrosequence analysis. The fact that (1) the mutation was present at a level substantially less than the wild-type allele in many heterozygous cases, (2) the wild-type allele was detectable in the great majority of homozygous patients, and (3) the age of homozygous cases (median, 63 years; range, 17-69 years) was not significantly different from heterozygous cases (median, 62 years; range, 5-88 years) strongly suggests that the mutation was acquired. Other studies have shown that V617F is absent in T cells and the great majority of buccal epithelial samples, clearly indicating that the mutation was acquired in these individuals.16-19 However, we cannot exclude the possibility that V617F might be inherited in occasional cases, although this seems unlikely as linkage to chromosome 9p has been excluded in MPD families.30,41
Although V617F accounts for an important subset of cases with CML-like diseases and atypical MPDs, the pathogenesis of the majority of these cases remains unknown. In our original series of 40 CML-like cases, only one-third of those that were negative for rare tyrosine kinase fusion genes were found to have the V617F JAK2 mutation or an activating NRAS mutation (no case had a JAK2 and RAS mutation). PV and IMF are both considered to be clonal diseases, suggesting that the molecular basis of roughly 20% and 60%, respectively, of these diseases remain to be elucidated. However, these figures may be overestimates since it is likely that some of the cases in our study had an unrecognized reactive condition. It is known that the clinical diagnostic precision varies between clinicians and it is possible therefore that the true incidence of V617F in PV and IMF is somewhat higher than we found in this study.42 On the other hand, it is likely that technical differences relating to the sensitivity of V617F detection are also a contributory factor to the published differences in the proportions of positive cases within MPD subtypes. As for ET, approximately half of cases have clonal disease and half are polyclonal, and therefore presumably reactive.43 Although we did not perform clonality assays, we found the V617F mutation in 41% of ET cases, suggesting that the great majority of clonal ET is V617F positive. We did not observe V617F in patients with CML or AML; however, the numbers of cases analyzed were relatively small and therefore we cannot exclude the possibility that a small subset of cases might carry this mutation. Interestingly, 6 of the patients in our series had chronic neutrophilic leukemia, 2 of which were V617F positive.
We found that homozygosity for V617F in all disease subtypes was closely associated with chromosome 9 UPD, confirming previous results and suggesting a selective advantage for 2 copies of mutant JAK2.17 We also found significant 9p homozygosity in 4 MPD cases without V617F, which could indicate a mutation in a different region of JAK2. Although gain of chromosome 9p is known to be associated with MPDs,44-46 UPD for this region is much more common. Recently, UPD for diverse chromosomal regions has been identified in acute leukemia47,48 and, although most of these regions have not yet been associated with specific acquired mutations, it is possible that reduction of oncogenic mutations to homozygosity by cytogenetically cryptic mitotic recombination is widespread in malignancy.
The fact that V617F was found in the great majority of cells and was homozygous in many cases suggests that it is probably the primary abnormality driving myeloproliferation. As such, it is clearly a very attractive target for signal transduction therapy with small molecule inhibitors, although at the time of writing no clinically available JAK2 inhibitors have been described. JAK family members play a crucial role in the immune system; for example, inherited JAK3 deficiency causes severe combined imunodeficiency,49,50 and JAK2 also plays an important role in cardiovascular signaling systems.51 Development of inhibitors that inhibit V617F without undesirable side effects may therefore be challenging.
Diagnosis of MPDs is often complex, expensive and in the case of ET, based solely on exclusion criteria. Since the detection of acquired V617F is simple to perform and unambiguously establishes the presence of a clonal disorder, we believe that JAK2 mutation testing will rapidly become a frontline test for individuals with a suspected diagnosis of an MPD.
Prepublished online as Blood First Edition Paper, May 26, 2005; DOI 10.1182/blood-2005-03-1320.
Supported by the Leukaemia Research Fund (United Kingdom), the Dr Mildred Scheel Stiftung, the Wessex Cancer Trust, the Cancer Research and Treatment Fund, Inc., and the European LeukemiaNet within the 6th European Community Framework Programme for Research and Technological Development.
We would like to thank Candice Price for assistance in providing samples and patient information.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal