Abstract
Lymphopenia and increasing viral load in the first 10 days of severe acute respiratory syndrome (SARS) suggested immune evasion by SARS-coronavirus (CoV). In this study, we focused on dendritic cells (DCs) which play important roles in linking the innate and adaptive immunity. SARS-CoV was shown to infect both immature and mature human monocyte-derived DCs by electron microscopy and immunofluorescence. The detection of negative strands of SARS-CoV RNA in DCs suggested viral replication. However, no increase in viral RNA was observed. Using cytopathic assays, no increase in virus titer was detected in infected DCs and cell-culture supernatant, confirming that virus replication was incomplete. No induction of apoptosis or maturation was detected in SARS-CoV–infected DCs. The SARS-CoV–infected DCs showed low expression of antiviral cytokines (interferon α [IFN-α], IFN-β, IFN-γ, and interleukin 12p40 [IL-12p40]), moderate up-regulation of proinflammatory cytokines (tumor necrosis factor α [TNF-α] and IL-6) but significant up-regulation of inflammatory chemokines (macrophage inflammatory protein 1α [MIP-1α], regulated on activation normal T cell expressed and secreted [RANTES]), interferon-inducible protein of 10 kDa [IP-10], and monocyte chemoattractant protein 1 [MCP-1]). The lack of antiviral cytokine response against a background of intense chemokine up-regulation could represent a mechanism of immune evasion by SARS-CoV.
Introduction
Coronaviruses (CoVs) comprise a large family of RNA viruses that infect a broad range of vertebrates, from mammalian to avian species.1 Prior to the emergence of severe acute respiratory syndrome (SARS) in 2002 to 2003, human CoVs were known to be associated mainly with relatively mild upper respiratory diseases such as the common cold. The novel SARS-CoV, however, caused severe, rapidly progressive atypical pneumonia with fever, myalgia, and diarrhea.2,3 The detection of virus in stool and urine in addition to the respiratory tract of patients with SARS further suggested that SARS is a systemic disease.4,5
At autopsy, white pulp atrophy was observed in the spleen, and there was lymphoid depletion in lymph nodes.6-8 Together with lymphopenia and increasing viral load in the first 10 days of disease,3,4,6 these clinical features strongly suggest an evasion of the immune system by SARS-CoV. As with other viral infections, such as measles, this lymphoid depletion may have pathogenic significance.
Dendritic cells (DCs) are antigen-presenting cells that play key roles in linking innate and adaptive immunity.9-11 Immature DCs reside in the respiratory tract for immune surveillance, and they respond dynamically to local tissue inflammation in the airways and the distal lung.12,13 They express a wide range of receptors, including c-type lectins14,15 and toll-like receptors,16,17 for the recognition of conserved pathogen patterns. Dendritic cells signal the presence of danger to cells of the adaptive immune response and modulate their responses via the secretion of proinflammatory and/or antiviral cytokines.18 In particular, DCs secrete cytokines to polarize T-helper (Th) cells toward the Th1 or Th2 subsets.10
The migration of DCs from tissues to lymph nodes is essential for antigen presentation and triggering of adaptive immune responses. The trafficking of DCs is regulated by chemokines that can be classified as homeostatic (constitutively expressed) or inflammatory (induced/augmented) according to their immune fuctions.19-21 Acute respiratory viruses commonly induce inflammatory chemokines, such as macrophage inflammatory protein 1α (MIP-1α), regulated upon activation, normal T cell expressed and secreted (RANTES), interferon-inducible protein of 10 kDa (IP-10), and monocyte chemotactic protein 1 (MCP-1), in local tissues.21 Dendritic cells are also a major source of these chemokines.20
On the basis of the function of DCs in immune surveillance, priming, and tolerance, we hypothesized that DCs play an important role in the immunopathology of SARS. In addition, the developmental status of the host immune cells may affect their responses to viral infection. Hence, we also compared the cytokine and chemokine gene expression in SARS-CoV–infected adult and cord blood (CB) DCs. This study provides evidence that SARS-CoV can infect DCs and alter their cytokines/chemokines production. Our results suggest possible mechanisms of immune escape and amplification of immunopathology in SARS.
Materials and methods
Samples
Adult blood samples were from the white cell fraction of blood donated to the Hong Kong Red Cross by healthy volunteers. Human umbilical CB samples were collected from the placenta of normal full-term uncomplicated pregnancies. Informed consent was obtained from the mothers prior to delivery. The protocol was approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (EC1473-00).
Cell separation
Blood mononuclear cells were isolated from whole blood by centrifugation, using Ficoll-Hypaque gradients (Pharmacia Biotech, Uppsala, Sweden), washed, and labeled with immunomagnetic antibodies. Positive selection was performed according to manufacturer's specification (Miltenyi Biotec, Bergisch Gladbach, Germany) as in previous experiments.22-26 Isolated CD14+ monocytes from the positive fraction were resuspended in RPMI 1640, supplemented with 50 IU/mL penicillin and 50 μg/mL streptomycin, and 10% fetal bovine serum (Invitrogen, Grand Island, NY). Cell viability, as measured by trypan blue exclusion, was greater than 95%. The purity of the isolated cells as measured by flow cytometry was constantly between 90% and 95%.
Generation of DCs in vitro
CD14+ monocytes were cultured in the presence of interleukin 4 (IL-4; 10 ng/mL; R&D Systems, Minneapolis, MN) and granulocyte-macrophage colony-stimulating factor (GM-CSF; 50 ng/mL; R&D Systems) for 7 days at 37°C in a humidified atmosphere containing 5% CO2 as in our previous studies.22-26 The cultures were fed with fresh medium and cytokines on day 3, and cell differentiation was monitored by light microscopy. For the generation of mature DCs, 10 μg/mL lipopolysaccharide (LPS; Sigma, St Louis, MO) was added for the last 2 days of the culture. On day 7, DCs were harvested, centrifuged, washed, and adjusted to 1 × 106 cells/mL before virus infection. The maturation of DCs was confirmed by flow cytometry on a panel of maturation markers, including CD40–fluorescein isothiocyanate (FITC), CD80-FITC, CD83-FITC, CD86-FITC, major histocompatability complex (MHC) class II–FITC, CD11c-phycoerythrin (PE), mannose receptor (MR) PE, and CD1a–phycoerythrin-cyanin 5 (PC5) (BD Pharmingen, San Diego, CA).
Virus preparation, titration, and infection
Laboratory procedures involving live viruses were performed in biosafety level-3 containment. SARS-CoV, strain HKU-398493 was cultured in fetal rhesus kidney-4 (FRhK-4) cells. The cell culture supernatant was harvested, centrifuged to remove cell fragments, divided into aliquots, and kept frozen at -70°C. SARS-CoV titer of the stock virus was determined by infection of FRhK-4 cells. Cytopathic changes on FRhK-4 cells were monitored every day up to 4 days, and virus titer was expressed as tissue culture infective dose (TCID50). This virus titration method was also used to determine infectious virus production in SARS-CoV–infected DCs.
Cells were inoculated by SARS-CoV at a multiplicity of infection (MOI) of 1. The virus was allowed to be adsorbed for 1 hour at 37°C, and unbound virus was washed off by excess volume of phosphate-buffered saline (PBS; time = 0 hour after infection). Mock-infected cells were treated in parallel, except that virus was not added. In generating positive controls for gene expression study, Influenza A virus, H1N1 (54/98) previously isolated from human beings and prepared solely in Madin-Darby canine kidney (MDCK) cells,27 was used to infect DCs.
Transmission electron microscopy
Electron microscopy was performed on SARS-CoV–infected immature and mature DCs at 0 hour, 6 hours, 12 hours, and 24 hours after infection. One million DCs were washed, fixed in 2.5% glutaraldehyde, and stored at 4°C for more than 8 hours before processing. The cell suspension was postfixed in osmium tetroxide and dehydrated in a series of ethanol. After dehydration, the pellets were embedded in agar, and ultrathin sections (silver interference color 90 nm) were cut using a diamond knife. The sections were double stained with uranyl acetate and lead citrate before being viewed on a Philips EM208S transmission electron microscope (Philips Electron Optics, Eindhoven, The Netherlands) at an accelerating voltage of 80 kV. More than 50 cells per section and 2 to 3 sections per condition/time point have been screened.
Indirect immunofluorescence assay
Mock- or virus-infected cells (∼ 104 cells) harvested at 12 hours and 24 hours after infection were air dried on spotted slides and fixed with acetone-ethanol (1:1). To determine the presence of SARS-CoV and viral protein, indirect immunofluorescence assays were performed using the heat-treated convalescent serum from a known patients with SARS as the source of anti–SARS-CoV antibodies (SARS-CoV antibody titer of 1:640) and FITC-conjugated anti–human immunoglobulin G (IgG) antibodies as secondary antibody (INOVA Diagnostics, San Diego, CA). Evans Blue was used as the counterstain. Fluorescent images were acquired using a Leica DMLB microscope and a Leica DC500 digital camera system with the Leica Image Manager software (Leica Microsystems AG, Wetzlar, Germany). Confocal microscopy was performed by Bio-Rad Radiance 2100 laser scanning confocal system equipped with Nikon E1000 microscope and the LaserSharp2000 software (Bio-Rad Laboratories, Hercules, CA).
Quantification of SARS-CoV RNA by real-time quantitative RT-PCR
Total RNA was extracted from approximately 5 × 105 cells harvested at 3 hours, 9 hours, 24 hours, day 3, and day 6 after infection by the RNeasy Mini Kit (Qiagen, Hilden, Germany). DNase-treated total RNA (2 μL) was reverse transcribed by either forward or reverse primers specific for SARS-CoV (Table 1) and Superscript II reverse transcriptase (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's recommendation. The forward primers transcribe the negative strands, whereas the reverse primers transcribe the positive strands into cDNA. The cDNA was diluted (1:20) and quantified by real-time reverse-transcriptase–polymerase chain reaction (PCR) using the Lightcycler Technology (Roche, Mannheim, Germany) as in our previous study.4 Detection of PCR product was based on SYBR green fluorescence signal. The standard curve was generated using serial dilutions of plasmids (∼ 10-1010 copies) containing cloned sequences involved.
PCR primers and probes
Genes . | Sequences . | Sizes, bp . |
|---|---|---|
| SARS-CoV | (F) 5′ TAC ACA CCT CAG CGT TG 3′ | 182 |
| (R) 5′ CAC GAA CGT GAC GAA T 3′ | ||
| β-actin | (F) 5′ CCC AAG GCC AAC CGC GAG AAG AT 3′ | 219 |
| (R) 5′ GTC CCG GCC AGC CAG GTC CAG 3′ | ||
| IFN-α | (F) 5′ CCT TCC TCC TGT CTG ATG GA 3′ | 67 |
| (P) 5′ (FAM) CAG ACA TGA CTT TGG ATT TCC CCA GG (TARMA) 3′ | ||
| (R) 5′ ACT GGT TGC CAT CAA ACT CC 3′ | ||
| IFN-β | (F) 5′ GCC GCA TTG ACC ATC T 3′ | 261 |
| (R) 5′ AGG AGT ACA GTC ACT GTG 3′ | ||
| IFN-γ | (F) 5′ CTA ATT ATT CGG TAA CTG ACT TGA 3′ | 75 |
| (P) 5′ (FAM) TCC AAC GCA AAG CAA TAC ATG AAC (TARMA) 3′ | ||
| (R) 5′ ACA GTT CAG CCA TCA CTT GGA 3′ | ||
| TNF-α | (F) 5′ GGC TCC AGG CGG TGC TTG TTC 3′ | 409 |
| (R) 5′ AGA CGG CGA TGC GGC TGA TG 3′ | ||
| IL-6 | (F) 5′ ATT CGG TAC ATC CTC GAC 3′ | 331 |
| (R) 5′ GGG GTG GTT ATT GCA TC 3′ | ||
| IL-12 | (F) 5′ CGG TCA TCT GCC GCA AA 3′ | 80 |
| (R) 5′ TGC CCA TTC GCT CCA AGA 3′ | ||
| MIP-1α | (F) 5′ CTC TGC ACC ATG GCT CTC TGC AAC 3′ | 98 |
| (R) 5′ TGT GGA ATC TGC CGG GAG GTG TAG 3′ | ||
| RANTES | (F) 5′ CCC CTC ACT ATC CTA CC 3′ | 285 |
| (R) 5′ TCA CGC CAT TCT CCT G 3′ | ||
| IP-10 | (F) 5′ CTG ACT CTA AGT GGC ATT 3′ | 208 |
| (R) 5′ TGA TGG CCT TCG ATT CTG 3′ | ||
| MCP-1 | (F) 5′ CAT TGT GGC CAA GGA GAT CTG 3′ | 91 |
| (R) 5′ CTT CGG AGT TTG GGT TTG CTT 3′ | ||
| ACE2 | (F) 5′ CTG GGA TCA GAG ATC GGA AGA 3′ | 288 |
| (R) 5′ CTC TCT CCT TGG CCA TGT TGT 3′ |
Genes . | Sequences . | Sizes, bp . |
|---|---|---|
| SARS-CoV | (F) 5′ TAC ACA CCT CAG CGT TG 3′ | 182 |
| (R) 5′ CAC GAA CGT GAC GAA T 3′ | ||
| β-actin | (F) 5′ CCC AAG GCC AAC CGC GAG AAG AT 3′ | 219 |
| (R) 5′ GTC CCG GCC AGC CAG GTC CAG 3′ | ||
| IFN-α | (F) 5′ CCT TCC TCC TGT CTG ATG GA 3′ | 67 |
| (P) 5′ (FAM) CAG ACA TGA CTT TGG ATT TCC CCA GG (TARMA) 3′ | ||
| (R) 5′ ACT GGT TGC CAT CAA ACT CC 3′ | ||
| IFN-β | (F) 5′ GCC GCA TTG ACC ATC T 3′ | 261 |
| (R) 5′ AGG AGT ACA GTC ACT GTG 3′ | ||
| IFN-γ | (F) 5′ CTA ATT ATT CGG TAA CTG ACT TGA 3′ | 75 |
| (P) 5′ (FAM) TCC AAC GCA AAG CAA TAC ATG AAC (TARMA) 3′ | ||
| (R) 5′ ACA GTT CAG CCA TCA CTT GGA 3′ | ||
| TNF-α | (F) 5′ GGC TCC AGG CGG TGC TTG TTC 3′ | 409 |
| (R) 5′ AGA CGG CGA TGC GGC TGA TG 3′ | ||
| IL-6 | (F) 5′ ATT CGG TAC ATC CTC GAC 3′ | 331 |
| (R) 5′ GGG GTG GTT ATT GCA TC 3′ | ||
| IL-12 | (F) 5′ CGG TCA TCT GCC GCA AA 3′ | 80 |
| (R) 5′ TGC CCA TTC GCT CCA AGA 3′ | ||
| MIP-1α | (F) 5′ CTC TGC ACC ATG GCT CTC TGC AAC 3′ | 98 |
| (R) 5′ TGT GGA ATC TGC CGG GAG GTG TAG 3′ | ||
| RANTES | (F) 5′ CCC CTC ACT ATC CTA CC 3′ | 285 |
| (R) 5′ TCA CGC CAT TCT CCT G 3′ | ||
| IP-10 | (F) 5′ CTG ACT CTA AGT GGC ATT 3′ | 208 |
| (R) 5′ TGA TGG CCT TCG ATT CTG 3′ | ||
| MCP-1 | (F) 5′ CAT TGT GGC CAA GGA GAT CTG 3′ | 91 |
| (R) 5′ CTT CGG AGT TTG GGT TTG CTT 3′ | ||
| ACE2 | (F) 5′ CTG GGA TCA GAG ATC GGA AGA 3′ | 288 |
| (R) 5′ CTC TCT CCT TGG CCA TGT TGT 3′ |
(F) indicates forward primers; (R), reverse primers; (P), Taqman probes; TNF, tumor necrosis factor; IFN, interferon; IL, interleukin; MIP, macrophage inflammatory protein; RANTES, regulated on activation, normal T cell expressed and secreted; IP-10, interferon-γ induced protein 10; MCP-1, monocyte chemoattractant protein 1, ACE2, angiotensin converting enzyme 2.
Active caspase-3 assay
Activated caspase-3 was selected as a biologic marker for apoptosis. Mock- or SARS-CoV–infected adult immature DCs were assayed using a polyclonal active caspase-3 antibody apoptosis kit according to the manufacturer's specification (BD Pharmingen). Briefly, cultured cells harvested at different time points were washed twice with PBS, then fixed, and permeabilized in a solution containing pH-buffered saline, saponin, and 4% (wt/vol) paraformaldehyde for 20 minutes on ice. Cells were then washed twice with a buffer containing fetal bovine serum, sodium azide, and saponin and stained by monoclonal antibody against active caspase-3. Stained cells were washed, resuspended, and analyzed by flow cytometry (COULTER EPICS ELITE; Beckman Coulter, Miami, FL). Ten thousand events per sample were collected into listmode files and analyzed by WINMDI 2.8 analysis software (The Scripps Research Institute, La Jolla, CA). For positive control, Jurkat T cells (ATCC TIB-152; American Type Culture Collection [ATCC], Manassas, VA) were induced to undergo apoptosis by 25 ng/mL anti-Fas antibodies (clone CH11; Upstate, Lake Placid, NY) for 3 or 24 hours, harvested, and stained in parallel with the mock- or SARS-CoV–infected DCs.
Determination of surface-marker expression on DCs by flow cytometry
Mock- or SARS-CoV–infected adult immature DCs (MOI = 1) were harvested at 48 hours after infection, fixed in 4% paraformaldehyde, and stored at 4°C for more than 1 hour before analysis. The fixed cells were washed and stained for a panel of maturation markers (including CD83-FITC, CD86-FITC, MHC class I–FITC, and MHC class II–FITC) and isotype control-FITC (BD Pharmingen). To determine whether SARS-CoV infection will impair DC maturation, LPS (10 μg/mL) was added to the SARS-CoV–infected immature DCs throughout the postinfection period. Mock-infected cells were included as control of the experiment.
Quantification of cytokine/chemokine RNA by real-time quantitative RT-PCR
Cells (∼ 1.5 × 105) were harvested at 3 hours and 9 hours after infection, and total RNA was extracted by TRIzol Reagent (Invitrogen Life Technologies). In later experiments, QiaShredder columns (Qiagen) were used to ensure adequate homogenization, and RNA was extracted by the RNeasy Mini Kit (Qiagen). Reverse transcription was performed on the DNase-treated total RNA using oligo (dT) primers and Superscript II reverse transcriptase (Invitrogen Life Technologies) according to the manufacturer's recommendation. The cDNA synthesized was diluted (1:50) and quantified by real-time PCR using the Lightcycler Technology (Roche, Mannheim, Germany) or Taqman Technology (Applied Biosystems, Foster City, CA). Specific primers (Table 1) were used and nonspecific reactions and primer-dimer artifacts have been minimized (as evaluated by gel electrophoresis). Detection of PCR product was based on SYBR green or Taqman fluorescence signal. Dissociation curve analysis was performed after SYBR green assays to ensure specific target detection. The β-actin gene was amplified as an internal control. Standard curves were generated using serial dilutions of plasmids (∼ 10-1010 copies) containing cloned sequences involved. Results were calculated as the number of targeted molecules per microliter of cDNA. To standardize results for variability in RNA and cDNA quantity and quality, we express the results as the number of target copies per 104 copies of β-actin gene.27
Statistical analysis
All data were expressed as mean ± SEM. All samples were paired and differences between groups analyzed by paired Student t test or the nonparametric equivalents.
Results
SARS-CoV could infect both immature and mature DCs from adults and CB
In the initial experiments, we determined whether SARS-CoV can infect DCs by electron microscopy and immunofluorescence staining. Because the route of entry of SARS-CoV into DCs has not been identified, we used both immature and LPS-treated mature DCs that have different expression of receptors and different endocytotic functions. We also used DCs derived from adult and cord blood monocytes to determine whether the developmental status of the host would affect virus entry.
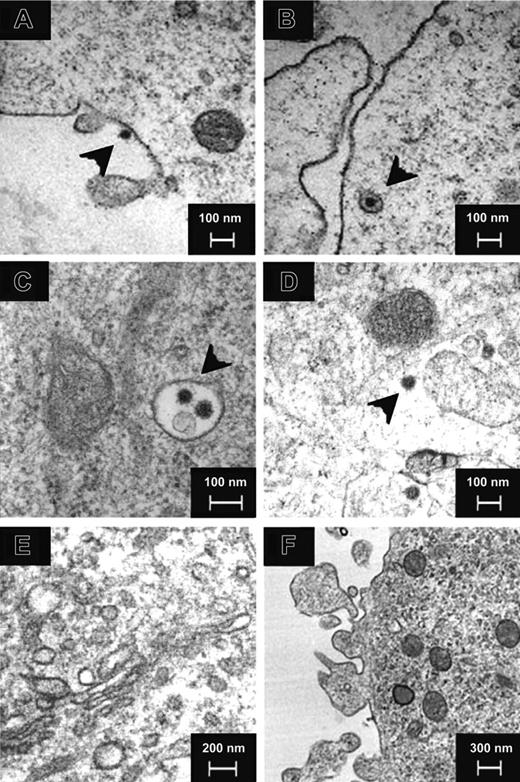
Electron microscopy showed SARS-CoV (black arrows) binding to DC (Figure 1A) and adsorbed in either an invagination of the plasma membrane or an endosome (Figure 1B) at 0 hour after infection. At 6 hours, 12 hours, and 24 hours after infection, viral particles were detected in endosomes (Figure 1C) and cytoplasm (Figure 1D) but not in the Golgi apparatus (Figure 1E). No virus budding (Figure 1F) was observed in all the cells examined (n > 200). Cytopathic effect was observed in some immature DCs but to a lesser extent in the mature DCs (data not shown). Similar finding was observed in adult and cord blood immature and mature DCs from 3 independent donors.
Electron microscopy of SARS-CoV–infected human DCs. Negative-contrast thin-section transmission electron micrograph of SARS-CoV–infected human DCs showed virus (black arrowheads) binding (A) and uptake (B) at 0 hours after infection. At 6 hours, 12 hours, and 24 hours after infection, virus particles were detected in endosomes (C) and cytoplasm (D) but not in the Golgi apparatus (E). No virus budding was observed (F) in all cells examined. Images are representative of SARS-CoV–infected human DCs from 3 independent adult or CB donors.
Electron microscopy of SARS-CoV–infected human DCs. Negative-contrast thin-section transmission electron micrograph of SARS-CoV–infected human DCs showed virus (black arrowheads) binding (A) and uptake (B) at 0 hours after infection. At 6 hours, 12 hours, and 24 hours after infection, virus particles were detected in endosomes (C) and cytoplasm (D) but not in the Golgi apparatus (E). No virus budding was observed (F) in all cells examined. Images are representative of SARS-CoV–infected human DCs from 3 independent adult or CB donors.
Using convalescent serum from patients with SARS as source of anti–SARS-CoV antibodies, positive immunofluorescence staining was detected in SARS-infected DCs but not the mock-infected DCs (Figure 2). The observations in adult and CB DCs were similar, and a representative case of CB DCs is included. Similar staining was detected in both immature and mature monocyte-derived DCs with greater than 90% of DCs being positive at both 12 hours (Figure 2B) and 24 hours (Figure 2C) after infection. Confocal microscopy showed that positive staining was not limited to the cell surface but inside cytoplasm (Figure 2D).
Incomplete replication of SARS-CoV was detected in adult immature DCs
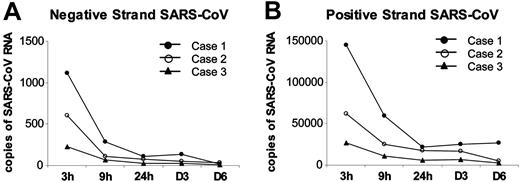
SARS-CoV is a positive single-stranded RNA virus, and the detection of negative-stranded RNA (the negative RNA template) may be an indication of viral replication.28 Using specific forward and reverse primers for SARS-CoV, both negative and positive strands of SARS-CoV were detected in infected DCs (Figure 3) but not in the mock-infected cells (data not shown; n = 3). The pattern of expression for the negative and positive strands was similar. There was a rapid decline in viral RNA from 3 hours to 9 hours after infection, but, because of sample variation, the difference did not yield statistical significance. No change in virus RNA expression was detected in later time points of 24 hours, day 3, and day 6.
Immunofluorescence assay for SARS-CoV detection in human DCs. Mock-infected human DCs were included as a control (A). Positive immunofluorescence staining was detected in human immature and mature DCs at 12 hours (B) and 24 hours (C) after infection with SARS-CoV (MOI = 1). Confocal microscopy showed positive staining in the cytoplasm of DCs (D). Images are representative of immature and mature DCs from 11 independent adult or CB donors.
Immunofluorescence assay for SARS-CoV detection in human DCs. Mock-infected human DCs were included as a control (A). Positive immunofluorescence staining was detected in human immature and mature DCs at 12 hours (B) and 24 hours (C) after infection with SARS-CoV (MOI = 1). Confocal microscopy showed positive staining in the cytoplasm of DCs (D). Images are representative of immature and mature DCs from 11 independent adult or CB donors.
Viral gene expression in SARS-CoV–infected human adult immature DCs by quantitative RT-PCR. Both negative (A) and positive strands (B) of SARS-CoV Replicase 1b mRNA were detected in SASR-CoV–infected adult immature DCs at 3 hours, 9 hours, 24 hours, day 3, and day 6 after infection (MOI = 1). Similar pattern of decreased viral gene expression was detected for the negative and positive strands in all 3 cases.
Viral gene expression in SARS-CoV–infected human adult immature DCs by quantitative RT-PCR. Both negative (A) and positive strands (B) of SARS-CoV Replicase 1b mRNA were detected in SASR-CoV–infected adult immature DCs at 3 hours, 9 hours, 24 hours, day 3, and day 6 after infection (MOI = 1). Similar pattern of decreased viral gene expression was detected for the negative and positive strands in all 3 cases.
Active caspase-3 assay for apoptosis in human adult immature DCs. Comparing with mock-infected adult immature DCs, no significant induction of active caspase-3–positive cells was observed at 6 hours, 12 hours, and 24 hours after infection (n = 4; P > .05). Data are shown as mean ± SEM of DCs from 4 independent donors. The percentage of active caspase-3–positive Jurkat cells in the positive control at 3 hours and 24 hours after addition of anti-Fas antibodies were 15% and 35%, respectively.
Active caspase-3 assay for apoptosis in human adult immature DCs. Comparing with mock-infected adult immature DCs, no significant induction of active caspase-3–positive cells was observed at 6 hours, 12 hours, and 24 hours after infection (n = 4; P > .05). Data are shown as mean ± SEM of DCs from 4 independent donors. The percentage of active caspase-3–positive Jurkat cells in the positive control at 3 hours and 24 hours after addition of anti-Fas antibodies were 15% and 35%, respectively.
We determined the presence of infectious SARS-CoV in adult immature DCs and their culture supernatant by half log titration cytopathic assay using FRhK-4 cells (n = 3). SARS-CoV–infected DCs harvested on days 1 to 6 after infection were washed and resuspended in 500 μL PBS. The cells were disrupted by freezing and thawing once at -70°C. The virus titer from the cell pellet decreased from 2 to less than 1 log TCID50/4 × 105 cells from day 1 to day 6. In cell culture supernatant, the virus titer on day 1 ranged from less than 1 to 2 log TCID50/100 μL. No increase in virus titer was detected in cell culture supernatant from day 1 to day 6.
SARS-CoV did not induce apoptosis or maturation in adult immature DCs
Similar percentages of active caspase-3–positive cells was observed in mock- and SARS-CoV–infected adult immature DCs at 6 hours, 12 hours, and 24 hours after infection (n = 4; P > .05; Figure 4). In the positive control, the percentage of active caspase-3–positive Jurkat cells were 15% and 35% at 3 hours and 24 hours after addition of anti-Fas antibodies, respectively.
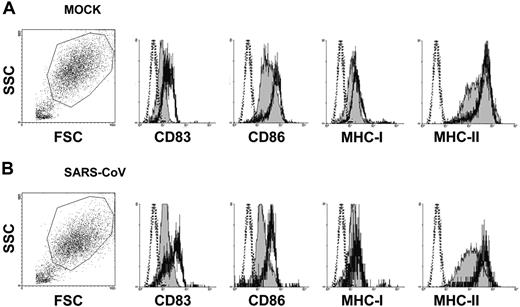
As shown by flow cytometric analysis (Figure 5), SARS-CoV alone did not up-regulate the expression of CD83, CD86, MHC class I, and MHC class II on adult immature DCs. However, SARS-CoV–infected cells can be stimulated by LPS (10 μg/mL) to up-regulate the expression of these molecules to similar levels as in the mock-infected controls.
SARS-CoV did not stimulate the gene expression of interferons or IL-12 in immature DCs
From our previous observation of induced proinflammatory cytokines in human macrophages by the avian influenza virus H5N1,27 we quantitated the mRNA expression of representative cytokines and chemokines in SARS-CoV–infected adult immature DCs. Low levels of interferon α (IFN-α), IFN-β, IFN-γ, and IL-12p40 expression (average in range of 0-30 copies per 104 β-actin) were observed in SARS-CoV–infected DCs (Figure 6). The IFN-β and IL-12p40 mRNA levels were marginally elevated from 3 hours to 9 hours after infection, but the differences did not reach statistical significance. The level of IFN-γ was significantly higher in SARS-CoV–infected CB DCs than adult DCs (Table 2).
CB immature DCs expressed significantly higher level of some cytokines and chemokine genes than adult immature DCs
. | 3 h after infection . | . | 9 h after infection . | . | ||
|---|---|---|---|---|---|---|
. | MOCK . | SARS-CoV . | MOCK . | SARS-CoV . | ||
| IFN-α | – | – | – | – | ||
| IFN-β | – | – | – | – | ||
| IFN-γ | – | .03 | – | – | ||
| IL-12p40 | .02 | – | – | – | ||
| TNF-α | – | – | – | – | ||
| IL-6 | .04 | – | – | .006 | ||
| MIP-1α | .03 | .003 | .01 | .003 | ||
| RANTES | – | – | – | – | ||
| IP-10 | .03 | – | – | .03 | ||
| MCP-1 | .03 | .005 | .01 | .02 | ||
. | 3 h after infection . | . | 9 h after infection . | . | ||
|---|---|---|---|---|---|---|
. | MOCK . | SARS-CoV . | MOCK . | SARS-CoV . | ||
| IFN-α | – | – | – | – | ||
| IFN-β | – | – | – | – | ||
| IFN-γ | – | .03 | – | – | ||
| IL-12p40 | .02 | – | – | – | ||
| TNF-α | – | – | – | – | ||
| IL-6 | .04 | – | – | .006 | ||
| MIP-1α | .03 | .003 | .01 | .003 | ||
| RANTES | – | – | – | – | ||
| IP-10 | .03 | – | – | .03 | ||
| MCP-1 | .03 | .005 | .01 | .02 | ||
P values are given showing CB > adult DCs. Adult DCs (n = 7), CB DCs (n = 5).
– indicates no statistically significant difference detected.
SARS-CoV stimulated moderate expression of proinflammatory genes in immature DCs
The production of proinflammatory cytokines tumor necrosis factor α (TNF-α) and IL-6 were in the range of 0 to 400 copies per 104 β-actin. Comparing the mock- and SARS-CoV–infected DCs, there was a moderate induction of TNF-α and IL-6 at both 3 hours and 9 hours after infection (Figure 7). The up-regulation of IL-6 in SARS-CoV–infected CB DCs was significantly higher than that in adult DCs (Table 2).
SARS-CoV up-regulated chemokine gene expression in immature DCs
The gene expression of inflammatory chemokines, MIP-1α, RANTES, IP-10, and MCP-1, were all significantly up-regulated in SARS-CoV–infected DCs (Figure 8). The induction of gene expression in DCs by SARS-CoV was strongest for IP-10 and MCP-1. The expressions of MIP-1α, IP-10, and MCP-1 genes in both mock- and SARS-CoV–infected CB DCs were significantly higher than that in adult DCs (Table 2). Similar finding was observed for RANTES, but because of sample variations, the difference between CB and adult DCs was not statistically significant.
Flow cytometry analysis of cell-surface molecule expression on human adult DCs. Mock- (A) or SARS-CoV–infected (B) adult immature DCs (MOI = 1) were harvested at 48 hours after infection and stained for flow cytometry analysis. Surface staining is shown by filled histogram, and isotype control is marked by the dotted line. SARS-CoV alone did not up-regulate the expression of CD83, CD86, MHC class I, and MHC class II. However, SARS-CoV–infected cells can be stimulated by LPS (10 μg/mL; thick line) to up-regulate the expression of these molecules to similar levels as in the mock-infected controls. Data shown are representative of adult immature DCs from 5 independent donors. FSC indicates forward scatter; SSC, side scatter.
Flow cytometry analysis of cell-surface molecule expression on human adult DCs. Mock- (A) or SARS-CoV–infected (B) adult immature DCs (MOI = 1) were harvested at 48 hours after infection and stained for flow cytometry analysis. Surface staining is shown by filled histogram, and isotype control is marked by the dotted line. SARS-CoV alone did not up-regulate the expression of CD83, CD86, MHC class I, and MHC class II. However, SARS-CoV–infected cells can be stimulated by LPS (10 μg/mL; thick line) to up-regulate the expression of these molecules to similar levels as in the mock-infected controls. Data shown are representative of adult immature DCs from 5 independent donors. FSC indicates forward scatter; SSC, side scatter.
Antiviral cytokine gene expression profile of SARS-CoV–infected human immature DCs by quantitative RT-PCR. Antiviral cytokine mRNA concentrations in adult (A) and CB (B) immature cells were assayed at 3 hours and 9 hours after infection with SARS-CoV (MOI = 1). Mock-infected cells were included as negative control. The concentrations were normalized to those of β-actin mRNA in the corresponding sample. There were low expressions of IFN-α, IFN-β, IFN-γ, and IL-12p40 genes in SARS-CoV–infected DCs. Data are shown as mean ± SEM (adult n = 7; CB n = 5).
Antiviral cytokine gene expression profile of SARS-CoV–infected human immature DCs by quantitative RT-PCR. Antiviral cytokine mRNA concentrations in adult (A) and CB (B) immature cells were assayed at 3 hours and 9 hours after infection with SARS-CoV (MOI = 1). Mock-infected cells were included as negative control. The concentrations were normalized to those of β-actin mRNA in the corresponding sample. There were low expressions of IFN-α, IFN-β, IFN-γ, and IL-12p40 genes in SARS-CoV–infected DCs. Data are shown as mean ± SEM (adult n = 7; CB n = 5).
Discussion
Severe acute respiratory syndrome is a recently described infectious disease that the human population has no prior immune experience.29 Therefore, the SARS outbreak in 2003 provides a unique opportunity for the study of human response to a novel virus. The clinical presentation of patients with SARS suggested that SARS-CoV might have specific mechanisms to escape from normal immune responses. In this study, we focused on DCs, which are the key antigen-presenting cells that play crucial roles in the antiviral immune response, including the priming of specific T-cell response.
The binding and entry of SARS-CoV were shown by electron microscopy (Figure 1) and further confirmed by immunofluorescence staining (Figure 2). Similar to other viruses, the uptake of SARS-CoV into DCs may be through macropinocytosis, receptor binding leading to endocytosis, or membrane fusion. Macropinocytosis is a nonspecific mechanism for virus internalization.30 In a previous study, we have demonstrated that mature DCs have lower endocytotic function than immature DCs.22 Our observation of similar infectivity of SARS-CoV in both immature and mature DCs from adult or CB suggested that SARS-CoV entry to DCs is not dependent on the efficiency of endocytosis.
Aminopeptidase N (CD13), which is the receptor responsible for the entry of another human CoV (229E) into intestinal, lung, and kidney epithelial cells,31 is present on DCs.32,33 Despite initial speculation that CD13 may be involved in SARS, there is no evidence to support its role. Angiotensin-converting enzyme 2 (ACE-2) has now been identified as a functional receptor for SARS-CoV,34 and the tissue distribution of ACE-2 has been studied extensively.35,36 In line with the report on the lack of ACE-2 protein in immune cells,35 we did not detect any gene expression of ACE-2 in purified monocytes or DCs (data not shown). Our results suggested that cell types lacking ACE-2 may also be infected by SARS-CoV, and other receptors may be involved in virus entry.
Proinflammatory cytokine gene expression profile of SARS-CoV–infected human immature DCs by quantitative RT-PCR. Proinflammatory cytokine mRNA concentrations in adult (A) and CB (B) immature cells were assayed at 3 hours and 9 hours after infection with SARS-CoV (MOI = 1). Mock-infected cells were included as negative control. The concentrations were normalized to those of β-actin mRNA in the corresponding sample. There were moderate up-regulation of TNF-α and IL-6 expression in SARS-CoV–infected DCs. Data are shown as mean ± SEM (adult n = 7; CB n = 5; *P < .05).
Proinflammatory cytokine gene expression profile of SARS-CoV–infected human immature DCs by quantitative RT-PCR. Proinflammatory cytokine mRNA concentrations in adult (A) and CB (B) immature cells were assayed at 3 hours and 9 hours after infection with SARS-CoV (MOI = 1). Mock-infected cells were included as negative control. The concentrations were normalized to those of β-actin mRNA in the corresponding sample. There were moderate up-regulation of TNF-α and IL-6 expression in SARS-CoV–infected DCs. Data are shown as mean ± SEM (adult n = 7; CB n = 5; *P < .05).
Chemokine gene expression profile of SARS-CoV–infected human immature DCs by quantitative RT-PCR. Chemokine mRNA concentrations in adult (A) and CB (B) immature cells were assayed at 3 hours and 9 hours after infection with SARS-CoV (MOI = 1). Mock-infected cells were included as negative control. The concentrations were normalized to those of β-actin mRNA in the corresponding sample. There was significant up-regulation of MIP-1α, RANTES, IP-10, and MCP-1 in SARS-CoV–infected DCs. Data are shown as mean ± SEM (adult n = 7; CB n = 5; *P < .05).
Chemokine gene expression profile of SARS-CoV–infected human immature DCs by quantitative RT-PCR. Chemokine mRNA concentrations in adult (A) and CB (B) immature cells were assayed at 3 hours and 9 hours after infection with SARS-CoV (MOI = 1). Mock-infected cells were included as negative control. The concentrations were normalized to those of β-actin mRNA in the corresponding sample. There was significant up-regulation of MIP-1α, RANTES, IP-10, and MCP-1 in SARS-CoV–infected DCs. Data are shown as mean ± SEM (adult n = 7; CB n = 5; *P < .05).
Initial postgenomic characterization of the SARS-CoV has revealed 23 potential N-linked glycosylation sites,37 and some of the sites of the surface spike (S) protein are of high-mannose structure.38 Hence, the uptake of SARS-CoV into DCs may also be mediated through binding to c-type lectin receptors, such as mannose receptor (CD206), DC–specific intracellular adhesion molecule-grabbing nonintegrin (SIGN; CD209), langerin (CD207), and DEC-205 (CD205).9 This hypothesis is supported by a recent report that retroviral vectors pseudotyped with the SARS-CoV S protein can bind to DC-SIGN and enter mature DCs.39 Cell-mediated transfer of the pseudovirus to Vero cells was also demonstrated, further supporting an important role played by DCs.
Viral infection usually results in a full replication cycle with production of progeny viruses. However, human CoV is known to be difficult to culture in vitro and only FRhK-43 and Vero E6 cells40 were reported to be permissive for SARS-CoV replication. In this study, the replication of SARS-CoV in DCs appeared to be incomplete; there was expression of the viral genome (including the negative and positive RNA templates; Figure 3), but there was no increase in viral copies over 6 days of culture, no virus budding, or production of infectious virus into the culture medium.
Human CoV (229E) has been shown to induce apoptosis in monocytes/macrophages,41 and some viruses, such as measles,42 also induce apoptosis of DCs. In this study, we did not observe significant cell death in SARS-CoV–infected DCs under light and electron microscopy or in active caspase-3 assays (Figure 4). This result suggested that the immunosuppressive effect of SARS-CoV may not be mediated through direct cytopathic effect on DCs.
Some viral infections, such as influenza,43 promote DC maturation that results in enhanced killing of the virus by the host. However, some viruses, such as measles, herpes simplex, dengue viruses, suppress DC maturation by inhibiting the expression of costimulatory and MHC molecules.12 For example, vaccinia virus inhibits the expression of CD83, CD86, and MHC class II molecules in immature DCs.44 The decrease in CD86 and MHC class II molecules may lead to antigenic tolerance and decreased antigen presentation, respectively. Although our results demonstrated that SARS-CoV did not up-regulate the expression of CD83, CD86, MHC class I, or MHC class II molecules on immature DCs, maturation of SARS-CoV–infected DCs can still be induced by LPS, suggesting SARS-CoV did not impair DC maturation (Figure 5).
Viruses enhance their own survival by interfering with normal innate immune responses of the host. Usually, type 1 interferons are effectively generated in response to viral infection and keep activated T cells alive.45 However, very low level of interferon gene expression was observed in SARS-CoV–infected DCs (Figure 6). In general, double-stranded RNA, when bound to toll-like receptor 3, triggers the production of interferons in DCs.46 The lack of activation of interferon production by SARS-CoV may involve mechanisms that interferes with downstream signaling molecules such as IFN regulatory factor-3 (IRF-3), TRIF (TIR [Toll/IL-1 receptor] domain–containing adapter inducing IFN), and putative kinases or proteases.47 Because DCs and interferons both play a role in the generation of antibody responses,48 the lack of interferon induction in DCs may contribute to the slow antibody production and progressive increase of viral load over the first 10 days of SARS observed clinically.4 Interestingly, SARS-CoV–infected CB DCs expressed slightly higher levels of interferon genes than adult DCs. Further investigation is needed to determine whether this observation is relevant to the less-severe disease presentation observed in pediatric SARS.
Interleukin 12 is the main cytokine secreted by DCs that regulates the differentiation of CD4+ T cells into Th1 cells and serves important function in cell-mediated immunity.49 Similar to the observation in measles virus–infected DCs,42 the lack of IL-12 production in SARS-CoV–infected DCs might suppress Th1 and favor Th2 responses. Furthermore, the immune escape mechanism operated in HIV14,15 and dengue virus50 via DC-SIGN leading to suppressed IL-12 production may also be implicated in SARS.
Both TNF-α and IL-6 are proinflammatory cytokines that regulate apoptosis, cell proliferation, differentiation, immunity, and inflammation.51 We have previously reported that the severity of avian flu may be due to high TNF-α production in macrophages infected by H5N1 viruses.27 Likewise, patients with SARS were treated with corticosteroids in the belief that local production of proinflammatory cytokines is responsible for the immunopathology in the lungs.52 However, we only detected a slight up-regulation of TNF-α and IL-6 mRNA expressions in SARS-CoV–infected DCs (Figure 7).
Chemokines are chemotactic messengers that play important roles in leukocyte recruitment.53 Up-regulation of chemokines has also led to chemokine-mediated host pathology in viral diseases.54 In concordance with the detection of high-plasma concentration of chemokines in patients with SARS,55 Glass et al56 reported that there is massive up-regulation of chemokines in the lungs of SARS-CoV–infected mice. In this study, we also observed significant up-regulation of representative inflammatory chemokines in SARS-CoV–infected DCs (Figure 8). MIP-1α, RANTES, IP-10, and MCP-1 are CXC chemokines without the ELR (glutamic acid-leucine-arginine) motif. They preferentially act on mononuclear cells, and their up-regulation has been detected in influenza A virus infection.57 These chemokines may be responsible for the recruitment and adhesion of inflammatory cells and the migration of leukocytes into the tissue.57 Our findings will need to be substantiated by in vivo experiments.
More important, chemokines are also implicated in the autocrine regulation of DC migration to draining lymph nodes.19,20,58 We hypothesize that migrating virus-infected DCs may facilitate the virus spread, skew T-cell responses through altered cytokine production, or induce apoptosis in T cells leading to immunosuppression.59 Interestingly, we detected significantly higher level of chemokine genes (20- to 100-fold) in CB DCs than in adults DCs, the relevance of which requires further study.
Overall, we concluded that the lack of antiviral cytokine response against a background of intense chemokine up-regulation could represent a mechanism of immune evasion by SARS-CoV.
Prepublished online as Blood First Edition Paper, April 28, 2005; DOI 10.1182/blood-2004-10-4166.
Supported partially by the Special SARS (severe acute respiratory syndrome) Research Fund and the Outstanding Researcher Award (Y.L.L.) from the University of Hong Kong; a Public Health Research Grant (AI95357) from the National Institute of Allergy and Infectious Diseases; and the Research Fund for the Control of Infectious Diseases (RFCID) from the Hong Kong Special Administrative Region (HKSAR) government.
H.K.W.L. and C.Y.C. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the staff of the Department of Microbiology and Electron Microscope Unit, The University of Hong Kong, for their technical support; and the staff of labor ward, Queen Mary Hospital, in facilitating the collection of cord blood.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal