Abstract
Exposure to ionizing radiation (IR) and certain chemotherapeutic agents not only causes acute bone marrow (BM) suppression but also leads to long-term residual hematopoietic injury. This latter effect has been attributed to damage to hematopoietic stem cell (HSC) self-renewal. Using a mouse model, we investigated whether IR induces senescence in HSCs, as induction of HSC senescence can lead to the defect in HSC self-renewal. It was found that exposure of C57BL/6 mice to a sublethal dose (6.5 Gy) of total body irradiation (TBI) resulted in a sustained quantitative and qualitative reduction of LKS+ HSCs. In addition, LKS+ HSCs from irradiated mice exhibited an increased expression of the 2 commonly used biomarkers of cellular senescence, p16Ink4a and SA-β-gal. In contrast, no such changes were observed in irradiated LKS- hematopoietic progenitor cells. These results provide the first direct evidence demonstrating that IR exposure can selectively induce HSC senescence. Of interest, the induction of HSC senescence was associated with a prolonged elevation of p21Cip1/Waf1, p19Arf, and p16Ink4a mRNA expression, while the expression of p27Kip1 and p18Ink4c mRNA was not increased following TBI. This suggests that p21Cip1/Waf1, p19Arf, and p16Ink4a may play an important role in IR-induced senescence in HSCs.
Introduction
Bone marrow (BM) suppression is the primary cause of death after accidental or intentional exposure to a moderate or high dose of total body irradiation (TBI) and the most common dose-limiting side effect of conventional cancer therapy using ionizing radiation (IR) and/or certain chemotherapeutic agents. It has been well established that acute myelosuppression induced by IR and/or chemotherapy is the result of induction of apoptosis in the rapidly proliferating hematopoietic progenitor cells (HPCs) and to a lesser degree in the relatively quiescent hematopoietic stem cells (HSCs).1,2 The acute myelosuppression is an immediate concern for the victims of nuclear accidents and terrorism and for patients undergoing cancer treatment, as it could cause high mortality and morbidity and worsen the outcome of cancer treatment. However, management of acute myelosuppression has been significantly improved in recent years by the use of various hematopoietic growth factors (HGFs) such as granulocyte-colony stimulating factor, granulocyte/macrophage-colony stimulating factor, or erythropoietin.3 These HGFs have the ability to promote the recovery of BM hematopoietic function primarily by stimulating HSC and HPC proliferation and differentiation.
Many patients exposed to a moderate or high-dose TBI or undergoing chemotherapy and/or radiotherapy also exhibit long-term residual damage to BM hematopoietic function manifested by a defect in HSC self-renewal and a decrease in HSC reserves.4,5 Unlike acute myelosuppression, the residual BM damage is latent but long lasting and shows little tendency for recovery. Because of this latency, its clinical implications have been largely overlooked. Moreover, the importance of the residual damage may be further obscured by the seemingly complete recovery of peripheral-blood cell counts, BM cellularity, and number of colony-forming units (CFUs) after the use of HGFs for ameliorating IR- and/or chemotherapy-induced acute hematopoietic toxicity. In fact, the use of HGFs may worsen IR- and/or chemotherapy-induced residual BM damage by promoting HSC and HPC proliferation and differentiation at the expense of HSC self-renewal.6 This could lead to an augmented exhaustion of HSCs and further compromise the long-term recovery of BM hematopoietic function.
Although the residual BM damage is latent, it can manifest as a hypoplastic syndrome at later times or under hematopoietic stress, such as subsequent cycles of consolidation cancer treatment or autologous BM transplantation.4,5 Unfortunately, the mechanisms involved in residual BM injury have not been clearly defined. It has been hypothesized that the defect in HSC self-renewal might be attributable in part to the induction of HSC senescence by IR and/or chemotherapeutic agents, because HSCs appear to have a finite ability for self-renewal and may undergo senescence or exhaustion after severe depletion.7-9 However, cellular and molecular evidence for HSC senescence in residual BM injury is lacking. Two recent observations call into question whether HSCs have a limited replicative capacity. First, the observation that a single transplanted stem cell is capable of completely repopulating the host's ablated hematopoietic system demonstrates the enormous power of HSC self-renewal.10 Second, the calculations of HSC self-renewal based on the competitive repopulating unit have shown that serial transplantation does not result in the reduction of the ability of HSC self-renewal as previously suggested.11 Therefore, it is important to re-evaluate whether IR and/or chemotherapeutic agents induce HSC senescence, particularly at the cellular and molecular levels. Our recent studies showed that a majority of murine BM hematopoietic cells including HSCs died by apoptosis after exposure to a moderate dose of IR in vitro. However, a subset of these cells survived IR damage up to 35 days in a long-term BM-cell culture but lost their clonogenic function.12 These surviving cells exhibited an increased SA-β-gal activity, a biomarker for senescent cells,13 and expressed elevated levels of the proteins (p16Ink4a and p19Arf) encoded by the Ink4a-Arf locus, whose expression has been implicated in the establishment and maintenance of senescence by direct inhibition of various cyclin-dependent kinases (CDKs).12,14,15 A similar result was also found in murine BM hematopoietic cells that were briefly incubated with busulfan.12 These data demonstrate that exposure to IR or treatment with busulfan can induce BM hematopoietic cell senescence in vitro. However, it is not known if these in vitro findings can be extrapolated into an in vivo setting where a whole animal is irradiated. Furthermore, it has yet to be determined if hematopoietic-cell senescence actually occurs at the HSC level. Therefore, the present study was undertaken to specifically examine whether IR induces HSC senescence in vivo using a murine TBI model.
Materials and methods
Reagents
Anti-Sca-1-PE, anti-c-kit-APC; biotin-conjugated rat anti-CD3ϵ, anti-CD45R/B220, anti-Gr-1, anti-Mac-1, and anti-Ter-119; purified rat anti-CD16/CD32; and streptavidin-FITC were purchased from BD-PharMingen (San Diego, CA). Anti-p16Ink4a antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Recombinant mouse thrombopoietin (TPO), stem-cell factor (SCF), and interleukin-3 (IL-3) were purchased from R & D Systems (Minneapolis, MN).
Mice
Male C57BL/6-Ly-5.2 and C57BL/6-Ly-5.1 mice were purchased from The Charles River Laboratories (Wilmington, MA) and The Jackson Laboratories (Bar Harbor, ME), respectively. Heterozygous breeding pairs of Ink4a/Arf (FVB/N.129-Cdkn2atm1Rdp, p16Ink4a-/-/p19Arf-/-) knock-out mice were obtained from the NCI Mouse Models of Human Cancers Consortium (MMHCC) and bred at the Medical University of South Carolina (MUSC) AAALAC certified animal facility. After genotyping by polymerase chain reaction (PCR), homozygous Ink4a/Arf KO mice and wild-type littermates were selected for the study. All the mice were housed 4 in a cage and received food and water ad libitum. All mice were used at approximately 8 to 12 weeks of age. The Institutional Animal Care and Use Committee of MUSC approved all experimental procedures used in this study.
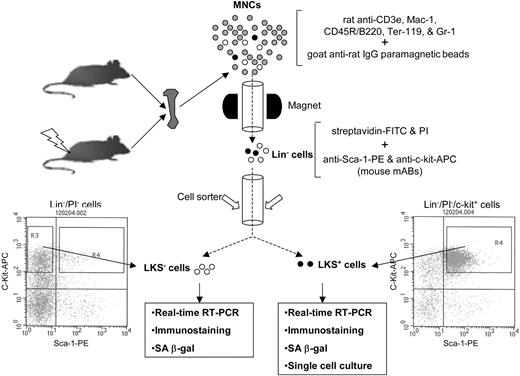
Schematic illustration of the isolation of Lin-, LKS+, and LKS- cells. BM-MNCs and Lin- cells were isolated as described previously.12 Lin- cells were labeled with streptavidin-FITC, anti-Sca-1-PE, and anti-c-Kit-APC after preincubation with anti-CD16/32 to block Fcγ receptors. After washing, the cells were resuspended in PBS containing 1 μg/mL propidium iodide (Molecular Probes) and then sorted immediately using a FACStarPlus cell sorter (Becton Dickinson). LKS- cells were sorted directly from R3 gate. LKS+ cells were first enriched by collecting c-Kit+ cells via presorting and then sorted from R4 gate.
Schematic illustration of the isolation of Lin-, LKS+, and LKS- cells. BM-MNCs and Lin- cells were isolated as described previously.12 Lin- cells were labeled with streptavidin-FITC, anti-Sca-1-PE, and anti-c-Kit-APC after preincubation with anti-CD16/32 to block Fcγ receptors. After washing, the cells were resuspended in PBS containing 1 μg/mL propidium iodide (Molecular Probes) and then sorted immediately using a FACStarPlus cell sorter (Becton Dickinson). LKS- cells were sorted directly from R3 gate. LKS+ cells were first enriched by collecting c-Kit+ cells via presorting and then sorted from R4 gate.
Ionizing radiation
Mice were exposed to various doses of IR in a JL Shepherd Model 143 137Cesium γ-irradiator (JL Shepherd, Glendale, CA) at a rate of 2.4 Gy/min. Mice were irradiated on a rotating platform.
Isolation of BM mononuclear cells (BM-MNCs), lineage negative hematopoietic (Lin-) cells, and LKS+ and LKS- cells
BM-MNCs and Lin- cells were isolated as described previously.12 Lin- cells were labeled with streptavidin-FITC, anti-Sca-1-PE, and anti-c-Kit-APC after preincubation with anti-CD16/32 to block the Fcγ receptors. After washing, the cells were resuspended in PBS containing 1 μg/mL propidium iodide (Molecular Probes, Eugene, OR) and sorted using a FACStarPlus cell sorter (Becton Dickinson, San Jose, CA) as illustrated in Figure 1.
Phenotypic analysis of BM-MNCs by flow cytometry
Briefly, 1 × 106 BM-MNCs were incubated with biotin-conjugated antibodies against CD3ϵ, CD45R/B220, Gr-1, Mac-1, and Ter-119 and then with streptavidin-FITC. After incubation with anti-CD16/CD32 antibody, the cells were stained with anti-Sca-1-PE and anti-c-kit-APC antibodies. For each sample, a minimum of 200 000 cells was analyzed on a FACS Caliber (Becton Dickinson) and the data were analyzed using CellQuest software (Becton Dickinson) after gating on viable cells.
Colony-forming cell (CFC) assay and cobblestone area-forming cell (CAFC) assay
The CFC assay was performed by culturing BM-MNCs in MethoCult GF M3434 methylcellulose medium (Stem Cell Technologies, Vancouver, BC). Colonies of CFU-granulocyte macrophage (GM) and burst-forming unit-erythroid (BFU-E) were scored on day 7 and those of CFU-granulocyte, -erythrocyte, -monocyte, and -megakaryocyte (GEMM) on day 12 of the incubation according to the manufacturer's protocol. The CAFC assay was performed as described elsewhere.12
Real-time reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated from BM-MNCs using TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer's protocol. RNA yield and quality were determined by measuring absorbencies at 260 nm and 280 nm, respectively. First-strand cDNA was synthesized from 2 μg total RNA using SuperScript II first-strand synthesis system (Invitrogen) according to the manufacturer's protocol. PCR primers and Taqman MGB probes for the cyclin-dependent kinase inhibitors (CDKIs) p21Cip1/Waf1, p27Kip1, p19Arf, p16Ink4a, and p18Ink4c and the housekeeping gene β2-microglobulin (B2m) were obtained from Applied Biosystems (Foster City, CA) (Tables 1, 2). The PCR primers and Taqman MGB probes for p19Arf and p16Ink4a were designed by Krishnamurthy et al to distinguish the 2 transcripts.16 β2-M cDNA was simultaneously amplified as an endogenous reference gene. All samples were analyzed in triplicate using an ABI Prism 7000 Sequence Detection System (Applied Biosystems). The threshold cycle (CT) values for each reaction were determined and averaged using TaqMan SDS analysis software (Applied Biosystems). The changes in CDKI gene expression were calculated by the comparative CT method (fold changes = 2[-ΔΔCT]) as described previously.17
List of primers and probes used for real-time RT-PCR analysis of p16Ink4a and p19Arf mRNA
Gene . | Cdkn2a (p16Ink4a) . | Cdkn2a (p19Arf) . |
|---|---|---|
| Forward primer | CGGTCGTACCC; CGATTCAG | TGAGGCTAGAGAG; GATCTTGAGAAG |
| Reverse primer | GCACCGTAGTT; GAGCAGAAGAG | GTGAACGTTGCCCA; TCATCATC |
| Probe (5′-FAM, 3′-Tamra) | AACGTTGCCCA; TCATCA | ACCTGGTCCAG; GATTC |
| Reference sequence | AF044336 | NM_009877 |
Gene . | Cdkn2a (p16Ink4a) . | Cdkn2a (p19Arf) . |
|---|---|---|
| Forward primer | CGGTCGTACCC; CGATTCAG | TGAGGCTAGAGAG; GATCTTGAGAAG |
| Reverse primer | GCACCGTAGTT; GAGCAGAAGAG | GTGAACGTTGCCCA; TCATCATC |
| Probe (5′-FAM, 3′-Tamra) | AACGTTGCCCA; TCATCA | ACCTGGTCCAG; GATTC |
| Reference sequence | AF044336 | NM_009877 |
All primers and probes were ordered from Applied Biosystems.
Primers and probes used for real-time RT-PCR analysis
Gene . | ABI assay ID . | Reference sequence . |
|---|---|---|
| Colkn1b | Mm00438168_ml | NM_009875 |
| Cdkn1a | Mm00432448_ml | NM_007669 |
| Cdkn2c | Mm00483243_ml | NM_004671 |
| B2m | Mm00437762_ml | NM_009735 |
Analysis of p16Ink4a mRNA expression in sorted LKS+ and LKS- cells
Total cellular RNA was extracted from about 15 000 sorted LKS+ cells and LKS- cells using the RNeasy Mini Kit (Qiagen Sciences, Germantown, MD) according to the manufacturer's instructions. The expression of p16Ink4a mRNA was determined by real-time RT-PCR.
Analysis of p16Ink4a expression by flow cytometry
Lin- cells were labeled with anti-Sca-1-PE and anti-c-Kit-APC antibodies and then fixed with BD-Cytofix/Cytoperm buffer (BD-PharMingen). After permeabilization with BD-CytopermPlus buffer, the cells were refixed with BD-Cytofix/Cytoperm buffer, washed, and then stained with either mouse p16Ink4a specific monoclonal antibody (F-12 mAB; Santa Cruz Biotechnology) or normal goat serum (as control) and FITC-conjugated goat anti-mouse IgG. The percentage of p16Ink4a positively stained cells was determined in LKS+ and LKS- cell gates using a FACS Caliber (Becton Dickinson).
Analysis of p16Ink4a expression by immunofluorescent microscopy
Sorted LKS+ cells and LKS- cells were cytospun onto slides, fixed, permeabilized, and immunostained with a mouse p16Ink4a specific monoclonal antibody and rhodamine-conjugated goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA) as described previously.12 After DNA was labeled with Hoechst 33342 (Molecular Probes), slides were mounted in Gel/Mount (Biomeda, Foster, CA). Cells were viewed and photographed using an Axioplan research microscope (Carl Zeiss, Jena, Germany) equipped with a 100 W mercury light source and a 25×/1.3 numeric aperture Plan-Neofluar objective. Images were captured with a Dage CCD100 integrating camera (Dage-MTI, Michigan City, IN) and a Flashpoint 128 capture board (Integral Technologies, Indianapolis, IN). Captured images were processed using Image Pro Plus software (Media Cybernetics, Silver Spring, MD) and displayed using Adobe Photoshop version 6.0 (Adobe Systems, San Jose, CA).
SA-β-gal activity analysis
SA-β-gal activity in sorted LKS+ cells and LKS- cells was determined using a SA-β-gal staining kit from Cell Signaling Technology (Beverly, MA) according to the manufacturer's instructions and our previously reported procedures.12
Single-cell culture
Single-cell cultures of sorted LKS+ cells were performed as described previously.18 Briefly, single LKS+ cells were deposited into wells of round-bottomed 96-well plates (Corning, Corning, NY) using a FACVantage cell sorter (Becton Dickinson). Each cell was incubated in Stempro-34 serum-free medium (Invitrogen) supplemented with 2 mM l-glutamine, 5 × 10-5 M 2-β-mercaptoethanol, 100 ng/mL mouse SCF, and 100 ng/mL mouse TPO (for the analysis of LKS+ cell proliferation) or plus 10% fetal bovine serum and 10 ng/mL mouse IL-3 (for clonogenic assay). The number of cells per well was monitored on days 1, 3, and 7, and the final evaluation of cell division and colony formation was conducted on day 14 of the culture. Only the wells containing one cell on day 1 of inspection were included in the analysis.
Competitive repopulation assay (CRA)
Five-hundred sorted LKS+ cells from 5 control or irradiated C57BL/6-Ly-5.1 mice (4 weeks after 6.5 Gy TBI) were mixed with 2 × 105 competitive BM-MNCs pooled from 3 C57BL/6-Ly-5.2 mice. They were transplanted into lethally irradiated (9.5 Gy TBI) C57BL/6-Ly-5.2 mice (10 recipients/group) by tail-vein injection. For analysis of engraftment, peripheral-blood samples were obtained from the retro-orbital plexus using heparin-coated micropipets (Drummond Scientific, Broomall, PA) at 1 and 2 months after transplantation from all the recipients. After red blood cells had been lysed by 0.15 M NH4Cl, the samples were stained with FITC-conjugated anti-Ly-5.1 and analyzed for donor-derived cells on a FACS Caliber (Becton Dickinson). Donor-derived cells (Ly-5.1) cells in T-cell, B-cell, granulocyte, and monocyte/macrophage lineages were also analyzed at 2 months after transplantation by staining with PE-conjugated anti-Thy-1.2, PE-conjugated anti-B220, PE-conjugated anti-Gr-1, and PE-conjugated anti-Mac-1, respectively.
Statistical analysis
The data were analyzed by analysis of variance (ANOVA). In the event that ANOVA justified post hoc comparisons between group means, these were conducted using the Student-Newman-Keuls test for multiple comparisons. For experiments in which only single experimental and control groups were used, group differences were examined by unpaired Student t test. Differences were considered significant at P < .05. All of these analyses were done using GraphPad Prism from GraphPad Software (San Diego, CA).
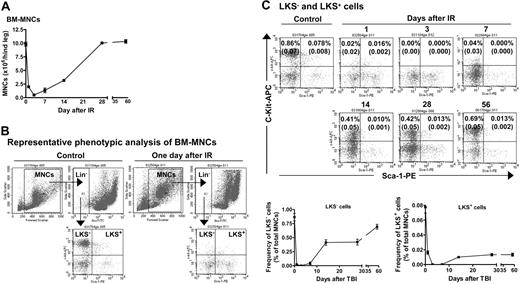
TBI induced hematopoietic suppression. (A) TBI induces a transient decrease in the number of BM-MNCs. Mice were exposed to 6.5 Gy TBI. At 1, 3, 7, 14, 28, and 56 days after TBI, BM-MNCs were isolated and enumerated. The results are expressed as the mean of BM-MNCs/hind leg ± SE (n = 3 mice/group). A group of 6 unirradiated mice was used as control (day 0). (B) Representative phenotypic analysis of BM-MNCs by flow cytometry. BM-MNCs (1 × 106) from control or irradiated mice were incubated with biotin-conjugated antibodies against CD3e, CD45R/B220, Gr-1, Mac-1, and Ter-119 and then with streptavidin-FITC. After incubation with anti-CD16/CD32 antibody, they were stained with anti-Sca-1-PE and anti-c-kit-APC antibodies. Cells incubated with appropriate conjugated rat antimouse isotypes were included as controls. For each sample, a minimum of 200 000 cells was analyzed on a FACS Caliber (Becton Dickinson) and the resultant data were analyzed using CellQuest software (Becton Dickinson) after gating on viable cells. (C) TBI induces a sustained reduction in the frequencies of LKS- and LKS+ cells in BM-MNCs. The frequencies of LKS- and LKS+ cells in BM-MNCs from control and irradiated mice were determined by flow cytometry as illustrated in panel B. Top panel: Representative flow cytometric analyses of LKS- and LKS+ cells in BM-MNCs from control or irradiated mice. The frequencies of LKS- and LKS+ cells are indicated in the LKS- and LKS+ cell gates as a percentage of total BM-MNCs. Bottom panel: Kinetics of LKS- and LKS+-cell recovery after TBI. The frequencies of LKS- and LKS+ cells are expressed as a mean percentage of BM-MNCs ± SE (n = 4 to 6). ANOVA analysis reveals that TBI induced a time-dependent reduction in the number of BM-MNCs and frequencies of LKS- and LKS+ cells (P < .001).
TBI induced hematopoietic suppression. (A) TBI induces a transient decrease in the number of BM-MNCs. Mice were exposed to 6.5 Gy TBI. At 1, 3, 7, 14, 28, and 56 days after TBI, BM-MNCs were isolated and enumerated. The results are expressed as the mean of BM-MNCs/hind leg ± SE (n = 3 mice/group). A group of 6 unirradiated mice was used as control (day 0). (B) Representative phenotypic analysis of BM-MNCs by flow cytometry. BM-MNCs (1 × 106) from control or irradiated mice were incubated with biotin-conjugated antibodies against CD3e, CD45R/B220, Gr-1, Mac-1, and Ter-119 and then with streptavidin-FITC. After incubation with anti-CD16/CD32 antibody, they were stained with anti-Sca-1-PE and anti-c-kit-APC antibodies. Cells incubated with appropriate conjugated rat antimouse isotypes were included as controls. For each sample, a minimum of 200 000 cells was analyzed on a FACS Caliber (Becton Dickinson) and the resultant data were analyzed using CellQuest software (Becton Dickinson) after gating on viable cells. (C) TBI induces a sustained reduction in the frequencies of LKS- and LKS+ cells in BM-MNCs. The frequencies of LKS- and LKS+ cells in BM-MNCs from control and irradiated mice were determined by flow cytometry as illustrated in panel B. Top panel: Representative flow cytometric analyses of LKS- and LKS+ cells in BM-MNCs from control or irradiated mice. The frequencies of LKS- and LKS+ cells are indicated in the LKS- and LKS+ cell gates as a percentage of total BM-MNCs. Bottom panel: Kinetics of LKS- and LKS+-cell recovery after TBI. The frequencies of LKS- and LKS+ cells are expressed as a mean percentage of BM-MNCs ± SE (n = 4 to 6). ANOVA analysis reveals that TBI induced a time-dependent reduction in the number of BM-MNCs and frequencies of LKS- and LKS+ cells (P < .001).
Results
TBI induced long-term hematopoietic damage
In a preliminary study, we found that exposure of mice to increasing doses (2, 4, and 6.5 Gy) of TBI reduced the number of BM-MNCs and the frequencies of CFUs and LKS- and LKS+ cells in BM-MNCs in a dose- and time-dependent manner (data not shown). To further characterize TBI-induced residual BM injury, mice were exposed to a sublethal dose (6.5 Gy) of TBI selected from this preliminary study. Changes in the number of BM-MNCs and frequencies of CFUs and LKS- and LKS+ cells in BM-MNCs were monitored for up to 8 weeks after TBI. As shown in Figure 2 and Table 3, exposure to 6.5 Gy TBI caused a rapid decrease in all of these parameters. The nadir for the frequency of CFUs was reached 1 day after TBI and that for the number of BM-MNCs and frequencies of LKS- and LKS+ cells was attained at 3 days after TBI. Thereafter, the number of BM-MNCs and frequencies of CFUs and LKS- and LKS+ cells rapidly recovered in a time-dependent manner. Twenty-eight days after TBI, the number of BM-MNCs returned to a normal level. In contrast, the frequencies of CFUs and LKS- and LKS+ cells remained below normal even at 56 days after TBI. Strikingly, 56 days after TBI the frequency of LKS+ cells in BM-MNCs from irradiated mice was only about 17% of that of control. These findings clearly demonstrate that exposure to IR causes long-term hematopoietic damage mainly in the HSC compartment.
Kinetics of recovery of various CFUs in BM-MNCs after TBI
Day after TBI . | CFU-GM . | BFU-E . | CFU-GEMM . |
|---|---|---|---|
| CTL | 66.8 ± 8.8 | 12.7 ± 2.3 | 3.3 ± 1.6 |
| 1 | 5.7 ± 1.0 | 0.2 ± 0.4 | nd |
| 3 | 8.0 ± 2.4 | 1.2 ± 1.2 | 0.3 ± 0.4 |
| 7 | 14.8 ± 8.5 | 1.5 ± 0.5 | 0.8 ± 0.9 |
| 14 | 27.0 ± 13.4 | 4.0 ± 3.2 | 1.5 ± 0.5 |
| 28 | 33.3 ± 8.7 | 8.7 ± 4.5 | 1.0 ± 1.0 |
| 56 | 40.4 ± 8.2 | 8.6 ± 3.6 | 1.7 ± 0.8 |
Day after TBI . | CFU-GM . | BFU-E . | CFU-GEMM . |
|---|---|---|---|
| CTL | 66.8 ± 8.8 | 12.7 ± 2.3 | 3.3 ± 1.6 |
| 1 | 5.7 ± 1.0 | 0.2 ± 0.4 | nd |
| 3 | 8.0 ± 2.4 | 1.2 ± 1.2 | 0.3 ± 0.4 |
| 7 | 14.8 ± 8.5 | 1.5 ± 0.5 | 0.8 ± 0.9 |
| 14 | 27.0 ± 13.4 | 4.0 ± 3.2 | 1.5 ± 0.5 |
| 28 | 33.3 ± 8.7 | 8.7 ± 4.5 | 1.0 ± 1.0 |
| 56 | 40.4 ± 8.2 | 8.6 ± 3.6 | 1.7 ± 0.8 |
Mice were exposed to 6.5 Gy TBI or unirradiated as control (CTL). At 1, 3, 7, 14, 28, and 56 days after TBI, BM-MNCs were isolated from irradiated and unirradiated mice. A CFC assay was performed. The results are presented as mean CFUs per 5 × 104 BM-MNCs ± SE (n = 3). ANOVA analysis revealed that TBI caused a significant reduction in the frequencies of CFU-GM, BFU-E, and CFU-GEMM in BM-MNCs in a time-dependent manner compared with the control values (P < .001). nd indicates not detectable.
TBI perturbed the expression of various CDKIs' mRNA in BM-MNCs
CDKIs are low-molecular-weight proteins that are capable of regulating cell-cycle progress via interaction with CDKs.14,15 Among various CDKIs, p21Cip1/Waf1, p27Kip1, p19Arf, p16Ink4a, and p18Ink4c have been extensively investigated in HPCs and HSCs and implicated in the regulation of HSC self-renewal and/or HPC expansion.19-24 Therefore, we examined whether IR-induced residual BM damage is associated with a perturbation of p21Cip1/Waf1, p27Kip1, p19Arf, p16Ink4a, and p18Ink4c mRNA expression. The expression of these CDKIs' mRNA and mRNA for the reference gene B2m was quantified by real-time RT-PCR as shown in Figure 3. The results of the analysis are summarized in Table 4 after the data were transformed to fold changes from control values for each of these CDKIs. As shown in Table 4, exposure of mice to TBI resulted in an immediate increase in mRNA expression for p21Cip1/Waf1 (16.55-fold increase) and p16Ink4a (4.27-fold increase) in BM-MNCs one day after TBI. The increase in p21Cip1/Waf1 mRNA gradually declined over a course of 4 weeks and remained modestly elevated (2.1-fold) at 28 days after TBI but became insignificant (< 2-fold) by day 56 after TBI. The expression of p16Ink4a mRNA expression remained elevated for up to 8 weeks after IR, with a pick time around day 14 (10.54-fold). The expression of p19Arf mRNA expression in irradiated BM-MNCs was initially reduced but was elevated 7 days after TBI (4.60-fold), peaked at 14 days after IR (8.23-fold), and remained significantly elevated out to 56 days (> 4-fold). In contrast, transient reductions in p27Kip1 and p18Ink4c mRNA expression were found in BM-MNCs shortly after IR. These results suggest that IR-induced long-term hematopoietic damage is primarily associated with the prolonged elevation of p21Cip1/Waf1, p19Arf, and p16Ink4a mRNA expression.
TBI-induced fold changes in CDKIs' mRNA expression in BM-MNCs
. | Days after TBI . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
. | 1 . | 3 . | 7 . | 14 . | 28 . | 56 . | |||||
| p21Cip1/Waf1 | 16.55 ± 4.55 | 10.37 ± 0.62 | 8.15 ± 1.65 | 3.60 ± 1.08 | 2.10 ± 0.40 | 1.38 ± 0.15 | |||||
| p27Kip1 | –2.75 ± 0.25 | –4.15 ± 1.15 | –1.20 ± 0.10 | 1.15 ± 0.05 | 1.70 ± 0.20 | 1.47 ± 0.15 | |||||
| p16Ink4a | 4.27 ± 1.33 | 9.59 ± 2.21 | 4.94 ± 0.77 | 10.54 ± 2.60 | 5.76 ± 1.44 | 4.63 ± 2.02 | |||||
| p19Arf | –3.83 ± 0.67 | 0.52 ± 0.58 | 4.60 ± 0.52 | 8.23 ± 2.72 | 4.28 ± 0.93 | 4.21 ± 0.66 | |||||
| p18Ink4c | –7.20 ± 0.30 | –5.00 ± 1.50 | 1.10 ± 0.10 | 1.00 ± 0.10 | 1.15 ± 0.05 | 1.35 ± 0.05 | |||||
. | Days after TBI . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
. | 1 . | 3 . | 7 . | 14 . | 28 . | 56 . | |||||
| p21Cip1/Waf1 | 16.55 ± 4.55 | 10.37 ± 0.62 | 8.15 ± 1.65 | 3.60 ± 1.08 | 2.10 ± 0.40 | 1.38 ± 0.15 | |||||
| p27Kip1 | –2.75 ± 0.25 | –4.15 ± 1.15 | –1.20 ± 0.10 | 1.15 ± 0.05 | 1.70 ± 0.20 | 1.47 ± 0.15 | |||||
| p16Ink4a | 4.27 ± 1.33 | 9.59 ± 2.21 | 4.94 ± 0.77 | 10.54 ± 2.60 | 5.76 ± 1.44 | 4.63 ± 2.02 | |||||
| p19Arf | –3.83 ± 0.67 | 0.52 ± 0.58 | 4.60 ± 0.52 | 8.23 ± 2.72 | 4.28 ± 0.93 | 4.21 ± 0.66 | |||||
| p18Ink4c | –7.20 ± 0.30 | –5.00 ± 1.50 | 1.10 ± 0.10 | 1.00 ± 0.10 | 1.15 ± 0.05 | 1.35 ± 0.05 | |||||
Mice were exposed to 6.5 Gy TBI or unirradiated as control. At 1, 3, 7, 14, 28, and 56 days after TBI, BM-MNCs were isolated from irradiated and unirradiated mice. RNA was extracted from these cells and analyzed by real-time RT-PCR. The relative quantity of individual CDKIs' mRNA was calculated for each sample after normalization with the expression of B2m mRNA and is presented as fold changes from control value ± SE (n = 3-8).
Analysis of mRNA expression for β2-M and CDKIs by real-time RT-PCR. (A) Dose-dependent amplification of β2-M mRNA by real-time RT-PCR. Top panel: The intensities of fluorescence signal of the FAM dye (delta Rn) are plotted on the y-axis and the cycle number is plotted on the x-axis. The amplification plots shifted to the right as RNA levels were reduced (from 100 ng to 10 pg, 1:10 diluted). The threshold cycle (CT) is defined as the cycle number at which the delta Rn of a sample crossed a threshold of 10 SD above baseline fluorescence. Bottom panel: The CT value is plotted on the y-axis and the amount of RNA input is plotted on the x-axis. The CT value decreases linearly with the RNA input. (B) Time-dependent induction of mRNA for CDKIs by TBI. Mice were exposed to 6.5 Gy TBI or unirradiated as control. At 1, 3, 7, 14, 28, and 56 days after TBI, BM-MNCs were isolated from irradiated and unirradiated mice and then RNA was extracted from these cells and analyzed by real-time RT-PCR to determine mRNA expression for various CDKIs. A representative analysis of CDKI mRNA expression by real-time RT-PCR is presented as mean ± SD of triplicate (ΔΔCT-CDKI = ΔCT-IR - ΔCT-CTL; ΔCT-IR = CT-IR/CDKI - CT-IR/β2-M; ΔCT-CTL = CT-CTL/CDKI - CT-CTL/β2-M).
Analysis of mRNA expression for β2-M and CDKIs by real-time RT-PCR. (A) Dose-dependent amplification of β2-M mRNA by real-time RT-PCR. Top panel: The intensities of fluorescence signal of the FAM dye (delta Rn) are plotted on the y-axis and the cycle number is plotted on the x-axis. The amplification plots shifted to the right as RNA levels were reduced (from 100 ng to 10 pg, 1:10 diluted). The threshold cycle (CT) is defined as the cycle number at which the delta Rn of a sample crossed a threshold of 10 SD above baseline fluorescence. Bottom panel: The CT value is plotted on the y-axis and the amount of RNA input is plotted on the x-axis. The CT value decreases linearly with the RNA input. (B) Time-dependent induction of mRNA for CDKIs by TBI. Mice were exposed to 6.5 Gy TBI or unirradiated as control. At 1, 3, 7, 14, 28, and 56 days after TBI, BM-MNCs were isolated from irradiated and unirradiated mice and then RNA was extracted from these cells and analyzed by real-time RT-PCR to determine mRNA expression for various CDKIs. A representative analysis of CDKI mRNA expression by real-time RT-PCR is presented as mean ± SD of triplicate (ΔΔCT-CDKI = ΔCT-IR - ΔCT-CTL; ΔCT-IR = CT-IR/CDKI - CT-IR/β2-M; ΔCT-CTL = CT-CTL/CDKI - CT-CTL/β2-M).
TBI selectively induces p16Ink4a expression in LKS+ cells
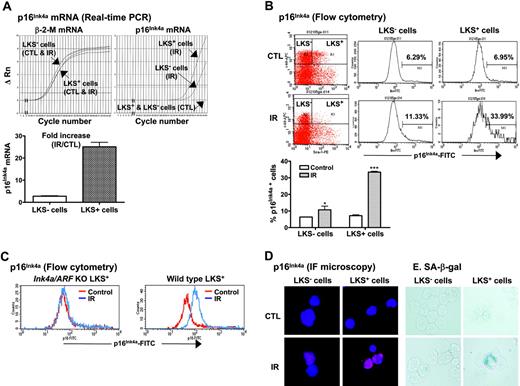
Increased expression of p16Ink4a has been implicated in the establishment and maintenance of cellular senescence and, more importantly, has been suggested to mediate HSC growth arrest and loss of HSC self-renewal in Bmi1 or Atm knock-out mice.14,15,21,22,25 Therefore, we next examined if IR induces p16Ink4a in HSCs. LKS- and LKS+ cells were isolated from BM-MNCs of control and irradiated mice as illustrated in Figure 1. The expression of p16Ink4a mRNA in LKS- and LKS+ cells was quantified by real-time RT-PCR and that of p16Ink4a protein by flow cytometry and immunofluorescent microscopy (Figure 4). As shown in Figure 4A, the expression of p16Ink4a mRNA was undetectable in both LKS- and LKS+ cells from control mice. TBI increased the expression of p16Ink4a mRNA in LKS- and LKS+ cells. However, the increase in p16Ink4a mRNA expression in irradiated LKS+ cells was significantly greater than that in irradiated LKS- cells (a minimal estimate of 25.1-fold increase for irradiated LKS+ cells vs 2.9-fold increase for irradiated LKS- cells). Subsequently, the increase in the expression of p16Ink4a in irradiated LKS- and LKS+ cells was confirmed at the level of protein by flow cytometry and immunofluorescent microscopy using the p16Ink4a-specific monoclonal antibody (Figure 4B,D). Again, LKS+ cells showed about a 5-fold increase in the percentage of the cells stained for p16Ink4a after TBI (irradiated LKS+ cells: 33.4% ± 0.3 vs unirradiated LKS+ cells: 7.2% ± 0.3; P < .001), whereas less than a 2-fold increase in p16Ink4a-stained cells was found in the irradiated LKS- population (irradiated LKS- cells: 10.7% ± 1.3 vs unirradiated LKS- cells: 6.4% ± 0.1; P < .05). However, no changes in p16Ink4a staining were observed in LKS+ and LKS- cells from irradiated Ink4a/Arf knock-out mice (Figure 4C), demonstrating the specificity of the assay. In addition, irradiated LKS+ cells exhibited much stronger immunofluorescent staining for p16Ink4a protein than irradiated LKS- cells (Figure 4D). These findings suggest that TBI can selectively induce p16Ink4a expression in HSCs.
TBI selectively increases p16Ink4a expression and SA-β-gal activity in LKS+ cells. (A) Quantification of p16Ink4a mRNA expression in LKS- and LKS+ cells from control (CTL) or irradiated (day 14 after TBI) mice. Top panel: Amplification profiles of representative real-time RT-PCR assays. Bottom panel: A minimal estimate of the fold increase in p16Ink4a mRNA expression is presented as the mean ± SE of 3 amplification assays. (B) Quantification of p16Ink4a-expressing cells in LKS- and LKS+ populations by flow cytometry. The expression of p16 Ink4a in LKS- and LKS+ cells from control (CTL) or irradiated (day 14 after TBI) mice was determined by flow cytometry as described in “Materials and methods.” Top panel: Representative flow cytometric analysis of the p16Ink4a-expressing cells in LKS- and LKS+ cell gates are shown. The numbers marked in the plots are the percentage of p16 Ink4a-positive cells. Bottom panel: The percentage of p16 Ink4a-positive cells is presented as the mean ± SE (n = 3). (C) Representative flow cytometric analysis of the p16Ink4a-expressing cells in LKS+ subpopulation from irradiated (day 14 after TBI) and control Ink4a/Arf KO or wild-type mice. (D) Representative immunofluorescent (IF) staining of p16Ink4a in sorted LKS- and LKS+ cells from irradiated and control mice. (E) Representative SA-β-gal staining in sorted LKS- and LKS+ cells from irradiated and control mice. *P < .05 and ***P < .001, compared with control unirradiated cells.
TBI selectively increases p16Ink4a expression and SA-β-gal activity in LKS+ cells. (A) Quantification of p16Ink4a mRNA expression in LKS- and LKS+ cells from control (CTL) or irradiated (day 14 after TBI) mice. Top panel: Amplification profiles of representative real-time RT-PCR assays. Bottom panel: A minimal estimate of the fold increase in p16Ink4a mRNA expression is presented as the mean ± SE of 3 amplification assays. (B) Quantification of p16Ink4a-expressing cells in LKS- and LKS+ populations by flow cytometry. The expression of p16 Ink4a in LKS- and LKS+ cells from control (CTL) or irradiated (day 14 after TBI) mice was determined by flow cytometry as described in “Materials and methods.” Top panel: Representative flow cytometric analysis of the p16Ink4a-expressing cells in LKS- and LKS+ cell gates are shown. The numbers marked in the plots are the percentage of p16 Ink4a-positive cells. Bottom panel: The percentage of p16 Ink4a-positive cells is presented as the mean ± SE (n = 3). (C) Representative flow cytometric analysis of the p16Ink4a-expressing cells in LKS+ subpopulation from irradiated (day 14 after TBI) and control Ink4a/Arf KO or wild-type mice. (D) Representative immunofluorescent (IF) staining of p16Ink4a in sorted LKS- and LKS+ cells from irradiated and control mice. (E) Representative SA-β-gal staining in sorted LKS- and LKS+ cells from irradiated and control mice. *P < .05 and ***P < .001, compared with control unirradiated cells.
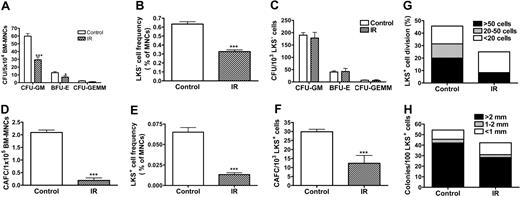
TBI selectively increases SA-β-gal activity in LKS+ cells, suppresses their replicative and clonogenic function in vitro, and impairs their capacity of long-term BM engraftment in vivo
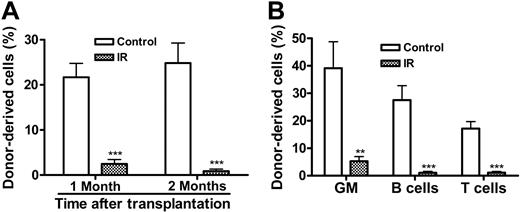
The increase in p16Ink4a expression in LKS+ cells after IR implies that IR may induce HSC senescence, since it has been suggested that p16Ink4a is not only a biomarker but also an effector of cellular senescence and mammalian aging.14-16,25 To test this hypothesis, we examined SA-β-gal staining in LKS- and LKS+ cells from mice with or without TBI, because SA-β-gal staining is a more widely used biomarker of cellular senescence.13 As shown in Figure 4E, SA-β-gal-positive staining was found only in irradiated LKS+ cells, while no positive SA-β-gal staining was observed in irradiated LKS- cells or in LKS- and LKS+ cells from control nonirradiated mice. Furthermore, in BM-MNCs, TBI decreased the frequencies of CFUs and day-35 CAFCs (Figure 5A,D), which measure the clonogenic function of HPCs and HSCs, respectively. Since TBI also decreased the frequencies of LKS- and LKS+ cells in BM-MNCs (Figure 5B,E), we determined whether the reduction in the frequencies of CFUs and day-35 CAFCs is the result of quantitative and/or qualitative reduction in HPCs and HSCs by expressing the frequencies of CFUs and day-35 CAFCs as a function of LKS- and LKS+ cells, respectively.12,26 As shown in Figure 5C, when the frequency of CFUs was expressed as a function of LKS- cells, it revealed that TBI had no effect on the clonogenic function of HPCs. In contrast, TBI significantly reduced the clonogenicity of HSCs when the day-35 CAFC frequency was expressed as a function of LKS+ cells (Figure 5F). This observation was further confirmed by the single-cell culture analysis using sorted LKS+ cells. As shown in Figure 5G, only about 25% of single sorted LKS+ cells from irradiated mice underwent at least one cell division after incubation with SCF/TPO, while about 45% of nonirradiated LKS+ cells did. Even though some of the irradiated LKS+ cells proliferated, the majority of the proliferating cells underwent fewer cell divisions than the corresponding nonirradiated LKS+ cells. Moreover, when IL-3 was added to the culture, fewer irradiated LKS+ cells formed colonies than nonirradiated LKS+ cells (irradiated LKS+ cells: 42.2% vs unirradiated LKS+ cells: 54.3%) (Figure 5H). Particularly, fewer irradiated LKS+ cells formed the colonies that were more than 2 mm than nonirradiated LKS+ cells (irradiated LKS+ cells: 28.2% vs unirradiated LKS+ cells: 42.1%). Furthermore, when LKS+ cells (500 cells) were isolated from irradiated mice BM at 1 month after TBI and then transplanted into lethally irradiated recipients along with 2 × 105 competitors in a CRA, about 2.50% and 0.89% of peripheral-blood leukocytes were derived from the irradiated donor cells at 1 and 2 months after transplantation, respectively (Figure 6). In contrast, transplantation of the same number of LKS+ cells from control mice resulted in 21.68% and 24.85% engraftment under the same experimental conditions. Taken together, these results strongly suggest that exposure to TBI can selectively cause long-term intrinsic damage to HSCs, probably by induction of HSC senescence.
TBI selectively suppresses LKS+-cell replicative and clonogenic function. (A-F) Mice were exposed to 6.5 Gy TBI or not irradiated as control (CTL). At 28 days after TBI, BM-MNCs were isolated from irradiated and control mice. The clonogenic function of HPCs and HSCs was measured by CFC assay and day-35 CAFC assay, respectively. The frequencies of LKS- and LKS+ cells in BM-MNCs were quantified by flow cytometry (B,E). The number of various CFUs and day-35 CAFCs is expressed as a function of BM-MNCs (A,D) or as a function of LKS- and LKS+ cells, respectively (C,F). The data are presented as mean ± SE (n = 5 for CFC assay and n = 3 for CAFC assay). (G-H) At 28 days after TBI, BM-MNCs were isolated from irradiated and control mice and single LKS+ cells were sorted into wells of rounded-bottom plates. After 14 days of culture with SCF/TPO, wells with a single seeded cell that had undergone at least one round of cell division were scored and graded by the number of cells produced. The results are presented as a percentage of LKS+ cells that divided (G). Similarly, single sorted cells were cultured with SCF/TPO/IL-3 for 14 days and the formation of hematopoietic-cell colonies (> 50 cells) was scored based on the size of the colonies. The data are expressed as a percentage of LKS+ cells that formed colony (H). *P < .05 and ***P < .001, compared with control.
TBI selectively suppresses LKS+-cell replicative and clonogenic function. (A-F) Mice were exposed to 6.5 Gy TBI or not irradiated as control (CTL). At 28 days after TBI, BM-MNCs were isolated from irradiated and control mice. The clonogenic function of HPCs and HSCs was measured by CFC assay and day-35 CAFC assay, respectively. The frequencies of LKS- and LKS+ cells in BM-MNCs were quantified by flow cytometry (B,E). The number of various CFUs and day-35 CAFCs is expressed as a function of BM-MNCs (A,D) or as a function of LKS- and LKS+ cells, respectively (C,F). The data are presented as mean ± SE (n = 5 for CFC assay and n = 3 for CAFC assay). (G-H) At 28 days after TBI, BM-MNCs were isolated from irradiated and control mice and single LKS+ cells were sorted into wells of rounded-bottom plates. After 14 days of culture with SCF/TPO, wells with a single seeded cell that had undergone at least one round of cell division were scored and graded by the number of cells produced. The results are presented as a percentage of LKS+ cells that divided (G). Similarly, single sorted cells were cultured with SCF/TPO/IL-3 for 14 days and the formation of hematopoietic-cell colonies (> 50 cells) was scored based on the size of the colonies. The data are expressed as a percentage of LKS+ cells that formed colony (H). *P < .05 and ***P < .001, compared with control.
Discussion
Exposure to moderate or high doses of TBI causes long-term hematopoietic injury.4,5 This injury has been attributed to quantitative and/or qualitative damage to HSCs by IR, particularly to a defect in HSC self-renewal.7-9 This assumption is supported by the data from the present study; these data provide the first direct evidence showing that exposure of mice to a moderate dose of TBI resulted in not only a sustained reduction in the frequency of BM LKS+ cells but also in the long-term inhibition of LKS+-cell clonogenic function. Although exposure to TBI also reduced the frequency of BM LKS- cells and various CFUs over a prolonged period, the clonogenic function of LKS- cells appeared normal when the CFUs were expressed as a function of LKS- cells. Lack of a defect in HPCs also has been reported recently in Bmi1 and ATM null mice, even though these mice exhibit a severe deficiency in HSC self-renewal.21,22 These findings highlight the importance of self-renewing HSCs in the maintenance of hematopoietic homeostasis, as Bmi1 and ATM null mice develop BM failure at a younger age than wild-type mice due to premature exhaustion of HSC reserves.21,22,27 Similarly, damage to HSC self-renewal by IR and/or chemotherapeutic agents may lead to hypoplastic syndrome at later times or under hematopoietic stress.4,5 This has been well documented in experimental animals but has yet to be proved in humans.4,5
TBI causes intrinsic damage to HSCs to inhibit long-term BM engraftment in a CRA. (A) Hematopoietic engraftment at 1 and 2 months after stem cell transplantation. The percentage of donor-derived peripheral-blood leukocytes from mice receiving a transplantation of irradiated or control LKS+ cells is expressed as mean ± SE (10 mice/group). (B) Multilineage reconstitution of myeloid (GM-granulocyte- monocyte/macrophage), T, and B cells was analyzed at 2 months after stem cell transplantation. The data are presented as mean ± SE (10 mice/group). ***P < .001; **P < .01.
TBI causes intrinsic damage to HSCs to inhibit long-term BM engraftment in a CRA. (A) Hematopoietic engraftment at 1 and 2 months after stem cell transplantation. The percentage of donor-derived peripheral-blood leukocytes from mice receiving a transplantation of irradiated or control LKS+ cells is expressed as mean ± SE (10 mice/group). (B) Multilineage reconstitution of myeloid (GM-granulocyte- monocyte/macrophage), T, and B cells was analyzed at 2 months after stem cell transplantation. The data are presented as mean ± SE (10 mice/group). ***P < .001; **P < .01.
Several CDKIs have been implicated in the regulation of HSC self-renewal. Among these CDKIs, p16Ink4a and p19Arf have been implicated in mediating HSC growth arrest and apoptosis, leading to the loss of HSC self-renewal.21,22 For instance, a defect in HSC self-renewal in Bmi1 or ATM knock-out mice was associated with increased expression of p16Ink4a and p19Arf mRNA in LKS+ cells.21,22 Repression of p16Ink4a and p19Arf expression and interruption of p16Ink4a-Rb pathway by retroviral transfection with Bmi-1 and human papilloma virus type 16 E7, respectively, restored the repopulating capacity of HSCs from Bmi1-/- or ATM-/- mice.21,22 In contrast, transfection of HSCs with the p16Ink4a retrovirus leads to either complete growth arrest or limited cell proliferation, and p19Arf retroviral transfection causes HSC apoptosis.22 The role of p21Cip1/Waf1 in regulation of HSC self-renewal appears more complex. Constitutive expression of p21Cip1/Waf1 can restrict the entry of HSCs into cell cycle and its absence could increase the proliferation of HSCs and eventually lead to an accelerated exhaustion of HSCs under hematopoietic stress,20 whereas increased expression of p21Cip1/Waf1 initiates the process of cellular senescence in response to genotoxic stress.25 It has been suggested that p18Ink4c may function as a negative regulator of HSC self-renewal, as its absence enhances HSC-cell renewal.24 Although p27Kip1 by itself may primarily restrict HPC proliferation and expansion,19 in cooperation with the MYC-antagonist MAD1 it can also negatively regulate HSC self-renewal and proliferation.23 Because of their obvious importance in regulating HSC function, we examined whether the residual BM damage induced by TBI is associated with an increased expression of one or more of these CDKIs. The results showed that the induction of residual BM damage was associated with a prolonged elevation of p21Cip1/Waf1, p19Arf, and p16Ink4a mRNA expression, while the expression of p27Kip1 and p18Ink4c mRNA was not elevated. Increased expression of p21Cip1/Waf1, p19Arf, and p16Ink4a was also found in prematurely senescent BM hematopoietic cells that were exposed to IR and followed by a long-term BM-cell culture in vitro.12
Increased expression of p21Cip1/Waf1, p19Arf, and p16Ink4a has been implicated in the induction of cellular senescence after extensive cell replication or exposure to stress.14,15,25 However, these CDKIs may play different roles in the initiation, establishment, and maintenance of cellular senescence.14,15,25 Cdkn1a is one of the target genes of p53. The expression of p21Cip1/Waf1 mRNA was elevated shortly following p53 phosphorylation and activation in BM cells after TBI (data not shown) but returned to nearly normal level at 2 months after TBI. This finding is in agreement with previous observations, indicating that p21Cip1/Waf1 may play an important role in the initiation of senescence but may not be required for the establishment and maintenance of senescence.14,15,25 In contrast, a sustained increase in p19Arf and p16Ink4a mRNA expression was observed in BM cells after TBI. Increase in p19Arf and p16Ink4a has been shown to play an important role in the establishment and maintenance of senescence via inhibiting CDKs, leading to the hypophosphorylation of Rb and formation of senescence-associated heterochromatic foci.14,15,25 In summary, our findings suggest that induction of p21Cip1/Waf1, p19Arf, and p16Ink4a may play an important role in IR-induced residual damage to HSCs, probably by induction of HSC senescence. However, the precise role of each of these CDKIs in IR-induced residual BM damage has yet to be defined, as a recent publication indicates that p19Arf and p16Ink4a do not play a significant role in the loss of HSC self-renewal following serial BM transplantation.28 In addition, it has yet to be determined whether these CDKIs may regulate HSC hematopoietic function by affecting HSC homing to and interacting with a niche and altering the fate decision of HSCs to self-renew versus proliferate/differentiate.
Intriguingly, exposure of mice to TBI resulted in a rapid but transient reduction in the expression of p27Kip1 and p18Ink4c mRNA in BM-MNCs; this reduction occurred just prior to the appearance of HSC regeneration and HPC expansion following TBI. However, it has yet to be determined whether exposure to TBI actually causes changes in p27Kip1 and p18Ink4c expression at the level of the proteins, because it has been reported that the expression of p27Kip1 and p18Ink4c proteins can be posttranscriptionally regulated through ubiquitination-mediated degradation and via coupled transcriptional and translational control, respectively.29,30 Since p27Kip1 and p18Ink4c function as negative regulators of HSC self-renewal and HPC expansion, it will be interesting to determine whether reduction in p27Kip1 and p18Ink4c expression can alleviate the restriction of HSC self-renewal and HPC expansion and whether down-regulation of p27Kip1 and p18Ink4c expression may facilitate HSC and HPC recovery following IR damage.19,23,24
Recently, p16Ink4a has been indicated as a new biomarker for cellular senescence and mammalian aging.16 In addition, its expression has been associated with the loss of HSC self-renewal in Bmi1 or ATM knock-out mice, probably via induction of HSC senescence.14,15,21,22 Therefore, we examined whether exposure to TBI can specifically induce p16Ink4a and senescence in HSCs. It was found that the induction of p16Ink4a by IR was primarily restricted to the HSC population, while only a modest increase in the expression of p16Ink4a mRNA and protein was found in irradiated LKS- cells. The irradiated LKS+ cells also showed increased SA-β-gal staining, a biomarker of cellular senescence.13 Moreover, LKS+ cells from irradiated mice exhibited a defect in clonogenic function and were less responsive to SCF/TPO and SCF/TPO/IL-3 stimulation for cell proliferation and clonogenic formation compared with unirradiated LKS+ cells. When irradiated LKS+ cells were transplanted into lethally irradiated recipients in a CRA, they contributed to a significantly lesser BM engraftment than nonirradiated LKS+ cells. In contrast, none of these changes were observed in irradiated LKS- cells. These data suggest that exposure to TBI can selectively induce p16Ink4a expression and senescence in HSCs.
Induction of senescence has been well documented in many different types of somatic cells and tumor cells after exposure to IR and various chemotherapeutic agents.25 However, this is the first study to demonstrate that induction of cellular senescence can also occur at the level of stem cells. It is yet to be determined whether exposure to IR or chemotherapeutic agents can induce stem-cell senescence in renewable tissues other than BM. Since stem cells are capable of self-renewal and produce progeny to replenish worn-out cells and to repair and even regenerate damaged tissues, it is understandable that induction of stem-cell senescence could contribute significantly to IR-induced long-term tissue injury, including the residual damage to the hematopoietic system. Thus, it will be interesting to determine whether inhibition of stem-cell senescence can be exploited to ameliorate IR-induced long-term tissue damage, particularly to BM. In addition, understanding how normal stem cells senesce after IR and chemotherapy will help us to elucidate the molecular mechanisms whereby cancer stem cells evade radiotherapy and/or chemotherapy.
Prepublished online as Blood First Edition Paper, September 8, 2005; DOI 10.1182/blood-2005-04-1418.
Supported in part by grants from the National Institutes of Health (NIH) to D.Z. (R01-CA78688, R01-CA86688, and CA102558), B.A.S. (R01-DC00713), and M.O. (R01-HL-69123) and by the Office of Research and Development, Medical Research Services, Department of Veterans Affairs to M.O. The work was performed in a renovated space supported by NIH grant C06RR14516.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Drs Aiping Bai and HaiQun Zeng for their technical assistance in animal studies and cell sorting.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal