Abstract
Recent reports suggested that parvovirus B19 (B19) might persist in immunocompetent individuals such as blood donors, but only cross-sectional data were available. Serial samples from a cohort of multitransfused patients with hemoglobinopathies and a cross-sectional population of pregnant women were tested for B19 markers. Of 76 red cell recipients, 6 (8%) had persistent viral DNA for 1 to 3 or more years, depending on the sensitivity of the genomic amplification assay. All patients also carried B19-specific immunoglobulin G (IgG). In contrast, 0.8% of 500 pregnant women carried both detectable B19 DNA and specific IgG. These results demonstrate that persistence of low levels of B19 DNA suggested by cross-sectional studies is frequent in multitransfused patients and that the virus may remain detectable several years after infection in nonimmunodeficient individuals. (Blood. 2005;106:2890-2895)
Introduction
Discovered in 1975, the B19 parvovirus (B19) is a small, nonenveloped erythrovirus that can cause a wide range of mild and self-limited clinical manifestations such as erythema infectiosum (fifth disease) and oligoarthritis.1 However, B19 infection can also cause acute anemia by aplastic crisis in patients with shortened red cell survival; chronic anemia in immunocompromised patients, such as those with human immunodeficiency virus infection (HIV), congenital immunodeficiency, malignancies for which they are undergoing chemotherapy, or those who have undergone organ transplantation2,3 ; and it can cause hydrops fetalis or intrauterine death in pregnant women.4
In the classical natural history, acute B19 infection occurring in immunologically competent individuals is controlled by neutralizing antibodies: a transient, high-level viremia is present for less than one week, then declines with the appearance of specific immunoglobulin M (IgM) antibodies, which persist for 8 to 10 weeks,5 and the appearance of lifelong specific IgG antibodies. However, a persistent infection may be observed in immunocompromised patients unable to produce neutralizing antibodies and to clear the virus, leading to chronic B19 carriage with or without anemia.6-8 Persistence of infection in the bone marrow has also been reported in immunocompetent individuals with or without symptoms.9-11
The primary route of transmission of B19 is respiratory (via aerosol droplets), but the infection may also be transmitted by transfusion of blood components; in particular, packed red cells from blood collected during the short preseroconversion viremic phase.12-14 Such an infection may have a serious clinical outcome in 3 categories of susceptible recipients: (1) patients with shortened red cell survival (eg, thalassemia major, sickle cell disease, and other hemolytic diseases); (2) pregnant women; and (3) immunocompromised patients (previously exposed to B19 or not). In immunocompetent recipients, B19 exposure by transfusion is mostly inconsequential, since a large proportion is immune. The risk of exposure to a window period with a high-load B19 viremia is relatively small as the frequency ranges between 1 in 3300 and 1 in 50 000 in blood donors according to seasonal epidemiologic circumstances and the sensitivity of the detection methods.15,16
Recent reports suggested the existence of persistently B19-infected blood donors17,18 and of 1% of blood donors carrying B19 DNA in the presence of specific IgG antibodies in a cross-sectional study.19 These studies, however, suggested but did not prove the existence nor determine the frequency of persistent B19 infection in immunocompetent individuals. For this reason, we have undertaken a study to estimate the frequency of B19 infection persistence in such individuals.
Patients, materials, and methods
Populations and study design
Longitudinal study. The population consisted of 76 nonimmunodeficient patients multitransfused with packed red cells (PRCs) for anemia related to a congenital hemoglobinopathy (thalassemia major or sickle cell disease). All lived in the Paris, France, area and were prospectively followed and transfused in our hospital. The mean duration of the follow-up was 6.9 years.1-21 There were 32 males and 44 females, with a mean age of 27.0 years (range, 1-49 years). During the follow-up of each patient, serum samples were taken at routine visits preceding each transfusion. Blood samples were collected by venipuncture and processed within 2 hours of collection. Serum was separated, aliquoted, and stored frozen at -80°C until tested. An informed consent was obtained from all patients. The protocol was submitted to the approval of the Ethical Committee of Amiens Hospital.
The protocol stated that B19 IgG and B19 DNA would be tested in all patients at baseline visit sample (1988 for the majority of the patients, later if the patient was first seen at a later date) and B19 IgG at the most recent visit (censor date, April 2004 if the patient was still alive, earlier if the patient was dead or lost to follow-up). In case of seroconversion to B19 IgG or presence of B19 DNA, IgG and IgM antibodies to B19 and B19 DNA were tested in a yearly sample until seroconversion and in all available samples afterward.
In order to confirm any result positive for B19 DNA and to detect low viral load in the samples collected after B19 primary infection, the sequential samples were tested 5 times (on a new aliquot) in another laboratory with a polymerase chain reaction (PCR) assay using primer pairs located in a different region of the genome. Another aliquot from the same samples was sent to a third laboratory for the determination of the viral load by real-time quantitative PCR (QPCR), as well as to confirm the results of the 2 previous laboratories.
The cumulated number of lifetime PRCs received by each patient of the longitudinal study was recorded. Two groups of recipients were identified: those receiving more than 100 PRCs and those receiving fewer than 100 PRCs. The prevalence of B19 IgG in the most recent samples was determined in each group.
Cross-sectional study. B19 markers were also studied in 500 pregnant women (mean age, 30.6 years; range, 16-47 years). Blood samples were collected by venipuncture during a systematic routine examination during pregnancy. Serum samples were processed within 2 hours of collection, aliquoted, and stored at -80°C until tested. They were screened for B19 IgM and IgG and for viral DNA by VP1 PCR assay (see “Biological assays”). Each sample positive for B19 DNA was retested on a new aliquot with the same assay. A third aliquot was sent to another laboratory to quantify viral DNA by QPCR and confirm the initial result.
Biologic assays
B19 serologic assays. Testing for B19 antibodies was performed using a commercial assay (Parvovirus B19 IgG or IgM Enzyme Immunoassay Third Generation; Biotrin, Dublin, Ireland), according to the manufacturer's instructions. Capture antigen was recombinant VP2 capsid protein. The ratio of optical density sample to optical density cutoff (S/CO) was calculated for each sample. As S/CO more than 1.01 for IgM and more than 1 for IgG test was considered positive. An S/CO less than 1.0 and less than 0.99 for IgM and IgG test, respectively, was considered negative.
PCR assays. Three procedures of genomic amplification were used, each in a different laboratory: VP1 PCR in laboratory A, NS1 PCR in laboratory B, NS1 QPCR in laboratory C. VP1 PCR and NS1 PCR were used for the screening of B19 DNA-positive samples and for a reciprocal control of specificity of results in 2 different regions of viral genome; QPCR was used to detect and quantify B19 DNA load in serial samples from patients positive for B19 DNA.
VP1 PCR. DNA was extracted from 200 μL serum using the High Pure Viral Nucleic Acid Kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions. Screening for B19 DNA was performed with a nested PCR using primers annealing in the VP1 gene. The first round of amplification was performed using primers 376 and 37720 and the Expand High Fidelity PCR system (Roche Diagnostics) in a 30-μL reaction containing 6 μL DNA. The second round was performed using primers NVP1 (5′ GTG GCA AAT GGT GGG AAA GTG A 3′) and NVP2 (5′ GAT AAG GCA TGG GGG TGG TCA 3′) and the Expand High Fidelity PCR system in a 30-μL reaction containing 2 μL amplified DNA with the first PCR. The program cycles for the 2 PCRs were as follows: one step of denaturation at 94°C for 10 minutes, 10 cycles at 94°C for 30 seconds, at 48°C for 30 seconds, at 68°C for 30 seconds, and 25 cycles at 94°C for 30 seconds, at 48°C for 30 seconds, at 68°C for 30 seconds with 5 seconds by extension/cycle. The final extension was performed at 72°C for 7 minutes. To confirm the positive results, the amplified products were hybridized with a specific biotinylated DNA probe (5′ AAT ATT AAAAGA TCA TTA TAA TAT TTC TTT AGA TAA TCC CC 3′) using the commercial kit Hybridowell Universal (Argene, Varilhes, France). Using the first International Standard for B19 DNA Nucleic Acid Testing (NAT) assays 99/800 (National Institute for Biological Standards and Control [NISBC], Potters Bar, United Kingdom), the limit of detection of the assay was estimated at 200 IU/mL.
NS1 PCR. DNA was extracted from 200 μL serum using the QIAamp DNA Mini kit (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions. Screening of samples for the presence of B19 DNA was performed with a PCR assay using primers e1905f and e1987r located in the NS1 gene (limit of detection: 100 IU/mL). To confirm the positive results, the PCR products were hybridized with a specific biotinylated DNA probe (e1954fp) using a commercial assay (DNA Enzyme Immunoassay Kit; Diasorin, Antony, France).21 The identification of erythrovirus genotype 1, or not 1 (2 or 3) was made by MfeI restriction of the NS1 PCR products.22
NS1 QPCR. The viral load DNA was quantified after DNA extraction from 200 μL serum using a High Pure Viral Nucleic Acid Kit (Roche Diagnostics) according to the manufacturer's instructions. Quantification of B19 DNA was performed with a QPCR assay using primers B19F and B19R located in the NS1 gene. A fluorogenic probe (B19P) was 5′ labeled with VIC and 3′ labeled with 6-carboxy-tetramethyl-rhodamine.19 For each run, the first International Standard for Parvovirus B19 DNA NAT assays 99/800 (NISBC) was used to construct a quantification reference curve. Each sample was tested in duplicate and results were averaged.
Interpretation of data
The 95% B19 DNA detection limit of the real-time QPCR of 50 IU/mL was previously established using the 99/800 standard and Probit software.19 This value does not constitute a cut-off, but a prediction of detection of quantifiable viral DNA. Below this value, signal follows the Poisson distribution, predicting that when the template concentration is low, detectable amplification may or may not take place. When amplification does take place, the value obtained can be plotted against a reference curve and quantified. Although the accuracy of the attributed DNA load is then less than when both replicates are taken into account and above the 95% confidence limit, the value obtained is indicative.
Statistical analysis
Chi-2 square with Yates correction was used when appropriate. The difference was considered as significant when P was less than or equal to .05.
Results
Longitudinal study
Of 76 multitransfused patients, 20 (26.3%) were negative for B19 IgG at the baseline control and at the most recent control, 55 (72.3%) were positive at both time points, and one seroconverted to B19 IgG. The mean age and the mean follow-up period of these 3 groups are shown in Table 1. The mean age of patients positive or negative for B19 IgG at the baseline sample was 32.6 years and 12.5 years, respectively (P = .02). The prevalence of detectable B19 DNA in this population of multitransfused patients positive for B19 IgG at a given time of follow-up was 10.7% (6/56). No positive B19 DNA result was found in B19 IgG-negative patients (0/20). The 6 cases of B19 DNA-positive patients are presented below. No clinical symptom suggestive of a B19 infection was reported during the follow-up. All were transfused since childhood.
Serologic and transfusion data of 76 multitransfused patients in the longitudinal study
B19 lgG at baseline/end of study . | . | . | . | . | Lifetime transfusions, no. (%) . | . | |
|---|---|---|---|---|---|---|---|
| . | No. patients . | No. male/no. female . | Mean age, y (range) . | Mean y of follow-up (range) . | Fewer than 100 transfusions* . | More than 100 transfusions† . | |
| Neg/neg | 20 | 10/10 | 12.5 (1-26) | 7.1 (1-18) | 1 (5.3) | 19 (33.3) | |
| Pos/pos | 55 | 22/33 | 32.6 (4-49) | 6.6 (1-16) | 18 (94.7) | 37‡ (64.9) | |
| Neg/pos | 1 | 0/1 | 13 (NA) | 21 (NA) | 0 (0) | 1 (1.8) | |
B19 lgG at baseline/end of study . | . | . | . | . | Lifetime transfusions, no. (%) . | . | |
|---|---|---|---|---|---|---|---|
| . | No. patients . | No. male/no. female . | Mean age, y (range) . | Mean y of follow-up (range) . | Fewer than 100 transfusions* . | More than 100 transfusions† . | |
| Neg/neg | 20 | 10/10 | 12.5 (1-26) | 7.1 (1-18) | 1 (5.3) | 19 (33.3) | |
| Pos/pos | 55 | 22/33 | 32.6 (4-49) | 6.6 (1-16) | 18 (94.7) | 37‡ (64.9) | |
| Neg/pos | 1 | 0/1 | 13 (NA) | 21 (NA) | 0 (0) | 1 (1.8) | |
NA indicates not applicable.
N = 19.
N = 57.
P = .02.
As shown in Table 1, among the 57 patients having received over 100 lifetime transfusions, 66.7% were positive for B19 IgG. Among the 19 patients having received fewer than 100 lifetime transfusions, 94.7% carried B19 IgG (P = .02). Among the 56 B19 IgG-positive patients of the study, the mean age at the last visit of the most transfused individuals and of the least transfused individuals was 32.8 years and 52.4 years, respectively (P = .02).
Case 1 (infection dated by diagnosis of seroconversion to B19)
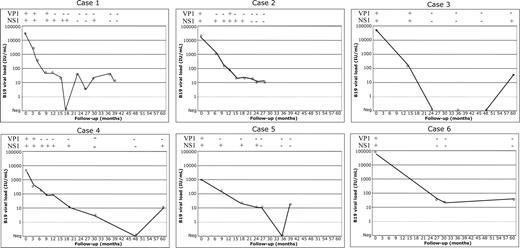
The screening of B19 IgG in the baseline sample and in the most recent sample of the 76 patients identified a case of seroconversion. The B19 IgG, B19 IgM, and B19 DNA status (determined on available sequential samples) of this seroconverter are shown in Figure 1 and Table 2. At entry, the patient was a 13-year-old girl with thalassemia major. The follow-up duration in our hospital was 21 years. Acute B19 infection was documented by the occurrence of B19 DNA in serum, followed one week later by the appearance of B19 IgM and IgG. A slight fever (38.5°C) and a purpura had been noted 7 days after a delivery and 5 days before the first sample positive for B19 DNA. The B19 IgM level regularly decreased over the first 4 months of infection and was no longer detectable fewer than 8 months after infection. The B19 IgG level remained detectable at a high and steady level. B19 DNA was consistently detectable between 2001 and the most recent sample tested in 2004. During the initial 4-month period, the viral load progressively decreased by 2 logs, reaching a level below 100 IU/mL. For the following 2.5 years, B19 DNA load remained consistently low but detectable in 18 of 22 samples tested with QPCR. In contrast, the other 2 less sensitive assays detected viral DNA for 8 and 16 months, respectively.
Evolution over time of parvovirus B19 markers in case 1
Sample date . | No. mo. of follow-up . | B19 IgM, S/CO* . | B19 IgG, S/CO . | VP1 PCR, × 1† . | NS1 PCR, × 5 . | QPCR, × 2 . | Viral load, IU/mL‡ . |
|---|---|---|---|---|---|---|---|
| 1998, 03 Apr | — | − | − | − | NA | NA | NA |
| 2001 | |||||||
| 25 Jan | — | − | − | + | NA | NA | NA |
| 08 Feb | 0 | 9.9 | 6.5 | + | +++++ | ++ | 12 850 |
| 28 Feb | 1 | 11.0 | 7.5 | + | +++++ | ++ | 3 155 |
| 26 Mar | 2 | 6.8 | 7.7 | + | +++++ | ++ | 1 545 |
| 02 May | 3 | 2.8 | 8.0 | + | +++++ | ++ | 1 285 |
| 29 May | 4 | 1.9 | 8.8 | + | +++++ | ++ | 205 |
| 04 Oct | 8 | − | 9.3 | + | +++−− | ++ | 77 |
| 05 Nov | 9 | − | 8.4 | + | +++−− | ++ | 50 |
| 21 Nov | 10 | − | 8.6 | − | +++−− | −− | 0 |
| 26 Dec | 11 | − | 8.6 | − | +++−− | ++ | 22 |
| 2002 | |||||||
| 28 Jan | 12 | − | 10.0 | − | ++−−− | +− | 51 |
| 06 Mar | 14 | − | 7.5 | + | +−−−− | ++ | 67 |
| 08 Apr | 15 | − | 8.4 | − | +−−−− | ++ | 12 |
| 13 May | 16 | − | 8.0 | − | +−−−− | −− | 0 |
| 11 Jun | 17 | − | 9.2 | − | ++−−− | −− | 0 |
| 09 Dec | 23 | − | 7.4 | − | −−−−− | ++ | 30 |
| 2003 | |||||||
| 20 Jan | 24 | − | 7.8 | − | −−−−− | ++ | 8 |
| 20 Mar | 26 | − | 7.5 | − | −−−−− | ++ | 10 |
| 18 Apr | 27 | − | 8.0 | − | −−−−− | +− | 5 |
| 20 Jun | 29 | − | 7.2 | − | −−−−− | +− | 2 |
| 11 Jul | 30 | − | 7.5 | − | +−−−− | ++ | 16 |
| 25 Aug | 31 | − | 7.5 | − | −−−−− | ++ | 37 |
| 26 Sep | 32 | − | 6.8 | − | −−−−− | +− | 20 |
| 24 Nov | 34 | − | 7.4 | − | −−−−− | +− | 21 |
| 22 Dec | 35 | − | 7.3 | − | −−−−− | +− | 9 |
| 2004 | |||||||
| 20 Feb | 37 | − | 7.7 | − | −−−−− | +− | 30 |
| 15 Mar | 38 | − | 7.0 | − | +−−−− | −− | 0 |
| 15 Apr | 39 | − | 7.8 | − | −−−−− | ++ | 12 |
Sample date . | No. mo. of follow-up . | B19 IgM, S/CO* . | B19 IgG, S/CO . | VP1 PCR, × 1† . | NS1 PCR, × 5 . | QPCR, × 2 . | Viral load, IU/mL‡ . |
|---|---|---|---|---|---|---|---|
| 1998, 03 Apr | — | − | − | − | NA | NA | NA |
| 2001 | |||||||
| 25 Jan | — | − | − | + | NA | NA | NA |
| 08 Feb | 0 | 9.9 | 6.5 | + | +++++ | ++ | 12 850 |
| 28 Feb | 1 | 11.0 | 7.5 | + | +++++ | ++ | 3 155 |
| 26 Mar | 2 | 6.8 | 7.7 | + | +++++ | ++ | 1 545 |
| 02 May | 3 | 2.8 | 8.0 | + | +++++ | ++ | 1 285 |
| 29 May | 4 | 1.9 | 8.8 | + | +++++ | ++ | 205 |
| 04 Oct | 8 | − | 9.3 | + | +++−− | ++ | 77 |
| 05 Nov | 9 | − | 8.4 | + | +++−− | ++ | 50 |
| 21 Nov | 10 | − | 8.6 | − | +++−− | −− | 0 |
| 26 Dec | 11 | − | 8.6 | − | +++−− | ++ | 22 |
| 2002 | |||||||
| 28 Jan | 12 | − | 10.0 | − | ++−−− | +− | 51 |
| 06 Mar | 14 | − | 7.5 | + | +−−−− | ++ | 67 |
| 08 Apr | 15 | − | 8.4 | − | +−−−− | ++ | 12 |
| 13 May | 16 | − | 8.0 | − | +−−−− | −− | 0 |
| 11 Jun | 17 | − | 9.2 | − | ++−−− | −− | 0 |
| 09 Dec | 23 | − | 7.4 | − | −−−−− | ++ | 30 |
| 2003 | |||||||
| 20 Jan | 24 | − | 7.8 | − | −−−−− | ++ | 8 |
| 20 Mar | 26 | − | 7.5 | − | −−−−− | ++ | 10 |
| 18 Apr | 27 | − | 8.0 | − | −−−−− | +− | 5 |
| 20 Jun | 29 | − | 7.2 | − | −−−−− | +− | 2 |
| 11 Jul | 30 | − | 7.5 | − | +−−−− | ++ | 16 |
| 25 Aug | 31 | − | 7.5 | − | −−−−− | ++ | 37 |
| 26 Sep | 32 | − | 6.8 | − | −−−−− | +− | 20 |
| 24 Nov | 34 | − | 7.4 | − | −−−−− | +− | 21 |
| 22 Dec | 35 | − | 7.3 | − | −−−−− | +− | 9 |
| 2004 | |||||||
| 20 Feb | 37 | − | 7.7 | − | −−−−− | +− | 30 |
| 15 Mar | 38 | − | 7.0 | − | +−−−− | −− | 0 |
| 15 Apr | 39 | − | 7.8 | − | −−−−− | ++ | 12 |
— indicates not applicable (follow-up began with first sample tested after QPCR); −, negative result; NA, sample not available; +, positive result.
Sample to cut-off.
Number of replicates.
Viral load was calculated by QPCR19 as the mean of 2 duplicate values or a single value when only one replicate was positive.
Follow-up of 6 cases of persistent B19 infection. B19 DNA in case 1 (seroconversion to B19), in cases 2 and 3 (detection of B19 IgM), and in cases 4 to 6 (unknown date of B19 infection) was quantified by QPCR. Serial samples were available over 28 to 60 months. Quantitative results are shown in the graphs; qualitative VP1 and NS1 PCR results of the corresponding samples are shown on top of each graph as + for positive and - for negative. ND indicates not done. Detailed results of all B19 markers for case 1 are shown in Table 2.
Follow-up of 6 cases of persistent B19 infection. B19 DNA in case 1 (seroconversion to B19), in cases 2 and 3 (detection of B19 IgM), and in cases 4 to 6 (unknown date of B19 infection) was quantified by QPCR. Serial samples were available over 28 to 60 months. Quantitative results are shown in the graphs; qualitative VP1 and NS1 PCR results of the corresponding samples are shown on top of each graph as + for positive and - for negative. ND indicates not done. Detailed results of all B19 markers for case 1 are shown in Table 2.
Cases 2 and 3 (detection of B19 IgM)
These 2 cases were characterized by the presence of B19 IgM and of B19 DNA in the first available sample. Case 2 was a 10-year-old boy with thalassemia major. A particularly low level of hemoglobin (50 g/L instead of the usual 80 g/L) was noted in the month preceding the first sample positive for B19 DNA that could be linked to a B19 erythroblastopenia. The duration of follow-up was 2 years. The disappearance of B19 IgM was paralleled by a 2-log decrease of viral load. From the second year, the viral load was lower than 50 IU/mL until the end of follow-up. Case 3 was a 14-year-old boy with thalassemia major. No clinical symptom evoking a B19 infection was noted. The duration of follow-up was 9 years. The viral load decreased 3 logs over the first year, then became undetectable during 32 months before becoming detectable (< 50 IU/mL) until the end of the follow-up. The follow-up of viral load in these 2 patients is shown in Figure 1.
Cases 4 to 6 (unknown date of infection)
The presence of B19 DNA (without B19 IgM) was found in the baseline samples from these patients. The duration of follow-up of these 3 patients was 10, 16, and 5 years, respectively. Case 4 was a 17-year-old boy with thalassemia major. No clinical symptom evoking a B19 infection was noted. Case 5 was a 5-year-old girl with sickle cell disease. A particularly low level of hemoglobin (35 g/L instead of the expected 80 g/L) was noted in the trimester preceding the first sample positive for B19 DNA. Case 6 was a 12-year-old male with thalassemia major. No clinical symptom evoking a B19 infection was noted. The evolution of B19 viral load of the 3 patients is shown in Figure 1.
There were 4 patients (cases 1, 3, 4, and 5) infected with genotype 1 erythrovirus. Case 2 was infected by a genotype other than genotype 1. Case 6 could not be genotyped.
Cross-sectional study
Among the 500 women of the cross-sectional study, 325 (65%) were positive for B19 IgG (mean S/CO: 5.07) and 5 (1%) were positive for B19 IgM (mean optical density [OD] ratio: 1.35). Four women (all positive for B19 IgG and negative for B19 IgM) were positive for B19 DNA by VP1 PCR assay. The B19 IgG (S/CO) and the B19 DNA levels (IU/mL) of these 4 women were 5.4/25, 7.5/22, 6.3/136 and 6.6/17. The prevalence of B19 DNA in IgG-seropositive pregnant women was 1.2%. The mean B19 IgG S/CO ratio of these 4 DNA-positive individuals compared with the 321 who were negative for viral DNA was not significantly different (P = .2). The 5 samples positive for B19 IgM did not contain detectable B19 DNA.
Comparison of assay sensitivity
A total of 75 samples from the 6 serially tested samples identified as B19 DNA carriers were tested with 3 different PCR methods with 95% sensitivity limits of 200 IU/mL, 100 IU/mL, and 50 IU/mL for VP1 PCR, NS1 PCR, and QPCR amplification, respectively. These 3 assays detected B19 DNA in 22.7%, 53.0%, and 74.7% of the tested samples, respectively. Tables 2 and 3 show the comparison of the 3 genomic amplication methods for B19 DNA. Thirty-four samples were positive by both NS1 PCR and QPCR, and 25 were discrepant, 5 and 20 being positive with the NS1 PCR and QPCR, respectively.
Comparison of 3 genomic amplification methods for the detection of parvovirus B19 DNA
. | . | No. positive*samples (%) . | . | . | ||
|---|---|---|---|---|---|---|
| Patient no. . | No. samples tested(%) . | VP1 PCR . | NS1 PCR . | QPCR . | ||
| 1 | 27 | 8 | 16 | 23 | ||
| 2 | 10 | 3 | 7 | 10 | ||
| 3 | 7 | 2 | 3 | 4 | ||
| 4 | 13 | 2 | 7 | 8 | ||
| 5 | 14 | 1 | 6 | 7 | ||
| 6 | 4 | 1 | 1 | 4 | ||
| Total | 75(100) | 17(23) | 40(53) | 56(75) | ||
. | . | No. positive*samples (%) . | . | . | ||
|---|---|---|---|---|---|---|
| Patient no. . | No. samples tested(%) . | VP1 PCR . | NS1 PCR . | QPCR . | ||
| 1 | 27 | 8 | 16 | 23 | ||
| 2 | 10 | 3 | 7 | 10 | ||
| 3 | 7 | 2 | 3 | 4 | ||
| 4 | 13 | 2 | 7 | 8 | ||
| 5 | 14 | 1 | 6 | 7 | ||
| 6 | 4 | 1 | 1 | 4 | ||
| Total | 75(100) | 17(23) | 40(53) | 56(75) | ||
A positive result was defined for PCR as at least 1 of 5 replicates positive and, for QPCR, 1 or 2 replicates positive.
Discussion
It was previously thought that many viruses (including B19) are fully removed from the host by the joint effect of the cellular and humoral immune response within a few weeks following acute infection, and that persistence of B19 and chronic infections were extremely rare.23 However, recent data suggested the possibility that chronic and symptomless B19 carriage may exist in immunocompetent individuals such as blood donors, despite the presence of specific IgG antibodies.19,24 However, these studies were conducted cross-sectionally and did not formally demonstrate persistence.
The present study conducted in a cohort of multitransfused patients with congenital hemoglobinopathy where a sample bank of long-term follow-up was available, identified one patient who seroconverted (case 1; Figure 1, Table 2) and 5 more who were carriers of B19 DNA at study entry. These 5 patients (cases 2-6) had, at entry, a recent infection with B19 indicated either by the presence of detectable IgM in 2 (cases 2 and 3) or a relatively high viral load (1000-80 000 IU/mL) that subsequently declined below 100 IU/mL (cases 4-6). It was felt that this was sufficient evidence to consider these 3 infections as recent and therefore compare them directly to the other 3 cases. The pattern of viremia is remarkably similar between patients (Figure 1), with an initial phase of viral load that declines below 100 IU/mL in the first year after infection followed by another 2 years during which viral load fluctuates between 10 IU/mL and 100 IU/mL. Beyond these initial 3 years, the viral load is consistently below 50 IU/mL and becomes undetectable persistently or occasionally. Of note is that in all 3 patients in whom a sample 5 years after infection was available, B19 DNA remained detectable.
The rate of persistent B19 infections in this study was 7.9% (6/76). Interestingly, this rate is significantly higher than what was previously reported in blood donors where, irrespective of the geographic origin, a prevalence of B19 DNA in IgG carriers was consistently found around 1% (0.6%-1.3%)19 or 0.8% in the present group of pregnant women (P < .001). Several elements may take part in explaining this discrepancy.
Cross-contaminations of samples can be completely ruled out, since the presence of B19 DNA was confirmed on different aliquots of multiple samples in 3 independent laboratories. Moreover, as shown in Table 3, the ability to identify prolonged B19 persistence is directly dependent on the sensitivity of the genomic detection assay used. Indeed, persistence would have lasted less than 15 months and less than 24 months with the VP1 PCR test and the NS1 assay, respectively. However, this does not necessarily imply that low levels of DNA would be detectable with more sensitive amplification assays in all B19 IgG carriers.
The ages of individuals included in studies might also play a role. Figure 1 suggests that beyond 3 years after infection, B19 DNA in persistently infected individuals is not always detectable. Assuming that most pregnant women were likely infected before age 20, since the median age of this group is 30.6 years, we can reasonably suppose that most of them probably had been infected for more than 3 years at the time of testing. Therefore, the detection of B19 DNA was sporadic or absent in this population. However, it can be noted that the relatively low sensitivity of the screening assay used in the study of pregnant women might have underestimated viral persistence, since long-term B19 persistence was not detected. This argument, however, did not apply in the previously reported studies of blood donors tested with the sensitive QPCR used in the present study.19
What could be the mechanism of chronic B19 carriage? It is unlikely that passive transmission of B19 by multitransfusion played a role, since the prevalence in donors is low and the viral load in blood donor population is expected to be low. Reinfection occurrence is also an unlikely phenomenon in view of the presence of high levels of IgG observed in the studied population, as indicated by the consistently high S/CO of the antibody assay. The possibility that fragments of viral DNA instead of full virions might remain in circulation is made unlikely by the detection of viral DNA with primer pairs amplifying 2 distant regions of the genome (NS1 and VP1).
Finally, the relative degree of immunomodulation known to be associated with repeated transfusion might have permitted persistence to establish more frequently in these recipients than in nontransfused individuals such as blood donors.25 It is therefore possible that the frequency of detectable B19 DNA and the profile of viral persistent infection in this cohort of patients might be higher or easier to detect because of a relative immunomodulatory effect induced by multitransfusion.
Previous studies have shown that B19 DNA can be detected in the bone marrow more frequently than in blood, suggesting that some low level of viral replication remains in sanctuaries from which it may occasionally spill into circulation in sufficient quantity to become detectable.26,27 This phenomenon could be at the origin of what was observed in several of our patients who, beyond 3 years after infection, carried or had undetectable viral DNA (Figure 1). It may also be considered that a majority of immunocompetent individuals who have recovered from the infection maintain a sufficiently efficient cellular and neutralizing humoral immune response to either eliminate the virus or keep it to an undetectable level in blood or in the bone marrow. This hypothesis is indirectly supported by the remarkably low frequency of reactivation of B19 infection in seropositive patients submitted to severe immunosuppression.28,29 Alternatively, among the possible mechanisms underlying persistence might be the selection of viral mutants remaining undetected by the host IgG as previously suggested.19 Genetic factors affecting either the specific receptor(s) of B19 or MHC-linked host immune response genes might equally be involved.30
What can be the source of B19 infection in the studied populations? The age of patients 2 to 6 at the time of infection (close to time of entry) ranged between 5 and 17 years, the seroconverting patient 1 being infected at age 31 years. Although B19 transmission by blood transfusion cannot be excluded because all patients were transfused since early childhood, primary infection during the first 2 decades of life is not surprising and suggests respiratory contamination. Moreover, the significant lowest prevalence of B19 IgG in the most transfused patients failed to suggest a contamination by blood products. The low frequency of seroconversion to B19 (1/21) in the seronegative recipients of blood components may be explained by a low risk of community-acquired infection, a low prevalence of circulating B19 DNA in blood products and, when present, a viral load below the threshold of infectivity in the presence of neutralizing antibodies present in the transfused blood products. Although these indirect elements suggest a low risk of B19 transmission by seropositive blood products, such might not be the case in severely immunodeficient recipients such as patients receiving immunosuppressive treatment for bone marrow or solid organ transplants.6,28,31
The serologic results obtained in the pregnant women were not always concordant with the molecular data. Indeed, the presence of B19 IgM at a low level without detectable B19 DNA was unexpected and might be explained by false reactivity related to the physiologic elevation of immune globulins in pregnant women. Contrary to this population, all multitransfused patients with IgM-positive samples were found to contain B19 DNA with the VP1 PCR assay.
The implications of these findings in blood transfusion safety are questionable. The identification of approximately 1% of blood donations containing B19 DNA might theoretically constitute a potential risk, especially for immunodeficient recipients of blood products. Nevertheless, further studies are needed to evidence the presence of neutralizing antibodies that could prevent the infection. Moreover, the minimal infectious dose in presence or in absence of B19 IgG has still to be determined.
Prepublished online as Blood First Edition Paper, June 23, 2005; DOI 10.1182/blood-2005-03-1053.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Véronique Faucher, Mélanie Mercier, Joelle Lerable, Micheline Thauvin, Martine Rocher, Dominique Rocherolle, and Armen Parysan for their helpful assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal