Abstract
T-cell large granular lymphocyte (T-LGL) leukemia is a clonal lymphoproliferation of cytotoxic T cells (CTLs) associated with cytopenias. T-LGL proliferation seems to be triggered/sustained by antigenic drive; it is likely that hematopoietic progenitors are the targets in this process. The antigen-specific portion of the T-cell receptor (TCR), the variable beta (VB)–chain complementarity-determining region 3 (CDR3), can serve as a molecular signature (clonotype) of a T-cell clone. We hypothesized that clonal CTL proliferation develops not randomly but in the context of an autoimmune response. We identified the clonotypic sequence of T-LGL clones in 60 patients, including 56 with known T-LGL and 4 with unspecified neutropenia. Our method also allowed for the measurement of clonal frequencies; a decrease in or loss of the pathogenic clonotype and restoration of the TCR repertoire was found after hematologic remission. We identified 2 patients with identical immunodominant CDR3 sequence. Moreover, we found similarity between multiple immunodominant clonotypes and codominant as well as a nonexpanded, “supporting” clonotypes. The data suggest a nonrandom clonal selection in T-LGL, possibly driven by a common antigen. In contrast, the physiologic clonal CTL repertoire is highly diverse and we were not able to detect any significant clonal sharing in 26 healthy controls.
Introduction
Physiologic and pathologic cellular immune responses to antigenic proteins can be polyclonal but can also be highly polarized for strong antigenic stimuli.1-7 Due to the diversity of peptide processing and HLA backgrounds8-10 responsible for antigen presentation, the patterns of clonal utilization are of complex nature.11-13 Proliferation of an individual T-cell clone may be difficult to discriminate in the context of the simultaneous polyclonal expansions but in some disorders, such as viral infections, highly polarized responses were described.14-16 Consequently, under certain circumstances an immunodominant clonal expansion may be so significant that it is detectable above the polyclonal background. It can result in the overrepresentation of the corresponding T-cell receptor (TCR) and consequently the entire α (A)– or β (B)–chain families.14,16-18 Complementarity-determining region 3 (CDR3) of the TCR VB chain is highly unique and can be used as a marker of individual T-cell clones and as a measure of the T-cell repertoire diversity.11,12,19 TCR α chain is less suitable for detection of αβ–T-cell clonality due to the paradigm of dual receptor T cell: up to 30% of rearranged human peripheral blood T cells express 2 distinct functional α chains as opposed to only one β chain.20 The structure of the VB CDR3 region results from somatic VDJ gene recombination and junctional diversification.11,12,19 Due to the NDN modification of β chain, VB CDR3 region is the most diverse region of the TCR.21 While TCR CDR3 region appears to most directly ligate to the antigenic peptide presented in the context of HLA, CDR1 and CDR2 loops have a major function in stabilizing the ligated CDR3 regions.22 In addition, VB CDR3 sequence covers over half of the peptide surface23 and appears to be highly important in determining ligand specificity.24 Finally, sequence substitution of VB CDR3 region can abolish the response, as confirmed by site-directed mutagenesis studies.25-28
Analysis of antigen-specific cells sorted using, eg, tetramers, demonstrates that a specific antigenic epitope can be recognized by a variety of unique TCR clonotypes.7,29,30 However, in the presence of identical HLA restriction element, TCRs with identical VB CDR3 regions strongly suggest recognition of the same antigenic peptides, assuming that VB clonality is associated with the usage of the same α chain.31-34
Highly significant clonal expansions occur in large granular lymphocyte (LGL) leukemia, a disease resulting from the proliferation of individual cytotoxic T-cell (CTL) clones.1,10,35,36 Traditionally, this condition is diagnosed through detection of an abnormal CD3+CD8+CD57+ cell population and the presence of a clonal TCR gene rearrangement.36-38 Moreover, the presence of LGL clones can often be uncovered using Vβ antibodies.39,40 Molecular identification of the clonal CTL expansions can establish the exact diagnosis of LGL leukemia but also possibly other diseases with underlying T-cell lymphoproliferations.10,19,41,42 The mechanism of transformation of the LGL clone is not well understood and the disease often does not behave as a typical malignancy; exuberant accumulation of malignant cells is often absent and the acquisition of a more malignant phenotype is rare. Instead, clonal CTLs in LGL leukemia while apparently dysregulated, appear to retain some physiologic properties of normal CTLs and resemble an exaggerated response to an immunodominant antigen. One of the theories postulates that the initial events in the genesis of LGL leukemia may involve an underlying immune response to exogenous and cross-reactive or intrinsic antigens. It is possible that, prior to the outgrowth of the immunodominant LGL clone, such a response may be polyclonal in nature. Clinical evidence supports this notion. LGL leukemia often occurs in the context of a specific autoimmune pathology.1,10 LGL-leukemia–associated paraneoplastic syndrome can involve isolated cytopenias.35,43-47 Some authors hypothesize that the mechanisms of cytopenia include secretion of Fas-L/soluble Fas receptors that are constitutively expressed by LGL cells and also found elevated in patient's serum.48,49 However, lineage-specific cytopenias cannot be easily explained by displacement of hematopoiesis or a global cytokine effect. Consequently, it is possible that the TCR specificity of the LGL clone determines the clinical presentation spectrum by highly specific recognition and killing of individual hematopoietic cell lineages. Thus, the transformation event may not be random and may occur in the context of a preexisting immune response (see for example Starkebaum et al47 ). The nature of this event remains unknown but may include blockade of apoptotic pathways or persisting upregulation of cell-cycle activating or antiapoptotic mechanisms. For example, constitutive activation of signal transducers and activators of transcription 3 (STAT3) has been described in LGL cells.50 It was also shown that patient's LGL cells, although resistant to Fas-mediated apoptosis, express high levels of Fas, similar to phytohemagglutinin (PHA)–activated normal T cells, suggesting an in vivo antigen activation.51 Of note is that the transforming events may involve the memory cell, which feeds into the mature effector CTL compartment.52
The extreme expansion of LGL clones can be easily characterized and there is no interference from other codominant clones contributing to a polyclonal T-cell response. Therefore, LGL leukemia can serve as a simplified model of natural and less exaggerated polyclonal CTL responses. Such processes may include, for example, typical aplastic anemia53 and classic autoimmune diseases such as rheumatoid arthritis and multiple sclerosis.54-57 Of note is that in all of these conditions, evolution of LGL leukemia has been described,45,47,58-60 and the LGL clone may have evolved from an initially polyclonal process. The TCR in LGL leukemia had been investigated at DNA sequence level in only a limited number of cases,54,59,61-63 and perhaps not surprisingly (due to diversity of the HLA background of patients) identical specificities were not found.
Here, we systematically studied the TCR repertoire in a large cohort of patients with LGL leukemia. For that purpose we used the rearranged VB CDR3 region as a clonal marker and focused our work on TCR β rather than α chain. We hypothesize that the evolution of the LGL clone may not be a random event but represents an overreactive response of an individual CTL clone in the context of an otherwise polyclonal reaction. Consequently, shared TCR specificities may be found not only between the expanded clonotypes within the TCR repertoire of an individual patient but also between immunodominant clones and those contained within polyclonal background. Moreover, if LGL leukemia reactivity is directed against an antigen also present in healthy individuals, identical clonotypes will be found in these individuals albeit at a very low level. We have studied disease-associated immunodominant clonotypes in a large cohort of LGL leukemia patients and compared their CDR3 sequences against a clonotypic database containing CDR3 regions from healthy individuals sequenced in our laboratory and CDR3 sequences described in the literature. Our results demonstrate that shared clonotypic sequences can be identified and may be used to track the presence of the corresponding CTL clones for diagnostic purposes.
Patients, materials, and methods
Patients and controls
We obtained peripheral blood specimens from 60 patients with known LGL leukemia, 13 patients with single-lineage cytopenias, and 26 healthy controls. Informed consent for sample collection was given by the individuals according to protocols approved by the Institutional Review Board of the Cleveland Clinic Foundation (Cleveland, OH), the National Heart, Lung, and Blood Institute, National Institutes of Health (Bethesda, MD), and the Penn State Cancer Institute (Hershey, PA). Previously, flow cytometry and CDR3 results were reported in part for 29 of these patients.63 The original diagnosis of LGL leukemia was established by clinical and laboratory parameters as suggested by Berliner et al,38 Semenzato et al,64 and Herling et al65 but we also included patients with otherwise unexplained cytopenias and those with an expansion in a specific CTL population who did not fulfill the current criteria for LGL leukemia.66 A diagnosis of myelodysplastic syndrome (MDS), when applicable, was established by bone marrow biopsy and peripheral blood counts and classified according to French American British (FAB) classification.67
Detection strategy
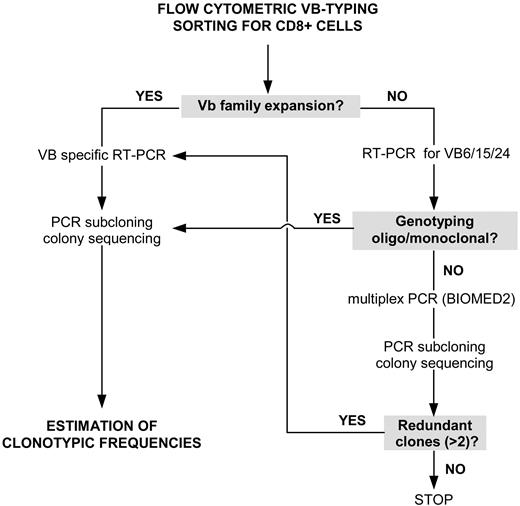
Our strategy for the detection of expanded (immunodominant) LGL clones initially included flow cytometric VB typing (Figure 1). When a significantly expanded VB family within the CD8+ CTL population was found, the respective CDR3 amplification product was subcloned and transformed into Escherichia coli. A pathologic VB expansion was defined as greater than mean + 2 standard deviations (SD) of the distribution found in healthy controls, but, in most of the cases, the expansion of VB families were greater than 20% of all CD8+ cells. On average, 25 colonies were sequenced in order to determine the nucleotide sequence of the immunodominant clones. The frequency at which a particular clone occurred was calculated as the % of identical sequences divided by the total number of clones analyzed.
Rational approach for detecting immunodominant clonotypes in patients with T-LGL leukemia. For explanations, see “Detection strategy.”
Rational approach for detecting immunodominant clonotypes in patients with T-LGL leukemia. For explanations, see “Detection strategy.”
In several patients the panel of VB antibodies used failed to identify a specific VB family expansion. In such cases, the VB families not covered by the antibody panel (VB6, 15, and 24) were subjected to reverse transcriptase–polymerase chain reaction (RT-PCR) and subcloned/sequenced if oligoclonal or monoclonal genotyping results were obtained. Samples negative by both flow cytometric VB typing and by RT-PCR for VB families 6, 15, and 24 were subjected to multiplex VB PCR using a BIOMED2 primer set,68 which amplifies all CDR3 regions. Samples with a monoclonal/oligoclonal pattern were subcloned and sequenced (Figure 1). Redundant clonotypes (> 2) detected through multiplex PCR were used to identify the VB family harboring the immunodominant clone; cloning and sequencing of specific VB CDR3 was used to determine the clonotypic frequency within a given VB class.
Flow cytometric VB typing
Fresh peripheral blood was stained for VB flow cytometry analysis to quantitate the contribution of each VB family to the CD4+ and CD8+ lymphocyte populations. The manufacturer's instructions (IOTest Beta Mark kit; Beckman-Coulter, Fullerton, CA) were modified as follows: 5 μL of phycoerythrin cyanin 5 (PC5)-conjugated CD4, 5 μL energy-coupled dye (ECD)–conjugated CD8 monoclonal antibodies (mAbs), and 20 μL of anti-VB antibody (VB1-5, VB7-9, VB11-14, VB16-18, VB20-23) were added. A 4-color protocol was used. The lymphocyte gate was set according to the size and forward scatter properties. VB family usage was determined within the CD4 and CD8 lymphocyte fractions using the appropriate gates. Initially, 9 normal samples were characterized to define the average size and standard deviations for the VB repertoire as detected by the antibodies. A significant expansion was defined as one that was mean plus 2 SD over the average VB family size.39 Because the antibody set contained in the IOBeta Mark kit does not cover the whole VB spectrum, VB PCR was performed for VB families 6, 15, and 24 on samples that were negative for expansion by flow cytometry. Samples negative for VB families 6, 15, and 24 and for VB families covered by the flow cytometry panel were subjected to multiplex PCR using BIOMED2 (see “RNA isolation and RT-PCR”).
Lymphocyte separation
Mononuclear cells were separated from peripheral blood by density gradient sedimentation (Mediatech, Herndon, VA). CD4+ and CD8+ T cells were isolated by flow cytometric sorting. Samples were stained with a CD8 mAb conjugated with fluorescein isothiocyanate (FITC; Pharmingen, San Diego, CA) and sorted on an Epics Altra high-speed flow cytometer (Beckman Coulter, Miami, FL).
RNA isolation and RT-PCR
Total RNA was extracted from CD8+ T cells with TRIZOL reagent (Invitrogen, Carlsbad, CA) and dissolved in a final volume of 20 μL of diethyl pyrocarbonate (DEPC) water. cDNA was generated from 6 μL of RNA by first-strand cDNA synthesis using either SuperScript II RT Kit or SuperScript III RT Kit (Invitrogen). cDNA was amplified using a nonlabeled or FAM-labeled constant region antisense primer (CB) and 22 different VB family–specific sense primers (including VB6, 15, and 24; pseudogenes VB10 and VB19 were not covered).11,12 PCR master mix was set up as follows: 2.5 μL of 10 × PCR buffer containing 15 mM magnesium acetate (Eppendorf, Hamburg, Germany), 2 μL of 2.5 mM deoxynucleoside triphosphates (dNTPs), 2 μL of 3 μM antisense unlabeled or FAM-labeled CB primer, 2 μL of 3 μM sense VB primer, 0.25 μL of Taq (5 U/μL; Eppendorf), and 1.5 to 2 μL of cDNA were mixed with dH2Otoa final volume of 25 μL. Thermocycling was performed either on an MJ thermocycler (MJ Research, Waltham, MA) or on a Mastercycler (Eppendorf). After an initial denaturation step at 95°C for 5 minutes and 10 touch-down cycles (denaturation at 95°C for 30 seconds; annealing at 63°C, –1°C per step, for 40 seconds and extension at 72°C for 50 seconds), the final 20 PCR cycles were performed (30 seconds at 95°C, 40 seconds at 54°C, 50 seconds at 72°C).
Alternatively we applied a 2-tube multiplex PCR that covers all VB TCR gene rearrangements (developed by the BIOMED-2 Concerted Action Group) as previously described.68
CDR3 region genotyping
One microliter of amplification products generated either with the BIOMED-2 VB multiplex primer set or the 22 VB/FAM-CB primers was mixed with 0.5 μL Genescan 400HD ROX size standard (Applied Biosystems, Foster City, CA) and 12 μL Hi-Di formamide (Applied Biosystems). The samples were run on an ABI Prism 310 Genetic Analyzer (Applied Biosystems) and analyzed using GeneScan Analysis Software v3.7 (Applied Biosystems). A set of numeric standards was adopted to differentiate normal from abnormal profiles. Abnormal (skewed) profiles were defined as those that deviated from a Gaussian distribution of intensity peaks (< 5 peaks, or contained one or several dominant peaks). Accuracy of an abnormal classification was verified by sequencing, as described by us previously.63
CDR3 region cloning and sequencing
PCR products were gel purified using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA) or Eppendorf Gel Extraction Kit (Eppendorf) following the manufacturer's instructions. Four microliters of the purified PCR product was ligated into the TA cloning vector pCR2.1 (Invitrogen) overnight at 14°C and heat-shock transformed into TOP10F E coli. Colony PCR was performed on an MJ thermocycler as follows: on average 25 bacterial colonies were picked and the inserts were amplified using M13F and M13R primers provided in the TA cloning kit (2 μL of 3-μM solution each) and 3.0 μL of 10 × PCR buffer (Invitrogen), 1 μL of 50 mM MgCl2, 2 μL dNTP (2.5 mM), 0.1 μL Taq polymerase (Invitrogen), in a total volume of 30 μL. After 5 minutes of denaturation at 94°C, 19 PCR cycles (30 seconds at 94°C, 30 seconds at 55°C, and 40 seconds at 72°C) were performed. Positive PCR products containing vector and insert were confirmed on a 1.2% agarose gel and purified using the Montage PCR96 Cleanup Kit (Millipore, Billerica, MA). Purified PCR products (5 μL) were sequenced using 1 μL of Big Dye Terminator v3.0 or v3.1 (Applied Biosystems), 1 μL of 3 μM M13F primer, and 3 μL dH2O. Cycle sequencing was performed on an MJ thermocycler (MJ Research) as follows: an initial denaturation at 96°C for 40 seconds was followed by 31 cycles (10 seconds at 96°C, 15 seconds at 55°C, 4 minutes at 60°C). Sequencing reactions were purified using the Montage SEQ96 Sequencing Reaction Cleanup Kit (Millipore) run on a 3100-Avant Genetic Analyzer (Applied Biosystems). A pathologically expanded, immunodominant clonotype was defined based on its frequency among cloned sequences within an expanded VB family. In this study, clonal diversity was defined as % of unique clonotypes out of a total number of clonotypes. We have defined the clonal redundancy as the frequency of the identical clonotypes within all clones sequenced (at least 12 clonotypes were sequenced per VB and patient). Based on the average size of the expansions in healthy individuals, pathologically expanded “major” clonotypes were defined as those occurring at the frequency greater than 12.5% of all sequenced clones (see “Identification strategy for immunodominant clones in LGL leukemia”). Minor, nonexpanded clonotypes were defined as CDR3 sequences occurring at the frequency of 1/N or greater but lesser than the size of the immunodominant (major) clonotypes.
Sequence analysis
Annotation nomenclature for VB, joining beta (JB), and CDR3 regions was adapted from the T cell receptor Factsbook69 and CDR3 region sequences were analyzed using the ImMunoGeneTics information system TCR alignment tool.70 For the purpose of this study we compared CDR3 regions rather than NDN regions. A CDR3 region includes invariant portions of VB and JB chains according to the nomenclature proposed on the ImMunoGeneTics website.71 The clonotypic database used for sequence comparisons contains 4893 sequences, 2273 generated in our laboratory and 2620 from other sources.
A 7– and 8–amino acid (AA) substitution code was applied for comparison of CDR3 sequences. Each amino acid residue was substituted with a category label corresponding to physicochemical properties of the individual amino acids. In the first system, the groups were as follows: GAVLI, aliphatic amino acids (B); FYW, aromatic amino acids (J); CM, sulfhydryl-group–containing amino acids (O); S, T, aliphatic, hydroxyl-group–containing amino acids (U); KRH, basic amino acids (X); DENQ, acidic amino acids (Z); P, proline (P).72 The second substitution code had the following amino acid groupings: WFY, aromatic (A); LIVPA, aliphatic (B); MC, hydrophobic (C); G, +/– hydrophilic (D); STNQ, hydrophilic (E); H, +/– charged (F); DE, acidic (G); KR, basic (H).
Results
Identification strategy for immunodominant clones in LGL leukemia
We studied 60 patients with LGL leukemia and 13 patients with neutropenia for whom drug-induced hematopoietic toxicity and other hematologic diseases were excluded. In 56 of the LGL patients we characterized a pathologically expanded CTL clone that was dominant within the respective TCR VB family and the entire CD8+ cell population. In addition, we were able to detect the presence of previously unknown immunodominant CTL clones in 4 patients with neutropenia (Tables 1, 2).
Patient characteristics, clinical presentation, and laboratory data
Patient no. . | Age, y . | Main clinical presentation . | Diagnosis . | Additional diagnosis . | TCR gamma rearrangement by PCR . | HLA-A . | HLA-B . | HLA-C . | DR . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 25 | Pan | LGL | — | Y | 26, 32 | 27, 44 | ND | 1, 4 |
| 2 | 72 | Pan | LGL | RA/PNH | Y | 2, 3 | 13, 35 | 4, 8 | 11, 13 |
| 3 | 58 | RCA/Neu | LGL | — | Y | 2, 2 | 58, 55 | ND | 1, 4 |
| 4 | 76 | RCA | LGL | — | Y | 2, 68 | 27, 51 | ND | 1, 13 |
| 5 | 62 | Neu | LGL | RA | Y | 1, 2 | 14, 51 | 2, 8 | 1, 13 |
| 6 | 66 | Neu | LGL | — | Y | NA | NA | NA | NA |
| 7 | 61 | Neu | LGL | — | Y | 3, 24 | 37, 40 | 3, 6 | 4, 10 |
| 8 | 36 | Neu | LGL | — | Y | 2, 11 | 18, 44 | 7, 16 | 11, 13 |
| 9 | 58 | Neu | LGL | MS | Y | 1, 32 | 8, 44 | 5, 7 | 4, 15 |
| 10 | 65 | Pan | LGL | — | Y | 1, 68 | 14, 37 | 6, 8 | 15, 13 |
| 11 | 42 | RCA | LGL | Hepatitis | Y | 1, 2 | 7, 37 | 6, 7 | N.D. |
| 12 | 61 | Neu | LGL | RhAr | Y | 1, 2 | 8, 15 | 3, 7 | 3, 4 |
| 13 | 63 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 14 | 77 | RCA | LGL | — | Y | ND | ND | ND | ND |
| 15 | 37 | Neu | LGL | SLE | N | ND | ND | ND | ND |
| 16 | 50 | RCA | LGL | — | Y | ND | ND | ND | ND |
| 17 | 64 | Pan | LGL | RARS | Y | 29, 08 | 40, 45 | ND | 11, 15 |
| 18 | 63 | RCA | LGL | UC | Y | 2, 2 | 8, 44 | 7, 7 | 3, 11 |
| 19 | 65 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 20 | 78 | RCA | LGL | RhAr | Y | ND | ND | ND | ND |
| 21 | 51 | Neu | LGL | — | Y | 2, 24 | 7, 73 | 7, 15 | 1, 4 |
| 22 | 76 | Pan | LGL | RA | Y | 24, 24 | 7, 35 | 4, 7 | 1, NA |
| 23 | 80 | RCA | LGL | RA | Y | 1, 29 | 13, 44 | 6, 16 | 4, 7 |
| 24 | 32 | Pan | LGL | Hepatitis | Y | 2, 24 | 40, 51 | 3, 3 | 4, 12 |
| 25 | 63 | RCA | LGL | RA | Y | 11, 33 | 51, 65 | 7, 8 | 1, 13 |
| 26 | 73 | Neu | LGL | RhAr | Y | 2, 29 | 44, 44 | 12, 16 | 4, 4 |
| 27 | 62 | Neu | LGL | Hepatitis | Y | 2, 2 | 7, 51 | 1, 7 | 11, 4 |
| 28 | 46 | Neu | LGL | RhAr | Y | 3, 24 | 37, 40 | 3, 6 | 4, 10 |
| 29 | 62 | Neu | LGL | — | Y | 23, 32 | 18, 35 | 4, 12 | 1, 13 |
| 30 | 78 | Neu | LGL | — | NA | ND | ND | ND | ND |
| 31 | 71 | RCA | LGL | — | Y | ND | ND | ND | ND |
| 32 | 57 | — | LGL | — | Y | ND | ND | ND | ND |
| 33 | 72 | RCA | LGL | — | Y | 1, 31 | 8, 39 | 7, 7 | 1, 3 |
| 34 | 71 | Neu | LGL | — | Y | 1, 2 | 8, 44 | 7, 7 | 16, 3 |
| 35 | 66 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 36 | 80 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 37 | 62 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 38 | NA | Neu | LGL | — | Y | ND | ND | ND | ND |
| 39 | NA | Neu | LGL | — | Y | ND | ND | ND | ND |
| 40 | NA | Neu | LGL | — | Y | ND | ND | ND | ND |
| 41 | NA | Neu | LGL | — | Y | ND | ND | ND | ND |
| 42 | 64 | RCA | LGL | AIHA | Y | ND | ND | ND | ND |
| 43 | NA | Neu | LGL | — | Y | ND | ND | ND | ND |
| 44 | 77 | Neu | LGL | RA | Y | ND | ND | ND | ND |
| 45 | 76 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 46 | NA | Neu | LGL | — | Y | ND | ND | ND | ND |
| 47 | 59 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 48 | NA | Neu | LGL | — | N | 3, 31 | 18, 51 | 12, 14 | 10, 15 |
| 49 | 70 | Neu | LGL | — | NA | ND | ND | ND | ND |
| 50 | 65 | Neu | AIN | — | N | 2, 11 | 7, 15 | 3, 7 | 15, 4 |
| 51 | 72 | Pan | LGL | RA | N | 1, 68 | 35, 44 | 4, 4 | 7, 8 |
| 52 | 51 | Neu | LGL | — | N | 11, 26 | 45, 51 | 3, 6 | 4, 9 |
| 53 | 29 | Neu | AIN | Hepatitis | N | ND | ND | ND | ND |
| 54 | 43 | Neu | AIN | Hepatitis | N | 3, 3 | 7, 35 | 4, 7 | 1, 15 |
| 55 | 64 | Neu | LGL | RhAr | Y | 24, 02 | 44, 51 | 2, 4 | 11, 13 |
| 56 | 39 | Pan | AIN | — | Y | 2, NA | 15, 44 | 3, 5 | 4, 13 |
| 57 | 56 | Neu | LGL | RhAr | Y | 1, 25 | 8, 18 | 7, 12 | 4, 11 |
| 58 | 60 | Pan | LGL | PNH | NA | 68, 68 | 14, 14 | 8, 8 | 1, 1 |
| 59 | 53 | Neu | LGL | AIH | Y | 1, 3 | 7, 37 | 6, 7 | 13, 15 |
| 60 | 51 | Pan | LGL | — | N | 11, 24 | 27, 22 | 1, 1 | 4, 15 |
Patient no. . | Age, y . | Main clinical presentation . | Diagnosis . | Additional diagnosis . | TCR gamma rearrangement by PCR . | HLA-A . | HLA-B . | HLA-C . | DR . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 25 | Pan | LGL | — | Y | 26, 32 | 27, 44 | ND | 1, 4 |
| 2 | 72 | Pan | LGL | RA/PNH | Y | 2, 3 | 13, 35 | 4, 8 | 11, 13 |
| 3 | 58 | RCA/Neu | LGL | — | Y | 2, 2 | 58, 55 | ND | 1, 4 |
| 4 | 76 | RCA | LGL | — | Y | 2, 68 | 27, 51 | ND | 1, 13 |
| 5 | 62 | Neu | LGL | RA | Y | 1, 2 | 14, 51 | 2, 8 | 1, 13 |
| 6 | 66 | Neu | LGL | — | Y | NA | NA | NA | NA |
| 7 | 61 | Neu | LGL | — | Y | 3, 24 | 37, 40 | 3, 6 | 4, 10 |
| 8 | 36 | Neu | LGL | — | Y | 2, 11 | 18, 44 | 7, 16 | 11, 13 |
| 9 | 58 | Neu | LGL | MS | Y | 1, 32 | 8, 44 | 5, 7 | 4, 15 |
| 10 | 65 | Pan | LGL | — | Y | 1, 68 | 14, 37 | 6, 8 | 15, 13 |
| 11 | 42 | RCA | LGL | Hepatitis | Y | 1, 2 | 7, 37 | 6, 7 | N.D. |
| 12 | 61 | Neu | LGL | RhAr | Y | 1, 2 | 8, 15 | 3, 7 | 3, 4 |
| 13 | 63 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 14 | 77 | RCA | LGL | — | Y | ND | ND | ND | ND |
| 15 | 37 | Neu | LGL | SLE | N | ND | ND | ND | ND |
| 16 | 50 | RCA | LGL | — | Y | ND | ND | ND | ND |
| 17 | 64 | Pan | LGL | RARS | Y | 29, 08 | 40, 45 | ND | 11, 15 |
| 18 | 63 | RCA | LGL | UC | Y | 2, 2 | 8, 44 | 7, 7 | 3, 11 |
| 19 | 65 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 20 | 78 | RCA | LGL | RhAr | Y | ND | ND | ND | ND |
| 21 | 51 | Neu | LGL | — | Y | 2, 24 | 7, 73 | 7, 15 | 1, 4 |
| 22 | 76 | Pan | LGL | RA | Y | 24, 24 | 7, 35 | 4, 7 | 1, NA |
| 23 | 80 | RCA | LGL | RA | Y | 1, 29 | 13, 44 | 6, 16 | 4, 7 |
| 24 | 32 | Pan | LGL | Hepatitis | Y | 2, 24 | 40, 51 | 3, 3 | 4, 12 |
| 25 | 63 | RCA | LGL | RA | Y | 11, 33 | 51, 65 | 7, 8 | 1, 13 |
| 26 | 73 | Neu | LGL | RhAr | Y | 2, 29 | 44, 44 | 12, 16 | 4, 4 |
| 27 | 62 | Neu | LGL | Hepatitis | Y | 2, 2 | 7, 51 | 1, 7 | 11, 4 |
| 28 | 46 | Neu | LGL | RhAr | Y | 3, 24 | 37, 40 | 3, 6 | 4, 10 |
| 29 | 62 | Neu | LGL | — | Y | 23, 32 | 18, 35 | 4, 12 | 1, 13 |
| 30 | 78 | Neu | LGL | — | NA | ND | ND | ND | ND |
| 31 | 71 | RCA | LGL | — | Y | ND | ND | ND | ND |
| 32 | 57 | — | LGL | — | Y | ND | ND | ND | ND |
| 33 | 72 | RCA | LGL | — | Y | 1, 31 | 8, 39 | 7, 7 | 1, 3 |
| 34 | 71 | Neu | LGL | — | Y | 1, 2 | 8, 44 | 7, 7 | 16, 3 |
| 35 | 66 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 36 | 80 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 37 | 62 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 38 | NA | Neu | LGL | — | Y | ND | ND | ND | ND |
| 39 | NA | Neu | LGL | — | Y | ND | ND | ND | ND |
| 40 | NA | Neu | LGL | — | Y | ND | ND | ND | ND |
| 41 | NA | Neu | LGL | — | Y | ND | ND | ND | ND |
| 42 | 64 | RCA | LGL | AIHA | Y | ND | ND | ND | ND |
| 43 | NA | Neu | LGL | — | Y | ND | ND | ND | ND |
| 44 | 77 | Neu | LGL | RA | Y | ND | ND | ND | ND |
| 45 | 76 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 46 | NA | Neu | LGL | — | Y | ND | ND | ND | ND |
| 47 | 59 | Neu | LGL | — | Y | ND | ND | ND | ND |
| 48 | NA | Neu | LGL | — | N | 3, 31 | 18, 51 | 12, 14 | 10, 15 |
| 49 | 70 | Neu | LGL | — | NA | ND | ND | ND | ND |
| 50 | 65 | Neu | AIN | — | N | 2, 11 | 7, 15 | 3, 7 | 15, 4 |
| 51 | 72 | Pan | LGL | RA | N | 1, 68 | 35, 44 | 4, 4 | 7, 8 |
| 52 | 51 | Neu | LGL | — | N | 11, 26 | 45, 51 | 3, 6 | 4, 9 |
| 53 | 29 | Neu | AIN | Hepatitis | N | ND | ND | ND | ND |
| 54 | 43 | Neu | AIN | Hepatitis | N | 3, 3 | 7, 35 | 4, 7 | 1, 15 |
| 55 | 64 | Neu | LGL | RhAr | Y | 24, 02 | 44, 51 | 2, 4 | 11, 13 |
| 56 | 39 | Pan | AIN | — | Y | 2, NA | 15, 44 | 3, 5 | 4, 13 |
| 57 | 56 | Neu | LGL | RhAr | Y | 1, 25 | 8, 18 | 7, 12 | 4, 11 |
| 58 | 60 | Pan | LGL | PNH | NA | 68, 68 | 14, 14 | 8, 8 | 1, 1 |
| 59 | 53 | Neu | LGL | AIH | Y | 1, 3 | 7, 37 | 6, 7 | 13, 15 |
| 60 | 51 | Pan | LGL | — | N | 11, 24 | 27, 22 | 1, 1 | 4, 15 |
Pan indicates pancytopenia; LGL, large granular lymphocyte leukemia; —, not applicable; Y, yes; ND, not done; RA, refractory anemia; PNH, paroxysmal nocturnal hemoglobinuria; RCA, red cell aplasia; Neu, neutropenia; NA, not available; MS, multiple sclerosis; RhAr, rheumatoid arthritis; SLE, systemic lupus erythematodus; N, no; RARS, RA with ringed sideroblasts; UC, ulcerative colitis; AIHA, autoimmune hemolytic anemia; and AIN, autoimmune neutropenia.
VB-JB restriction of immunodominant clones in studied patients
Patient no. . | VB . | % VB family expansion . | JB . |
|---|---|---|---|
| 1 | 14 | 90 | JB2.1 |
| 2 | 3 | 88 | JB2.4 |
| 3 | 22 | 89 | JB2.7 |
| 4 | 17 | 83 | JB2.3 |
| 5 | 9 | 64 | JB1.1 |
| 6 | 8 | 26 | JB1.5 |
| 7 | 6 | NA | JB2.1 |
| 8 | 23 | 15 | JB1.1, JB2.7* |
| 8 | 14 | 16 | JB2.7 |
| 9 | 3 | 63 | JB1.1 |
| 10 | 7 | 76 | JB2.7 |
| 11 | 20 | 23 | JB2.1, JB2.7* |
| 12 | 14 | 65 | JB2.7 |
| 13 | 12 | 24 | JB2.6 |
| 14 | 2 | 40 | JB2.7 |
| 15 | 18 | 8 | JB1.1, JB2.3* |
| 16 | 2 | 90 | JB2.7 |
| 17 | 2 | 20 | JB2.1 |
| 17 | 21 | 31 | JB2.1 |
| 18 | 17 | 45 | JB2.3 |
| 18 | 3 | 25 | JB2.1 |
| 19 | 2 | 38 | JB2.3 |
| 20 | 14 | 22 | JB1.2 |
| 21 | 13 | 7 | JB1.6 |
| 22 | 3 | 26 | JB1.5 |
| 23 | 3 | 33 | JB2.7 |
| 24 | 2 | 80 | JB2.7 |
| 25 | 15 | NA | JB2.1, JB2.3* |
| 26 | 9 | 25 | JB2.5 |
| 27 | 13 | 43 | JB1.1 |
| 27 | 18 | 25 | JB2.7 |
| 27 | 22 | NA | JB2.7 |
| 28 | 13 | 65 | JB2.7 |
| 29 | 5.3 | 25 | JB2.7 |
| 30 | 15 | NA | JB1.2, JB2.3* |
| 31 | 5 | NA | JB1.2 |
| 32 | 15 | NA | JB2.5, JB2.1, JB2.5* |
| 33 | 18 | 27 | JB2.7 |
| 34 | 3 | 10 | JB2.7 |
| 34 | 22 | 60 | JB1.2 |
| 35 | 7 | NA | JB1.6 |
| 36 | 9 | 78 | JB2.4 |
| 37 | 13 | 80 | JB2.5 |
| 38 | 12 | 93 | JB2.1 |
| 39 | 7 | 46 | JB2.2 |
| 40 | 24 | NA | JB2.2 |
| 40 | 13 | NA | JB1.2 |
| 41 | 15 | NA | JB2.7 |
| 41 | 24 | NA | JB2.5 |
| 42 | 6 | NA | JB2.5 |
| 42 | 15 | NA | JB2.4 |
| 43 | 6 | NA | JB2.3 |
| 43 | 24 | NA | JB2.1 |
| 43 | 12 | NA | JB2.3 |
| 44 | 9 | 81 | JB2.3 |
| 44 | 21 | 11 | JB1.1 |
| 45 | 24 | NA | JB1.1 |
| 46 | 6 | NA | JB2.1, JB2.7* |
| 46 | 17 | 8 | JB2.1 |
| 47 | 6 | NA | JB2.7 |
| 47 | 24 | NA | JB2.3 |
| 48 | 13 | 93 | JB1.1 |
| 49 | 1 | 70 | JB1.5 |
| 50 | 13 | 6 | JB2.7 |
| 51 | 12 | 8 | JB2.1 |
| 51 | 6 | NA | JB1.1 |
| 52 | 1 | NA | JB1.1 |
| 53 | 18 | 42 | JB2.7 |
| 54 | 13 | 6 | JB2.7, JB2.5* |
| 55 | 13 | 7 | JB2.3 |
| 55 | 18 | 78 | JB2.5 |
| 56 | 4 | 22 | JB2.1 |
| 57 | 13 | 89 | JB2.5 |
| 58 | 3 | NA | JB2.7 |
| 58 | 13 | NA | JB1.5 |
| 59 | 1 | 95 | JB1.6 |
| 60 | 14 | 28 | JB2.1 |
Patient no. . | VB . | % VB family expansion . | JB . |
|---|---|---|---|
| 1 | 14 | 90 | JB2.1 |
| 2 | 3 | 88 | JB2.4 |
| 3 | 22 | 89 | JB2.7 |
| 4 | 17 | 83 | JB2.3 |
| 5 | 9 | 64 | JB1.1 |
| 6 | 8 | 26 | JB1.5 |
| 7 | 6 | NA | JB2.1 |
| 8 | 23 | 15 | JB1.1, JB2.7* |
| 8 | 14 | 16 | JB2.7 |
| 9 | 3 | 63 | JB1.1 |
| 10 | 7 | 76 | JB2.7 |
| 11 | 20 | 23 | JB2.1, JB2.7* |
| 12 | 14 | 65 | JB2.7 |
| 13 | 12 | 24 | JB2.6 |
| 14 | 2 | 40 | JB2.7 |
| 15 | 18 | 8 | JB1.1, JB2.3* |
| 16 | 2 | 90 | JB2.7 |
| 17 | 2 | 20 | JB2.1 |
| 17 | 21 | 31 | JB2.1 |
| 18 | 17 | 45 | JB2.3 |
| 18 | 3 | 25 | JB2.1 |
| 19 | 2 | 38 | JB2.3 |
| 20 | 14 | 22 | JB1.2 |
| 21 | 13 | 7 | JB1.6 |
| 22 | 3 | 26 | JB1.5 |
| 23 | 3 | 33 | JB2.7 |
| 24 | 2 | 80 | JB2.7 |
| 25 | 15 | NA | JB2.1, JB2.3* |
| 26 | 9 | 25 | JB2.5 |
| 27 | 13 | 43 | JB1.1 |
| 27 | 18 | 25 | JB2.7 |
| 27 | 22 | NA | JB2.7 |
| 28 | 13 | 65 | JB2.7 |
| 29 | 5.3 | 25 | JB2.7 |
| 30 | 15 | NA | JB1.2, JB2.3* |
| 31 | 5 | NA | JB1.2 |
| 32 | 15 | NA | JB2.5, JB2.1, JB2.5* |
| 33 | 18 | 27 | JB2.7 |
| 34 | 3 | 10 | JB2.7 |
| 34 | 22 | 60 | JB1.2 |
| 35 | 7 | NA | JB1.6 |
| 36 | 9 | 78 | JB2.4 |
| 37 | 13 | 80 | JB2.5 |
| 38 | 12 | 93 | JB2.1 |
| 39 | 7 | 46 | JB2.2 |
| 40 | 24 | NA | JB2.2 |
| 40 | 13 | NA | JB1.2 |
| 41 | 15 | NA | JB2.7 |
| 41 | 24 | NA | JB2.5 |
| 42 | 6 | NA | JB2.5 |
| 42 | 15 | NA | JB2.4 |
| 43 | 6 | NA | JB2.3 |
| 43 | 24 | NA | JB2.1 |
| 43 | 12 | NA | JB2.3 |
| 44 | 9 | 81 | JB2.3 |
| 44 | 21 | 11 | JB1.1 |
| 45 | 24 | NA | JB1.1 |
| 46 | 6 | NA | JB2.1, JB2.7* |
| 46 | 17 | 8 | JB2.1 |
| 47 | 6 | NA | JB2.7 |
| 47 | 24 | NA | JB2.3 |
| 48 | 13 | 93 | JB1.1 |
| 49 | 1 | 70 | JB1.5 |
| 50 | 13 | 6 | JB2.7 |
| 51 | 12 | 8 | JB2.1 |
| 51 | 6 | NA | JB1.1 |
| 52 | 1 | NA | JB1.1 |
| 53 | 18 | 42 | JB2.7 |
| 54 | 13 | 6 | JB2.7, JB2.5* |
| 55 | 13 | 7 | JB2.3 |
| 55 | 18 | 78 | JB2.5 |
| 56 | 4 | 22 | JB2.1 |
| 57 | 13 | 89 | JB2.5 |
| 58 | 3 | NA | JB2.7 |
| 58 | 13 | NA | JB1.5 |
| 59 | 1 | 95 | JB1.6 |
| 60 | 14 | 28 | JB2.1 |
VB indicates variable beta chain (VB family) of the TCR; JB, joining beta chain of the expanded clonotypes; NA, not available.
Cases in which 2 (3 in patient 32) immunodominant clonotypes with identical VB but different JB families were detected.
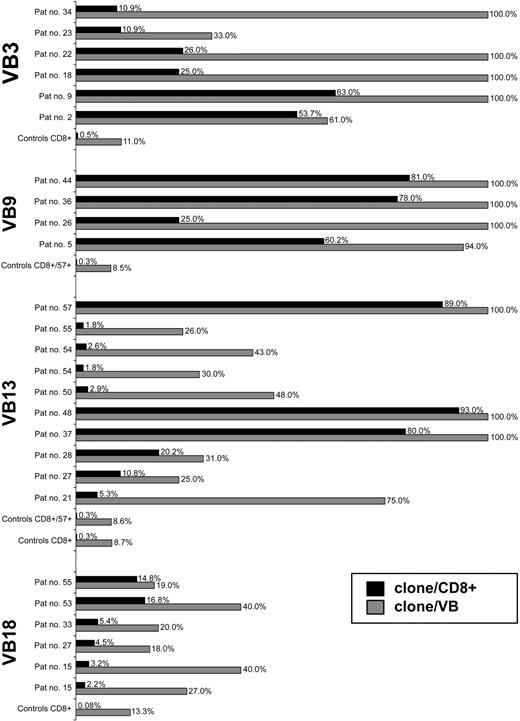
Based on the sequencing of a large number of CDR3 amplicons in controls (57 ± 25 [mean ± standard deviation; SD] colonies per VB family and sample; n = 26), we defined a clonal frequency of greater than 12.5% (mean+2 × SD of the largest clones found in healthy controls) as pathologic. On average, expansions in patients were 64% ± 30% of the CDR3 repertoire within a given VB family (Figure 2). A total of 86 immunodominant LGL clonotypes were identified (Figure 3). Their frequency was 18% to 100% (65% ± 30%) of a given VB family or 1.8% to 95% (35.4% ± 31%) of the total CD8 population (Figure 3). By comparison, the most expanded VB13 clones in healthy controls contribute to only 0.7% of the CD8 repertoire (Figure 2). Remarkably, in 21 patients we found more than one immunodominant clone, suggesting that some LGL lymphoproliferations are biclonal (Tables 1, 2; Figure 3).
Structural analysis of immunodominant LGL clones
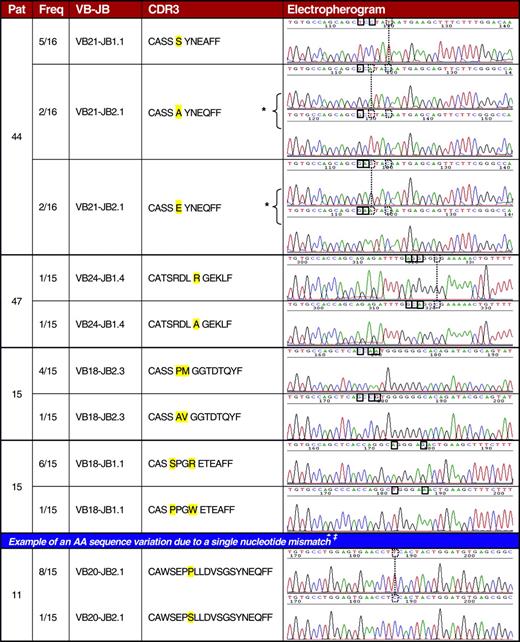
Given the immense variability of physiologic TCR repertoire, detection of identical clonotypes shared by patients with LGL leukemia could suggest a nonrandom nature of clonal transformation. Translation of nucleotide sequences into AA code resulted in 86 LGL CDR3 clonotypes (Figure 3). These sequences were also compared with LGL-specific immunodominant CDR3 sequences (n = 5) described in literature.59,61,62 We found 2 patients (nos. 50 and 54 in Table 3) with identical immunodominant clonotype. Interestingly, this clonotype was also shared with a third patient (no. 27) in whom it was not significantly expanded. Analysis of a much larger number of CDR3 sequences in healthy controls demonstrates that such a finding is not likely to be a totally random event. The presence of this sequence was confirmed through multiple independent amplifications and sequencing. As documented by HLA typing, these 3 patients (nos. 27, 50, 54) share the class I antigen B7. In addition, we found sequence identity between an immunodominant and a nonimmunodominant (or minor) clonotype and between 2 minor clonotypes in 2 LGL patient pairs (nos. 27 and 54; nos. 27 and 50 in Table 3). In 2 other patients, a major clonotype showed a striking homology to a minor clonotype also present within the TCR repertoire (nos. 27 and 49 in Table 3). Not only were identical CDR3 sequences shared between the patients but a high degree of similarity was also found between a large number of minor (nonexpanded) clonotypes (Table 3). Codominant clonotypic sequences within the TCR repertoire of individual patients were found that differ only in one or a very few nucleotides. In some instances, these nucleotide mismatches did not result in a new AA sequence (Figure 4), consistent with selection of high-affinity T-cell receptors during the initial polyclonal response. By comparison, when up to 172 clones were sequenced per VB family in 26 healthy donors, only one shared clonotype was found in 2 donors and only a total of 11 sequences were detected to show a similar level of homology between the healthy controls as described in LGLs (Table 4).
Similarity between LGL-associated clonotypes
Patient . | VB . | CDR3 . | JB . | % clonal frequency . |
|---|---|---|---|---|
| 27 | 13 | ASSVPGGEQYF | 2-7 | Minor |
| 50 | 13 | ASSVPGGEQYF | 2-7 | 48 |
| 54 | 13 | ASSVPGGEQYF | 2-7 | 43 |
| 27 | 13 | ASSYYREPQHFGD | 1-5 | Minor |
| 50 | 13 | ASSYYREPQHFGD | 1-5 | Minor |
| 27 | 13 | ASSYTFQGETQY | 2-5 | Minor |
| 54 | 13 | ASSYTFQGETQY | 2-5 | 29 |
| 27 | 18 | ASSPGTGLDGYTF | 1-2 | Minor |
| 49 | 1 | ASSPGTGRNQPQHFGD | 1-5 | 89 |
| 50 | 13 | ASRDGAGEQYF | 2-7 | Minor |
| 28 | 13 | ASRDRAGN | 2-6 | Minor |
| 44 | 21 | ASRSGTGAHEQYF | 2-7 | Minor |
| 28 | 13 | ASRSGTGSYNEQFF | 2-1 | Minor |
| 23 | 3 | ASSGEETQYF | 2-3 | Minor |
| 33 | 18 | ASSGETDTQYF | 2-3 | Minor |
| 62* | 13 | ASSLAGGPDTQYF | 2-3 | Minor |
| 47 | 6 | ASSLAGTGGGEQYF | 2-7 | Minor |
| 47 | 6 | ASSLAGTSSTDTQYF | 2-3 | Minor |
| 35 | 6 | ASSLGSYNSPQHFDG | 1-5 | Minor |
| 63* | 21 | ASSLGSYNSPLH | 1-6 | Minor |
| 62* | 13 | ASSLLKPTSDTQYF | 2-3 | Minor |
| 12 | 14 | ASSLLTKTGSYEQYF | 2-7 | 81 |
| 35 | 6 | ASSLVGDPGELF | 2-2 | Minor |
| 47 | 6 | ASSLVGGRPSYNEQFF | 2-1 | Minor |
| 27 | 18 | ASSPPVRARDTQYF | 2-3 | Minor |
| 33 | 18 | ASSPPVRSRAQY | 2-5 | Minor |
| 18 | 17 | ASSWGGAGELFF | 2-2 | Minor |
| 29 | 5 | ASSWGGRGEQFF | 2-1 | Minor |
| 65* | 15 | ATSDLGQGASGELF | 2-2 | Minor |
| 47 | 15 | ATSDLGQQETQY | 2-5 | Minor |
| 42 | 15 | ATSDLGEKVGNEQFF | 2-1 | Minor |
| 47 | 24 | ATSRDLAGEKLF | 1-4 | Minor |
| 35 | 24 | ATSRDLASGRETQYF | 2-3 | Minor |
| 47 | 24 | ATSRDLRGEKLF | 1-4 | Minor |
| 41 | 24 | ATSRDLRTQETQY | 2-5 | Minor |
Patient . | VB . | CDR3 . | JB . | % clonal frequency . |
|---|---|---|---|---|
| 27 | 13 | ASSVPGGEQYF | 2-7 | Minor |
| 50 | 13 | ASSVPGGEQYF | 2-7 | 48 |
| 54 | 13 | ASSVPGGEQYF | 2-7 | 43 |
| 27 | 13 | ASSYYREPQHFGD | 1-5 | Minor |
| 50 | 13 | ASSYYREPQHFGD | 1-5 | Minor |
| 27 | 13 | ASSYTFQGETQY | 2-5 | Minor |
| 54 | 13 | ASSYTFQGETQY | 2-5 | 29 |
| 27 | 18 | ASSPGTGLDGYTF | 1-2 | Minor |
| 49 | 1 | ASSPGTGRNQPQHFGD | 1-5 | 89 |
| 50 | 13 | ASRDGAGEQYF | 2-7 | Minor |
| 28 | 13 | ASRDRAGN | 2-6 | Minor |
| 44 | 21 | ASRSGTGAHEQYF | 2-7 | Minor |
| 28 | 13 | ASRSGTGSYNEQFF | 2-1 | Minor |
| 23 | 3 | ASSGEETQYF | 2-3 | Minor |
| 33 | 18 | ASSGETDTQYF | 2-3 | Minor |
| 62* | 13 | ASSLAGGPDTQYF | 2-3 | Minor |
| 47 | 6 | ASSLAGTGGGEQYF | 2-7 | Minor |
| 47 | 6 | ASSLAGTSSTDTQYF | 2-3 | Minor |
| 35 | 6 | ASSLGSYNSPQHFDG | 1-5 | Minor |
| 63* | 21 | ASSLGSYNSPLH | 1-6 | Minor |
| 62* | 13 | ASSLLKPTSDTQYF | 2-3 | Minor |
| 12 | 14 | ASSLLTKTGSYEQYF | 2-7 | 81 |
| 35 | 6 | ASSLVGDPGELF | 2-2 | Minor |
| 47 | 6 | ASSLVGGRPSYNEQFF | 2-1 | Minor |
| 27 | 18 | ASSPPVRARDTQYF | 2-3 | Minor |
| 33 | 18 | ASSPPVRSRAQY | 2-5 | Minor |
| 18 | 17 | ASSWGGAGELFF | 2-2 | Minor |
| 29 | 5 | ASSWGGRGEQFF | 2-1 | Minor |
| 65* | 15 | ATSDLGQGASGELF | 2-2 | Minor |
| 47 | 15 | ATSDLGQQETQY | 2-5 | Minor |
| 42 | 15 | ATSDLGEKVGNEQFF | 2-1 | Minor |
| 47 | 24 | ATSRDLAGEKLF | 1-4 | Minor |
| 35 | 24 | ATSRDLASGRETQYF | 2-3 | Minor |
| 47 | 24 | ATSRDLRGEKLF | 1-4 | Minor |
| 41 | 24 | ATSRDLRTQETQY | 2-5 | Minor |
VB indicates variable beta family; CDR3, complementarity-determining region 3; JB, joining beta family with first 3 variant AAs; minor, nonexpanded (single) clonotype.
Italics indicate identical AAs.
Patients in whom a major “immunodominant” clonotype was not determined, therefore not included in Table 1.
Identical clonotypes in LGL and other conditions
Patient . | Diagnosis . | VB-JB . | CDR3 . | % clonal frequency . |
|---|---|---|---|---|
| 27 | LGL | VB13-JB2.7 | ASSVPGGEQYF | Minor |
| 50 | LGL | VB13-JB2.7 | ASSVPGGEQYF | 48 |
| 54 | LGL | VB13-JB2.7 | ASSVPGGEQYF | 43 |
| 409 | HD | VB13-JB2.7 | ASSVPGGEQYF | Minor |
| 418 | HD | VB13-JB2.7 | ASSVPGGEQYF | Minor |
| 27 | LGL | VB18-JB2.3 | ASSAVGGTDTQYF | Minor |
| 142 | AHSCT | VB18-JB2.3 | ASSAVGGTDTQYF | 40 |
| 573 | AHSCT | VB18-JB2.3 | ASSAVGGTDTQYF | 25 |
| 217 | AHSCT | VB17-JB2.3 | ASSIAVRLASTDTQYF | 30 |
| 4 | LGL | VB17-JB2.3 | ASSIAVRLASTDTQYF | 94 |
| McFarland et al73 | HIV-1 | VB13-JB1.2 | ASSPITGTGSYGYTF | 7 |
| 27 | LGL | VB13-JB1.2 | ASSPITGTGSYGYTF | Minor |
| Beck et al74 | GvHD | VB13-JB2.2 | ASSSETSGRNTGELFF | 25 |
| 27 | LGL | VB13-JB2.2 | ASSSETSGRNTGELFF | Minor |
| 27 | LGL | VB13-JB2.7 | ASSYGQEHYEQYF | Minor |
| 419 | HD | VB13-JB2.7 | ASSYGQEHYEQYF | Minor |
| 418 | HD | VB13-JB2.1 | ASSYSGSHNEQFF | Minor |
| 54 | LGL | VB13-JB2.1 | ASSYSGSHNEQFF | 21 |
| 258 | AHSCT | VB15-JB2.5 | ATSDLTWGETQY | 25 |
| 574 | AHSCT | VB15-JB2.5 | ATSDLTWGETQY | 60 |
| 242 | AHSCT | VB15-JB2.5 | ATSDLTWGETQY | 27 |
| 32 | LGL | VB15-JB2.5 | ATSDLTWGETQY | 26 |
Patient . | Diagnosis . | VB-JB . | CDR3 . | % clonal frequency . |
|---|---|---|---|---|
| 27 | LGL | VB13-JB2.7 | ASSVPGGEQYF | Minor |
| 50 | LGL | VB13-JB2.7 | ASSVPGGEQYF | 48 |
| 54 | LGL | VB13-JB2.7 | ASSVPGGEQYF | 43 |
| 409 | HD | VB13-JB2.7 | ASSVPGGEQYF | Minor |
| 418 | HD | VB13-JB2.7 | ASSVPGGEQYF | Minor |
| 27 | LGL | VB18-JB2.3 | ASSAVGGTDTQYF | Minor |
| 142 | AHSCT | VB18-JB2.3 | ASSAVGGTDTQYF | 40 |
| 573 | AHSCT | VB18-JB2.3 | ASSAVGGTDTQYF | 25 |
| 217 | AHSCT | VB17-JB2.3 | ASSIAVRLASTDTQYF | 30 |
| 4 | LGL | VB17-JB2.3 | ASSIAVRLASTDTQYF | 94 |
| McFarland et al73 | HIV-1 | VB13-JB1.2 | ASSPITGTGSYGYTF | 7 |
| 27 | LGL | VB13-JB1.2 | ASSPITGTGSYGYTF | Minor |
| Beck et al74 | GvHD | VB13-JB2.2 | ASSSETSGRNTGELFF | 25 |
| 27 | LGL | VB13-JB2.2 | ASSSETSGRNTGELFF | Minor |
| 27 | LGL | VB13-JB2.7 | ASSYGQEHYEQYF | Minor |
| 419 | HD | VB13-JB2.7 | ASSYGQEHYEQYF | Minor |
| 418 | HD | VB13-JB2.1 | ASSYSGSHNEQFF | Minor |
| 54 | LGL | VB13-JB2.1 | ASSYSGSHNEQFF | 21 |
| 258 | AHSCT | VB15-JB2.5 | ATSDLTWGETQY | 25 |
| 574 | AHSCT | VB15-JB2.5 | ATSDLTWGETQY | 60 |
| 242 | AHSCT | VB15-JB2.5 | ATSDLTWGETQY | 27 |
| 32 | LGL | VB15-JB2.5 | ATSDLTWGETQY | 26 |
VB-JB indicates restriction of the variable beta and joining beta region of the TCR; LGL, large granular lymphocytic leukemia; minor, nonexpanded clonotype; HD, healthy donor; AHSCT, allogeneic hematopoietic stem cell transplantation; and GvHD, graft-versus-host disease.
We have also employed more redundant AA codes (8-AA and 7-AA instead of 20-AA code), allowing for the comparison of sequences based on the physical and chemical properties of the AAs (Tables 5, 6). Although the rules governing the recognition of the antigenic peptide by the TCR are not fully known, the VB CDR3 region appears to be most intricately involved in the contact of the target peptide and B chain. Thus similarity of the structure of the VB CDR3 region if occurring in the context of the common HLA restriction element may indicate shared specificity. However, coincidental similarity cannot be excluded. The comparison reveals an even higher degree of sequence homology than found using 20-AA translation. In contrast, analogous analysis performed on CD8+ and on CD8+CD57+ cell populations derived from healthy donors resulted in identification of a significantly lesser CDR3 sequence homology in 20-, 8-, and 7-AA coding (data not shown).
Examples of a comparison of LGL-associated clonotypes based on the physicochemical AA properties (8-AA substitution)
Patient . | VB-JB . | CDR3/20AA code . | . | CDR3/8AA substitution . | . | ||
|---|---|---|---|---|---|---|---|
| 53 | VB18-JB2.7 | ASS-PPVGDR | -SYEQYF | BEE-BBBDGH | -EAGEAA | ||
| 4 | VB17-JB2.3 | ASS-IAVRLA | -STDTQYF | BEE-BBBHBB | -EEGEEAA | ||
| 50 | VB13-JB2.7 | ASS-VPGG | -EQYF | BEE-BBDD | -GEAA | ||
| 54 | VB13-JB2.7 | ASS-VPGG | -EQYF | BEE-BBDD | -GEAA | ||
| 18 | VB3-JB2.1 | ASS-PLGAGV | -YNEQFF | BEE-BBDBDB | -AEGEAA | ||
| 27 | VB18-JB2.7 | ASS-PAGGSG | -SYEQYF | BEE-BBD DED | -EAGEAA | ||
| 12 | VB14-JB2.7 | ASS-LLTKTG | -SYEQYF | BEE-BBEHED | -EAGEAA | ||
| 29 | VB5-JB2.7 | ASS-PLRG | -SYEQYF | BEE-BB H D | -EAGEAA | ||
| 55 | VB18-JB2.5 | ASS-PNI | -ETQY | BEE-BEB | -GEEA | ||
| 19 | VB2-JB2.3 | ASS-PTLRDRGR | -TDTQYF | BEE-BEBHGHDH | -EGEEAA | ||
| 46 | VB17-JB2.1 | ASS-TFG | -EQFF | BEE-EAD | -GEAA | ||
| 44 | VB21-JB1.1 | ASS-SY | -NEAFF | BEE-EA | -EGBAA | ||
Patient . | VB-JB . | CDR3/20AA code . | . | CDR3/8AA substitution . | . | ||
|---|---|---|---|---|---|---|---|
| 53 | VB18-JB2.7 | ASS-PPVGDR | -SYEQYF | BEE-BBBDGH | -EAGEAA | ||
| 4 | VB17-JB2.3 | ASS-IAVRLA | -STDTQYF | BEE-BBBHBB | -EEGEEAA | ||
| 50 | VB13-JB2.7 | ASS-VPGG | -EQYF | BEE-BBDD | -GEAA | ||
| 54 | VB13-JB2.7 | ASS-VPGG | -EQYF | BEE-BBDD | -GEAA | ||
| 18 | VB3-JB2.1 | ASS-PLGAGV | -YNEQFF | BEE-BBDBDB | -AEGEAA | ||
| 27 | VB18-JB2.7 | ASS-PAGGSG | -SYEQYF | BEE-BBD DED | -EAGEAA | ||
| 12 | VB14-JB2.7 | ASS-LLTKTG | -SYEQYF | BEE-BBEHED | -EAGEAA | ||
| 29 | VB5-JB2.7 | ASS-PLRG | -SYEQYF | BEE-BB H D | -EAGEAA | ||
| 55 | VB18-JB2.5 | ASS-PNI | -ETQY | BEE-BEB | -GEEA | ||
| 19 | VB2-JB2.3 | ASS-PTLRDRGR | -TDTQYF | BEE-BEBHGHDH | -EGEEAA | ||
| 46 | VB17-JB2.1 | ASS-TFG | -EQFF | BEE-EAD | -GEAA | ||
| 44 | VB21-JB1.1 | ASS-SY | -NEAFF | BEE-EA | -EGBAA | ||
A redundant classification scheme was applied for the comparison of the TCR-CDR3 region, based on the structural, chemical, charge, and functional properties of the amino acids. For substitution code, see “Sequence analysis.”
VB-JB indicates VB and JB regions used by the immunodominant clonotype; CDR3/20AA code and CDR3/8AA, CDR3 region sequence in 20-AA and 8-AA code, respectively; and Jun, invariant amino acids of the JB region. Italic amino acids indicate the beginning of the JB region and include the invariant JB-specific amino acids. Underlined type indicates AA differences.
Examples of a comparison of LGL-associated clonotypes based on the physicochemical AA properties (7-AA substitution)
Patient . | VB-JB . | CDR3/20AA code . | . | CDR3/7AA substitution . | . | ||
|---|---|---|---|---|---|---|---|
| 7 | VB6-JB2.1 | ASS-LASI | -SYNEQFF | BUU-BBUB | -UJZZZJJ | ||
| 41 | VB24-JB2.5 | ATS-LGTL | -QETQYF | BUU-BBUB | -ZZUZJJ | ||
| 8 | VB23-JB1.1 | ASS-LGNR | -EVAFF | BUU-BBZX | -ZBBJJ | ||
| 52 | VB1-JB1.1 | ASS-AGQD | -TEAFF | BUU-BBZZ | -UZBJJBZ | ||
| 58 | VB13-JB1.5 | ASS-YGGG | -QPQHFGD | BUU-JBBB | -ZPZXJ | ||
| 55 | VB13-JB2.3 | ASS-YVGV | -DTQYF | BUU-JBBB | -ZUZJJ | ||
| 18 | VB3-JB2.1 | ASS-PLGAGV | -YNEQFF | BUU-PBBBBB | -JZZZJJ | ||
| 27 | VB18-JB2.7 | ASS-PAGGSG | -SYEQYF | BUU-PBBBUB | -UJZZJJ | ||
| 46 | VB17-JB2.1 | ASS-TFG | -EQFF | BUU-UJB | -ZZJJ | ||
| 44 | VB21-JB1.1 | ASS-SY | -NEAFF | BUU-UJ | -ZZBJJ | ||
| 14 | VB2-JB2.7 | SAS-GLLAGGA | -SYEQYF | UBU-BBBBBBB | -UJZZJJ | ||
| 24 | VB2-JB2.7 | SAS-FLTGLAG | -EQYF | UBU-JBUBBBB | -ZZJJ | ||
Patient . | VB-JB . | CDR3/20AA code . | . | CDR3/7AA substitution . | . | ||
|---|---|---|---|---|---|---|---|
| 7 | VB6-JB2.1 | ASS-LASI | -SYNEQFF | BUU-BBUB | -UJZZZJJ | ||
| 41 | VB24-JB2.5 | ATS-LGTL | -QETQYF | BUU-BBUB | -ZZUZJJ | ||
| 8 | VB23-JB1.1 | ASS-LGNR | -EVAFF | BUU-BBZX | -ZBBJJ | ||
| 52 | VB1-JB1.1 | ASS-AGQD | -TEAFF | BUU-BBZZ | -UZBJJBZ | ||
| 58 | VB13-JB1.5 | ASS-YGGG | -QPQHFGD | BUU-JBBB | -ZPZXJ | ||
| 55 | VB13-JB2.3 | ASS-YVGV | -DTQYF | BUU-JBBB | -ZUZJJ | ||
| 18 | VB3-JB2.1 | ASS-PLGAGV | -YNEQFF | BUU-PBBBBB | -JZZZJJ | ||
| 27 | VB18-JB2.7 | ASS-PAGGSG | -SYEQYF | BUU-PBBBUB | -UJZZJJ | ||
| 46 | VB17-JB2.1 | ASS-TFG | -EQFF | BUU-UJB | -ZZJJ | ||
| 44 | VB21-JB1.1 | ASS-SY | -NEAFF | BUU-UJ | -ZZBJJ | ||
| 14 | VB2-JB2.7 | SAS-GLLAGGA | -SYEQYF | UBU-BBBBBBB | -UJZZJJ | ||
| 24 | VB2-JB2.7 | SAS-FLTGLAG | -EQYF | UBU-JBUBBBB | -ZZJJ | ||
A redundant classification scheme was applied for the comparison of the TCR-CDR3 region, based on the structural, chemical, charge, and functional properties of the amino acids. For substitution code, see “Sequence analysis.”
VB-JB indicates VB and JB regions used by the immunodominant clonotype; CDR3/20AA code and CDR3/7AA, CDR3 region sequence in 20-AA and 7-AA code, respectively; and Jun, invariant amino acids of the JB region. Italic amino acids indicate the beginning of the JB region and include the invariant JB-specific amino acids. Underlined type indicates AA differences.
Clonotypic background
In addition to the major clone, other clonotypes occurring at low frequencies may be present. These clonotypes are likely randomly selected due to, for example, polyclonal expansion. If LGL clones arise in the context of initially polyclonal CTL responses, it is possible that structurally similar clonotypes will be present. Consequently, we compared AA sequences of the major (immunodominant) and minor clonotypes identified in each patient. We found a high degree of homology between major and minor clonotypes within the repertoire of individual patients (Figure 4). Single base differences were also detected but they were not included in this homology analysis, as such clonotypes may or may not be a result of polymerase reading errors. However, all the results were based on sequencing of a large numbers of clones, and clonotypes that varied in one or only a few AAs were found at multiple occasions.
Clonotypic dynamics and disease progress
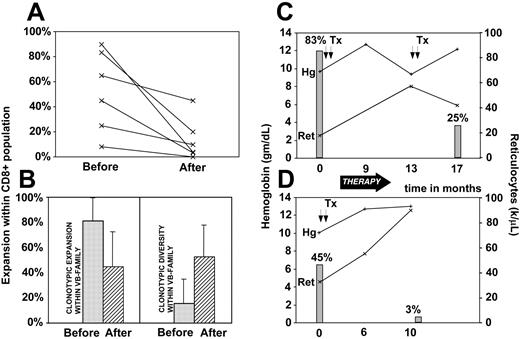
Immunodominant clonotypes can be used as tumor markers and we have investigated the changes in the contribution of the LGL clone in correlation with disease activity (Figure 5). The size of the pathologic clone was calculated by multiplication of the contribution of the VB family to the total CD8 spectrum and the relative size of the clone within a given VB family. For most patients, a marked decrease in the frequency of the immunodominant clone was observed with successful immunosuppressive therapy. In some patients the drop appeared to be less significant while in others the pathogenic clone was not detectable following the therapy (Figure 5A). Decrease in the contribution of the immunodominant clone to the CD8 repertoire was associated with the appearance of minor clones that were previously “diluted out” by the expanded clone and not detectable. This process resulted in increased clonal diversity (defined as percentage of different clones out of a total number of clones) and decreased redundancy (defined as percentage expansion of the largest clone; Figure 5B). Clinical correlation between the size of immunodominant clones and hematologic parameters is shown for 2 patients with LGL leukemia associated with pure red cell aplasia (Figure 5C-D). When we analyzed the structure of clonotypes that appeared after therapy we found that in some instances they showed similarity to the original immunodominant clones (data not shown).
Clonotype sharing with other diseases
We also investigated minor and immunodominant clonotype sharing between LGL patients and patients with other diseases (n = 156). We found 3 expanded LGL-associated clonotypes to also be present in patients studied after allogeneic bone marrow transplantation (Table 4). In addition, sharing of minor clonotypes was also found between LGL and bone marrow transplantation patients. Similarly, a minor LGL clonotype was detected in a patient with HIV. Two of the immunodominant LGL CDR3 sequences were found in healthy individuals tested and furthermore 2 minor LGL-associated clonotypes were shared with healthy donors.
Discussion
Our study addresses several important aspects of the physiologic and aberrant cellular immune response. LGL leukemia, albeit rare, can act as a model system to study CTL expansions, as the clonal CTLs show a behavior that is highly reminiscent of that of their normal counterparts. While flow cytometric40 and molecular75 VB typing has previously been applied to relatively large cohorts of LGL patients, LGL-like clonotypic expansions were described only in very small series including 10 patients with Felty syndrome (9 clonotypes),59 3 patients with MDS and erythroid hypoplasia (3 clonotypes),62 32 patients with rheumatoid arthritis (9 sequences),54 and in a donor/recipient pair after BMT (5 sequences).61
We have applied a rational strategy not only to identify individual LGL-leukemia–associated immunodominant clonotypes but also to quantitate their contribution to the total CTL repertoire during therapy. We demonstrate that either flow cytometric detection of phenotypically abnormal CTL populations or VB CDR3 genotyping may leave a significant number of CTL proliferations unidentified. By combining these methods and sequencing expanded clones, we found several patients with significant expansions of clonal CTLs that were unsuspected based on traditional clinical tests. It is likely that clinically manifested cases of LGL leukemia represent the extreme clonal/oligoclonal lymphoproliferations that, to a lesser extent, may also be present in certain patients with otherwise unexplained cytopenias. In addition, some of the expansions, when asymptomatic, can represent benign CTL reactions also referred to as T-cell clonopathy of undetermined significance.76
Clonotypic expansions in patients and healthy controls. The size of pathogenic LGL clones was compared with the most expanded clones found for corresponding VB families in healthy individuals. The figure depicts selective examples for individual patients/healthy controls and VB families. ▦ indicates percent clonotypic expansion within a given VB family (clone/VB). □ indicates percent expansion within the entire CD8+ cell population (% clonotypic expansion within a VB family multiplied by VB family contribution to CD8+ population; clone/CD8+). VB3-restricted clonotypic expansions were found in 6 patients and compared with the average size of the most redundant (expanded) clonotypes within VB3 in 9 healthy controls (a total of 356 CDR3 sequences). VB9-restricted expansions were found in 4 patients and compared with the average size of the most redundant clones for VB9 family studied in 2 healthy donors (a total of 45 CDR3 sequences). VB13-restricted expansions were found in a total of 10 patients and compared with the average size of the most redundant clones for VB13 family detected in 8 healthy controls (a total of 608 CDR3 sequences). A total of 6 patients were found to have VB18-specific immunodominant clonotypes and were compared with the average size of the most redundant clones for VB18 family derived from 8 healthy individuals (a total of 447 CDR3 sequences). In addition, repertoires of mature effector CTL cells (CD8+CD57+) derived from healthy individuals (n = 2) were studied. Pat no. indicates patient number.
Clonotypic expansions in patients and healthy controls. The size of pathogenic LGL clones was compared with the most expanded clones found for corresponding VB families in healthy individuals. The figure depicts selective examples for individual patients/healthy controls and VB families. ▦ indicates percent clonotypic expansion within a given VB family (clone/VB). □ indicates percent expansion within the entire CD8+ cell population (% clonotypic expansion within a VB family multiplied by VB family contribution to CD8+ population; clone/CD8+). VB3-restricted clonotypic expansions were found in 6 patients and compared with the average size of the most redundant (expanded) clonotypes within VB3 in 9 healthy controls (a total of 356 CDR3 sequences). VB9-restricted expansions were found in 4 patients and compared with the average size of the most redundant clones for VB9 family studied in 2 healthy donors (a total of 45 CDR3 sequences). VB13-restricted expansions were found in a total of 10 patients and compared with the average size of the most redundant clones for VB13 family detected in 8 healthy controls (a total of 608 CDR3 sequences). A total of 6 patients were found to have VB18-specific immunodominant clonotypes and were compared with the average size of the most redundant clones for VB18 family derived from 8 healthy individuals (a total of 447 CDR3 sequences). In addition, repertoires of mature effector CTL cells (CD8+CD57+) derived from healthy individuals (n = 2) were studied. Pat no. indicates patient number.
Our molecular clonotypic analysis was based on the B chain. The VB CDR3 region appears to interact with the antigenic peptide but A chain may also play an important role, especially in the initial phase of clonal high-affinity TCR selection.77 As expected, VB clonality was accompanied by VA clonality in analyzed patients. For 3 of our patients we have obtained analogous results showing that their clones express TCR phenotypes: VB1/JB1.6 and VA9-2/JA41*01 (patient no. 59), VB9/JB2.5 and VA26-1/JA9*01 (patient no. 26), VB13/JB2.5 and VA1-2/JA11*01 (patient no. 57; M.W.W., unpublished results, August 2004).
CDR3 motifs and expansions of immunodominant T-cell clones within VB family and CD8+ population. Pat indicates patient number; CDR3, sequence of the complementarity determining region 3; inv, first 3 invariant AAs of the JB region; VB-JB, restriction of the variable and joining β region of the TCR; imm clone/VB family, expansion of immunodominant, disease-associated T-cell clone within a given TCR VB family; imm clone/CD8+, expansion of immunodominant clone within the entire CD8+ population (clonal expansion within given VB family multiplied by expansion of this VB family within CD8+ cell population); and *, VB family expansion not available.
CDR3 motifs and expansions of immunodominant T-cell clones within VB family and CD8+ population. Pat indicates patient number; CDR3, sequence of the complementarity determining region 3; inv, first 3 invariant AAs of the JB region; VB-JB, restriction of the variable and joining β region of the TCR; imm clone/VB family, expansion of immunodominant, disease-associated T-cell clone within a given TCR VB family; imm clone/CD8+, expansion of immunodominant clone within the entire CD8+ population (clonal expansion within given VB family multiplied by expansion of this VB family within CD8+ cell population); and *, VB family expansion not available.
Our study was initiated based on the theory that the unique and often very lineage-restricted cytopenias associated with LGL leukemia may be a result of the lineage specificity of the target antigens recognized by clonal TCR. Consequently, it may be possible that in patients with similar presentations and matching HLA restriction elements, identical or highly homologous clonotypes will be found. Clearly, given the estimated physiologic variability of CDR3 regions within the VB chain,13 such an event may be rare but it would prove the nonrandom origin of clonal transformation in LGL leukemia. Based on an analogous theory, similarity of the rearranged immunoglobulin G (IgG) genes in B-cell clones found in chronic lymphocytic leukemia (CLL) suggests a nonrandom nature of transformation.78 Clearly, the restriction of TCR by HLA makes the principle of the T-cell recognition more complex. Nevertheless, numerous reports confirmed homology of clonotypes recognizing specific antigens in individuals matched for HLA elements.7,29,30,32,34 A specific TCR can recognize epitopes derived from the same protein and presented in the context of different yet genotypically similar HLA types.79
In analogy to these findings, identical expanded clonotypes were found in 2 patients. Based on the extent of the physiologic TCR repertoire and the extremely low frequency of shared clonotypes identified between healthy controls, the coincidental finding of common clonotypes is unlikely although it cannot be totally excluded. It suggests that these clones may not evolve randomly but occur in the context of an initially oligoclonal/polyclonal immune response directed against identical or highly similar antigenic targets. This conclusion is also supported by the identification of sequence sharing between the 2 other immunodominant clonotypes and nonexpanded clonotypes found in other LGL leukemia cases. It is likely that these minor clonotypes are the remnants of an initial polyclonal response. The low frequency of these clones may be due to “dilution” by expanding semiautonomous LGL clones. Consequently, the LGL clone may have evolved not from a total possible pool of CTL memory cells but from a selected population participating in a polyclonal immune response. Detection of high levels of similarity between clonotypes within a given VB repertoire of individual patients also supports our conclusions that several clones recognizing similar or identical peptides exist prior to the expansion of immunodominant clones. Occasional presence of clonotypes that are identical in their AA sequences but distinct in their nucleotide sequences may have resulted from selection of individual clones with identical antigenic affinity. In such cases the effective frequency of the corresponding TCR may be a sum of 2 independently rearranged clones. In the previous report,63 ClustalW analysis80 was used to assess the similarity between the clonotypic sequences. Since peptide binding is affected through the physical AA properties and the tertiary structure of CDR3, we believe that the analysis of physicochemical properties of AAs performed in the current study is more appropriate than linear homology comparisons in 20-AA code.
Sequencing of clonotypes prior to and following successful cytotoxic therapy also revealed that the clonotypes found after retraction of the LGL clone show similarity to the immunodominant clonotype. All of these observations have to be viewed in the context of the extremely low frequency of identical clonotypes (both expanded as well as minor) shared by healthy individuals in whom a relatively low homology between randomly sequenced clonotypes was also found. Association of LGL leukemia with typical pathologies is consistent with clonotype sharing. The initial autoimmune process leading to the specific manifestations may have provided a signal for the expansion of a clone that through a certain, yet not specified, molecular event induces uncontrolled proliferation. In our previous study, a smaller cohort of patients was analyzed and several similar and 2 identical clonotypes were identified.63 Based on these preliminary results, we expected a larger number of shared clonotypes to be found. Due to the higher sampling size, the frequency of clonotype sharing established in the current report is more precise.
Identification of highly homologous or identical clonotypes is remarkable. One could argue that PCR-based carryover could explain such a striking result. To avoid this possibility, in addition to general contamination precautions, in each PCR set, different VB families were amplified and sequenced. For analysis, the VB-specific primer sequence was used as an internal control to verify patient-specific sequences. Moreover, the finding of homologies between sequences reported in the literature and detected in our laboratory further illustrates that the isolation of identical sequences is not due to a contamination artifact. Identical sequences from different patients found in our laboratory were confirmed by repeated independent sequencing. Many of the immunodominant clonotypes in individual patients were invariably found on multiple time points throughout the clinical course and patients with common clonotypes shared at least one HLA allele that could produce a display of identical antigenic peptides. Finally, in our preliminary studies we have established optimal conditions for transformation to exclude that a higher frequency of certain clones is a result of bacterial division rather than preponderance of the specific amplification product in the sample. By minimizing the posttransformation phenotypic expression time, outgrowth of bacterial clones is avoided and the sequence distribution of the transformation product likely represents the composition of the PCR amplicons. The results obtained from healthy adult donors and cord blood controls clearly demonstrate that replicate sequences are only rarely encountered.
Examples of TCR repertoire similarities within individual LGL patients. Freq indicates clonotypic frequency as the number of identical clonotype sequences divided by total number of clones; and VB-JB, restriction of a given variable and joining beta chain of the TCR. Solid boxes indicate nucleotide exchanges resulting in AA exchange (the affected AAs are highlighted in yellow); dashed boxes and lines, nucleotide exchanges that do not lead to AA exchanges. ‡Total of 11 similar exchanges were found and were not included in the analysis, due to the possibility of Taq-polymerase–generated errors. Of interest, in patient 44 an immunodominant clonotype has 2 “supporting” clonotypes that differ in only 1 amino acid of the CDR3 sequence. Those 2 clonotypes were translated from 2 different nucleotide sequences each (*). In both cases the underlying sequences differed in 2 nucleotides that did not affect the AA sequence. Although we postulate that single nucleotide exchanges reflect Taq-polymerase reading errors, in this case the accumulation of nucleotide substitutions indicates that the 5 shown clones in patient 44 may have resulted from independent rearrangement events.
Examples of TCR repertoire similarities within individual LGL patients. Freq indicates clonotypic frequency as the number of identical clonotype sequences divided by total number of clones; and VB-JB, restriction of a given variable and joining beta chain of the TCR. Solid boxes indicate nucleotide exchanges resulting in AA exchange (the affected AAs are highlighted in yellow); dashed boxes and lines, nucleotide exchanges that do not lead to AA exchanges. ‡Total of 11 similar exchanges were found and were not included in the analysis, due to the possibility of Taq-polymerase–generated errors. Of interest, in patient 44 an immunodominant clonotype has 2 “supporting” clonotypes that differ in only 1 amino acid of the CDR3 sequence. Those 2 clonotypes were translated from 2 different nucleotide sequences each (*). In both cases the underlying sequences differed in 2 nucleotides that did not affect the AA sequence. Although we postulate that single nucleotide exchanges reflect Taq-polymerase reading errors, in this case the accumulation of nucleotide substitutions indicates that the 5 shown clones in patient 44 may have resulted from independent rearrangement events.
In addition to information about clonotype sharing and structure, our study provides an estimate of the frequency of clonal CTLs and the degree of decrease in the TCR repertoire variability. In some extreme cases, up to 95% of peripheral CTL repertoire is clonal, a surprising finding in view of the lack of immunodeficiency among patients with LGL leukemia. Under normal circumstances, even the most predominant clones that most likely correspond with the immune responses to ubiquitous antigens comprise only up to 1.4% of the whole repertoire, suggesting that the finding of an extreme expansion of an individual clone is a significant pathologic sign. The individual expansions varied in their contribution to the TCR repertoire among the patients and did not correlate with the severity of cytopenia. It is possible that killing efficiency may also depend on (1) the affinity of the clonal TCR to the antigen and (2) the frequency and distribution of the antigens in target tissues. However, hematologic responses were associated with a decrease in clonal dominance. (In cases in which the initially expanded clonotype was not found by sequencing, its persistence was confirmed by clonotypic PCR, consistent with the high relapse rate of LGL leukemia.)
We have compared LGL clonotypes with each other and with clonotypes found in healthy controls. Clonotypes specific for malignant clones were not encountered to a great extent in healthy individuals. Previously, we have demonstrated using clonotypic PCR that some of the LGL clonotypes may also be found at very low levels in controls. When the disease-associated clonotypes found in our study (and those described by others) were cross-referenced against a clonotypic database containing nearly 5000 clonotypes sequenced in our laboratory and described in the literature in association with various conditions, identical clonotypes derived from patients who underwent allogeneic BMT and some LGL leukemia patients were found. This finding is not surprising, as clonal T-cell expansions have been reported in BMT patients61,81,82 and a common mechanism may be involved. T-cell clonality after BMT results from the physiologic immune reconstitution that is shaped by various triggering antigens, including infectious (eg, cytomegalovirus [CMV]) and allogeneic epitopes. Shared/homologous clonotypes in LGL and after allogeneic BMT may provide insight into the nature of target antigens; for example, minor HLA antigen may drive the corresponding clonotypes after BMT,83 and theoretically such self -“minor” antigens could be targeted in LGL leukemia. This theory is particularly attractive as clonotype sharing was found in the context of the presence of matching HLA antigens.
Clonotypic expansions and disease course. Changes in the size of LGL clone were recorded prior to and after oral cytoxan therapy resulting in remission in 6 patients. (A) Clonotype expansions within CD8 cell population (percent of VB family within CD8+ population multiplied by % of identical sequences within a VB family). (B) Comparisons of redundancy (defined as percent of the pathogenic clone within VB family) before (▦) and after (▨) therapy and diversity (defined as percent of different clones within all clones sequenced). (C-D) Examples of the changes of clonal size expressed as percent of the total CD8 spectrum during the course of disease in 2 patients with LGL leukemia associated with pure red cell aplasia (▦). Lines represent the hemoglobin levels (+) and absolute reticulocyte counts (×). Tx and arrows indicate transfusions; Hg, hemoglobin; and Ret, reticulocytes. Error bars represent 2 SD.
Clonotypic expansions and disease course. Changes in the size of LGL clone were recorded prior to and after oral cytoxan therapy resulting in remission in 6 patients. (A) Clonotype expansions within CD8 cell population (percent of VB family within CD8+ population multiplied by % of identical sequences within a VB family). (B) Comparisons of redundancy (defined as percent of the pathogenic clone within VB family) before (▦) and after (▨) therapy and diversity (defined as percent of different clones within all clones sequenced). (C-D) Examples of the changes of clonal size expressed as percent of the total CD8 spectrum during the course of disease in 2 patients with LGL leukemia associated with pure red cell aplasia (▦). Lines represent the hemoglobin levels (+) and absolute reticulocyte counts (×). Tx and arrows indicate transfusions; Hg, hemoglobin; and Ret, reticulocytes. Error bars represent 2 SD.
Identification of immunodominant clonotypes has not only several pathophysiologic implications but also clinical applications. Clonotypic sequences may be used to monitor the frequency of malignant T-cell clones using, for example, quantitative PCR. Such an approach may also work for polyclonal responses in which clonotypes derived from most significantly expanded T-cell clones may serve as markers for specific autoimmune processes or for responses to vaccines or infectious agents. Finally, the availability of the clonotypic sequences may allow for the measurement of natural and pathologic anti-idiotypic responses as well as clonotype-based anti-idiotypic vaccination as a potential therapeutic modality for T-cell malignancies.
Prepublished online as Blood First Edition Paper, May 24, 2005; DOI 10.1182/blood-2004-10-4045.
Supported in part by grants RO1-HL043429 (J.P.M.), U54-RR019397 (J.P.M.), and RO1-CA113792 (J.P.M.); a grant from the Aplastic Anemia and MDS International Foundation (J.P.M.); and a generous gift from the Trotter family.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
M.W.W. would like to thank his former mentors Dr Martin Digweed and Dr Ilja Demuth for their exceptional skills in training young scientists and Dr Hans-Dieter Volk for his continued guidance.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal