Identification of malignant Sézary cells by T-cell receptor (TCR) clonality studies is routinely used for the diagnosis of Sézary syndrome, but T-cell clones expressed in a single patient have never been accurately characterized. We previously reported that CD158k expression delineates Sézary syndrome malignant cells, and, more recently, we identified vimentin at the surface membranes of Sézary cells and normal activated lymphocytes. In the present study, T-cell clones from 13 patients with Sézary syndrome were identified by immunoscopy and further characterized in the blood according to their TCR Vβ, CD158k, and vimentin cell-surface expression. We found in most patients a unique malignant T-cell clone that coexpressed CD158k and vimentin and that, when patients were tested, was also present in the skin. However, in some patients we detected the presence of a nonmalignant circulating clone expressing high amounts of vimentin and lacking CD158k. These results indicate that clonal expansion may originate from circulating malignant and nonmalignant CD4+ T cell populations in patients with Sézary syndrome. Identification of the malignant cells in Sézary syndrome cannot be achieved by T-cell clonality studies or by TCR Vβ monoclonal antibody (mAb) analysis alone; it also relies on CD158k phenotyping.
Introduction
Sézary syndrome (SS) is an erythrodermic, aggressive, cutaneous T-cell lymphoma characterized by a malignant T-cell clone that localizes in the blood and skin. In the absence of a specific marker for malignant Sézary cells, a diagnosis of SS relies on the circulating Sézary cell count and on the molecular detection of a circulating T-cell clone. Given that the cytomorphology of Sézary cells is not specific to SS,1,2 identification of a specific marker of malignant Sézary cells has been a challenging issue. Besides a T-cell memory phenotype, CD4+CD45RO+, Sézary cells were shown to lack CD263-5 and to have diminished CD3 expression,6 whereas the loss of CD73,7 appears not to be a specific feature of the malignant T-cell clone.5,8 More recently, it has been shown that Sézary cells specifically overexpress T-plastin, which, however, cannot be used as a cell-surface marker because it is located in the cytoskeleton, in association with actin.9 New markers of Sézary cells were described, including KIR3DL2/CD158k, the first positive cell-surface marker of Sézary cells. In patients with SS, KIR3DL2/CD158k is dimly expressed by circulating and cutaneous CD4+ lymphocytes.10-12 In healthy persons, CD158k is absent in CD4+ lymphocytes, and only minor subsets of T CD8+ and natural killer (NK) lymphocytes express high amount of this antigen.13 SC5 monoclonal antibody (mAb) was shown to react with an unknown cell-surface antigen expressed by normal activated lymphocytes and Sézary cells, whereas it failed to stain most circulating T lymphocytes in healthy individuals.14 We found that SC5 mAb specifically recognizes vimentin (VIM) at the cell surface of viable activated T lymphocytes and is effectively expressed by Sézary cells.15
The finding of an expanded T-cell clone in the blood of patients with SS is commonly considered to be a diagnostic marker. TCR Vβ phenotyping with specific mAbs may identify a “clonotypic” lymphocyte subset as an expanded population bearing a single TCR Vβ chain, which may be of diagnostic value.3,16-18 This approach, however, is limited by the lack of available mAbs reacting with some TCR VDJβ gene products in the blood4,16,17,19,20 and in skin samples.21 Moreover, there is no clear-cut limit of percentage of TCR Vβ expansion associated with malignancy. In our experience, the percentage of malignant Sézary cells followed by TCR Vβ usage varies from 5% to 95% of the CD4+ population repertoire. In addition, the expression of surface TCR may be artificially down-regulated on Sézary cells. Therefore, identifying T-cell clones in patients with SS relies on molecular analysis of the TCRβ or TCRγ chains at the DNA and RNA levels. Various polymerase chain reaction (PCR)–based techniques—including PCR-β,16 PCR-γ,22 PCR–denaturing gradient gel electrophoresis-γ (PCR–DGGE-γ),23 PCR–polyacrylamide gel electrophoresis-γ (PCR–PAGE-γ),24 and PCR–single-strand conformation polymorphism-γ (PCR–SSCP-γ)25 —have been validated. Immunoscopy based on TCR Vβ mRNA spectratype analysis and sequencing of the CDR3 region is a sensitive technique that has been applied for the detection and monitoring of T-cell clones in different conditions, including SS. It has been even suggested that immunoscopy may allow specific monitoring of the malignant T-cell clones in patients with SS.26 The diagnostic value of TCR clonality studies in cutaneous T-cell lymphoma (CTCL) is usually admitted; however, assignment of malignancy to T-cell clones remains subjective. It is thought that a proportion of circulating T-cell clones is nonmalignant,27 but T-cell clones in SS have never been accurately characterized. To rectify this, we identified circulating and cutaneous T-cell clones of patients with Sézary syndrome with the use of immunoscopy and determined their significance by phenotyping with monoclonal antibodies specific to the TCR VDJβ gene products, CD158k and VIM.
Patients, materials, and methods
Patients
Clinical characteristics of patients are summarized in Table 1. Thirteen patients, 8 men and 5 women whose mean age was 66 years (range, 44-79 years), were included in the study. In all patients, a diagnosis of Sézary syndrome was established based on clinical, histologic, and biologic criteria. All had pruriginous erythroderma with diffuse adenopathies, atypical circulating Sézary cells (more than 1000 cells/μL), and circulating T-cell clones found by TCR-γ chain analysis with the use of PCR-DGGE. As a control for molecular techniques, blood from a healthy donor was also collected. This study was approved by the Comité Consultatif de Protection des Personnes dans la Recherche Biomedicale institutional ethics committee. Informed consent was provided according to the Declaration of Helsinki.
Clinical characteristics of the 13 patients studied
Patient . | Sex/age, y . | Symptoms . | Sézary cells, % . | Previous treatment . | Current treatment . | Outcome . |
|---|---|---|---|---|---|---|
| 1 | F/70 | Erythemato-squamous plaques, then erythroderma | 30 | TM, CS, Ch, PUVA | Ch + CS | PR |
| 2 | M/79 | Erythroderma | 70 | — | TM + Ch + MTX | PR, then DOD |
| 3 | M/75 | Erythroderma | 70 | TM, CS | Ch + TM + CS | PR, then DOD |
| 4 | F/44 | Erythroderma | 80 | TM, CS | Cytapheresis + IFN | PR, then relapse |
| 5 | M/64 | Erythroderma, pruritus, xerosis | 80 | Ch, CS | TM + ECP + IFN | DOD |
| 6 | M/62 | Erythroderma, pruritus | 50 | PUVA | ECP then ECP + IFN | LTF |
| 7 | M/62 | Erythroderma, PPK | 95 | TM, CS, cytapheresis | ECP | PR, then relapse |
| 8 | F/71 | Erythroderma | 90 | TM, CS | TM | LTF |
| 9 | M/54 | Erythroderma | 10 | IFN | IFN | PR |
| 10 | M/66 | Erythroderma | 80 | ECP | Al | CR |
| 11 | F/75 | Annular erythematous lesions, then erythroderma, pruritus, xerosis, | 35 | Cytapheresis, TM, PUVA, IFN | ECP, then ECP + IFN | CR |
| 12 | M/60 | Erythroderma, pruritus, PPK, nail dystrophy | 50 | Cytapheresis, TM, PUVA, IFN | ECP, then ECP + IFN | Relapse |
| 13 | F/74 | Erythroderma | 60 | TM, CS, Ch | Cytapheresis | DOD |
Patient . | Sex/age, y . | Symptoms . | Sézary cells, % . | Previous treatment . | Current treatment . | Outcome . |
|---|---|---|---|---|---|---|
| 1 | F/70 | Erythemato-squamous plaques, then erythroderma | 30 | TM, CS, Ch, PUVA | Ch + CS | PR |
| 2 | M/79 | Erythroderma | 70 | — | TM + Ch + MTX | PR, then DOD |
| 3 | M/75 | Erythroderma | 70 | TM, CS | Ch + TM + CS | PR, then DOD |
| 4 | F/44 | Erythroderma | 80 | TM, CS | Cytapheresis + IFN | PR, then relapse |
| 5 | M/64 | Erythroderma, pruritus, xerosis | 80 | Ch, CS | TM + ECP + IFN | DOD |
| 6 | M/62 | Erythroderma, pruritus | 50 | PUVA | ECP then ECP + IFN | LTF |
| 7 | M/62 | Erythroderma, PPK | 95 | TM, CS, cytapheresis | ECP | PR, then relapse |
| 8 | F/71 | Erythroderma | 90 | TM, CS | TM | LTF |
| 9 | M/54 | Erythroderma | 10 | IFN | IFN | PR |
| 10 | M/66 | Erythroderma | 80 | ECP | Al | CR |
| 11 | F/75 | Annular erythematous lesions, then erythroderma, pruritus, xerosis, | 35 | Cytapheresis, TM, PUVA, IFN | ECP, then ECP + IFN | CR |
| 12 | M/60 | Erythroderma, pruritus, PPK, nail dystrophy | 50 | Cytapheresis, TM, PUVA, IFN | ECP, then ECP + IFN | Relapse |
| 13 | F/74 | Erythroderma | 60 | TM, CS, Ch | Cytapheresis | DOD |
TM indicates topical mechlorethamine; Ch, chlorambucil; PUVA, psoralen UVA photochemotherapy; —, none; CS, corticosteroid; ECP, extracorporeal photopheresis; IFN, interferon; MTX, methotrexate; Al, alemtuzumab; CR, complete remission; PR, partial remission; DOD, died of disease; LTF, lost to follow-up.
Peripheral blood mononuclear cells, tissue samples, and Sézary cell lines
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized venous blood by density gradient centrifugation over Lymphoprep (PAA Laboratories, Linz, Austria) and frozen at –80°C in culture medium consisting of RPMI 1640 supplemented with 30% fetal calf serum (FCS) and 10% dimethyl sulfoxide for subsequent storage in liquid nitrogen. The Sézary cell line HUT78 was cultured in RPMI 1640 supplemented with 10% FCS. The Pno cell line was developed and expanded from a patient with SS, as previously described.28 Frozen skin samples of erythrodermic skin were obtained from patients 1, 2, and 3 and stored at –80°C.
Total mRNA and cDNA preparation from blood and skin samples
Total RNA was extracted from 5 to 10 × 106 PBMCs from blood samples and from 105 to 30 × 105 sorted cells using the Trizol reagent (Invitrogen, Cergy-Pontoise, France) and chloroform/isopropanol precipitation. For RNA extraction from skin samples, 20 50-μm–thick frozen sections were transferred to Trizol and immediately homogenized twice for 2 minutes with a Mixer Mill MM301 (Verder, Erahny sur Oise, France) before chloroform/isopropanol precipitation. A carrier for RNA precipitation was used from skin specimens and from sorted cells. Total mRNA (5-15 μg) from PBMCs, sorted PBMCs, and skin samples were reverse transcribed with the use of oligo-dT primers and Powerscript reverse transcriptase (RT Clontech, Palo Alto, CA).
RT-PCR amplification of CD158k/KIR3DL2 and CD158e/KIR3DL1
PCR reactions were performed on 1 μg total cDNA from PBMCs or skin specimens. PCR was performed using the primers for CD158k/KIR3DL2 (sense, 5′-CCTCTTCTTTCTCCTTTATCG-3′; antisense, 5′-CAGCCCTGTCTCAAAACCA-3′) flanking a 330-bp segment of CD158k complete mRNA. Alternatively, the previously reported primers allowing amplification of a 368-bp fragment were used.29 Reverse transcription-PCR (RT-PCR) for KIR3DL1 was performed using previously described primers.29 RT-PCR for β-actin, used as a positive control, was performed with previously reported primers,30 allowing amplification of a 245-bp segment.
T-cell clonality studies with immunoscopy
Analysis of T-cell clonality was conducted on cDNA from PBMCs and skin specimens by TCR Vβ CDR3 size spectratyping and sequencing (immunoscopy), as previously described.31 Identification of T-cell clones first relied on GeneScan Analysis (Applied Biosystems, Foster City, CA) showing a monoclonal signature, as a single peak, in a given TCR Vβ family. T-cell clones were definitively characterized by direct sequencing of the TCR Vβ CDR3 region. TCR Vβ families were considered to have a restricted profile when 4 or fewer consecutive peaks were observed on GeneScan Analysis (Applied Biosystems). TCR Vβ-Cβ and TCR Vβ-Jβ segments were amplified using consensus sense primers for all 24 Vβ families, a unique antisense primer for the Cβ regions, and antisense primers for all 13 Jβ families.31 For GeneScan Analysis (Applied Biosystems), antisense Fam-conjugated primers were used. Fluorescent PCR products were applied to a polyacrylamide sequencing gel (POP6; Applied Biosystems), and electrophoresis was performed on an automated ABI Prism 3100 sequencer (Applied Biosystems). Products were analyzed with the GeneScan software (Applied Biosystems). For direct sequencing of the TCR Vβ CDR3 region, amplification of TCR Vβ-Cβ products was performed using high-fidelity polymerase (Invitrogen, Carlsbad, CA), and PCR products were purified with the QIAEXII purification kit (Qiagen, Valencia, CA). Purified products were directly sequenced in both directions with the same Vβ and Cβ primers and with the use of the ABI BigDye terminator sequencing kit (version 3) and the ABI Prism 3100 sequencer (Applied Biosystems).
Flow cytometry analyses and cell sorting
One- and 2-color flow cytometry analyses were performed on PBMCs using phycoerythrin (PE)– or fluorescein isothiocyanate (FITC)–conjugated mouse mAbs. For indirect staining, goat antimouse mAbs were used. mAbs directed against TCR Vβ families, CD3, and CD4 were purchased from Immunotech/Beckman Coulter (Marseille, France). Anti–TCRαβ (IP26, IgG1) and SC5 (IgM) mAbs were raised and produced in our laboratory and used as purified ascites fluid. The anti–CD158k/KIR3DL2 Q66 (IgM) mAb was obtained through exchanges during the Seventh International Workshop on white cell differentiation antigens (Harrogate, United Kingdom, 2000),32 and the anti–CD158e/CD158k AZ158 (IgG2a) mAb was obtained purified from Innate Pharma (Marseille, France). PBMCs (3 × 105 to 5 × 105) were incubated with primary and secondary mAbs for 20 minutes at 4°C. Flow cytometry analysis was performed using the EPICS XL (Beckman-Coulter, Miami, FL) on the gated lymphocyte population. For cell sorting, fluorescence staining was applied to 107 cells, and cells were sorted using the ELITE cell sorter (Beckman-Coulter).
Immunohistochemistry
For immunostaining procedures, 5-μm–thick frozen sections were applied on Superfrost plus slides (CML, Angers, France), air dried overnight, and fixed in acetone for 10 minutes before storage at –20°C. Slides were postfixed in acetone for 5 minutes and air dried before use. After rehydration, immunostaining was performed with the use of a biotin/avidin system conjugated to alkaline phosphatase (Vectastain ABC-AP kit; Vector Laboratories, Burlingame, CA). Alkaline phosphatase reaction was revealed by Naphthol-Fast Red (Sigma-Aldrich, Saint Quentin Fallavier, France), and sections were counterstained in blue with hematoxylin. Primary antibodies to CD3, CD4, and CD8 (DAKO SA, Glostrup, Denmark) were used at a 1:50 dilution. For KIR3DL2/CD158k immunostaining, slides were incubated for 4 hours at room temperature with AZ158 mAb, as previously described.11
Epidermal and dermal lymphocytic infiltrates were separately analyzed. In the epidermis, numbers of CD3+, CD4+, CD8+, and AZ158+ cells per millimeter were evaluated based on total epidermal length, measured within the stratum granulosum according to a morphometry system (Pathfinder with Lightvision software; IMSTAR, Paris, France). In the dermis, semiquantitative analysis was performed in which we categorized the CD4/CD3, CD8/CD3, and AZ158/CD3 ratios into 4 groups: group 1 (0%-25%), group 2 (greater than 25%-50%), group 3 (greater than 50%-75%), and group 4 (greater than 75%-100%).
Results
CD158k and VIM
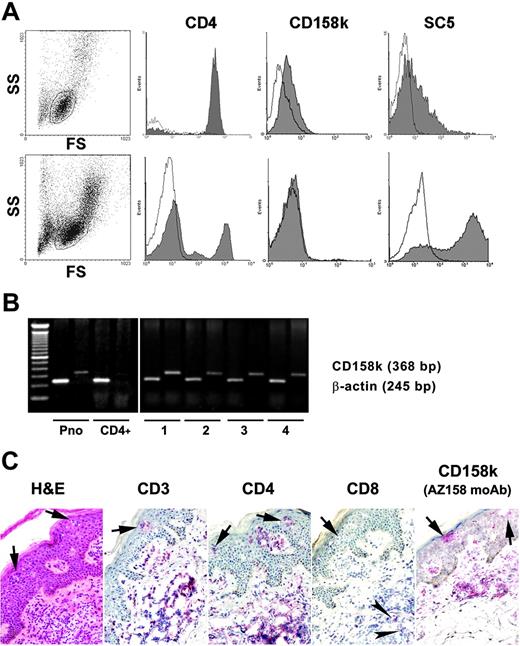
CD158k and VIM were expressed by Sézary cells obtained from 13 patients, but only the former receptor appeared to be associated with malignancy. A CD158kdim population was present in all 13 patients with SS, as shown in Figure 1A (upper panel) for patient 11, accounting for 8% to 85% of the gated lymphocytes (Table 2). In addition, CD158k RT-PCR findings were positive in all patients, including those with predominantly CD4+ circulating lymphocyte subsets, at levels similar to those of the previously reported Sézary cell line Pno,28 but they were virtually negative in most CD4+ PBMCs from healthy donors (Figure 1B). In all patients, a lymphocyte subset was stained with SC5 mAb, which recognizes external cell membrane VIM, ranging from 10% to 95% (Table 2). It was usually detected through flow cytometry as a broad peak (Figure 1A, upper panel). Importantly, several findings provided clear evidence that CD158k is a suitable marker of malignant Sézary cells. First, the percentage of Sézary cells, evaluated by cytomorphology, appeared to be correlated with the proportion of cells that expressed CD158kdim (Spearman correlation test: ρ= 0.765; P = .004) (Table 2). Second, we observed a loss of CD158k expression in 2 patients who experienced clinical remission. Patient 11 had complete remission under extracorporeal photopheresis. At this time, the circulating Sézary cell count was virtually negative, and we observed a marked decrease in CD4+ lymphocyte count, the absence of detectable CD158kdim cells, and the presence of approximately 3% of normal circulating NK lymphocytes expressing high amounts of CD158k (Figure 1A, lower panel; Table 2). In contrast to CD158k, anti-SC5 mAb reactivity was increased at the time of clinical remission. Similarly, in patient 10, loss of the CD158k subset associated with the enhancement of up to 95% anti-SC5 mAb–reactive cells was observed during the course of treatment with anti-CD52 mAb, while clinical improvement was occurring. Complete remission was obtained in this patient, and total disappearance of circulating Sézary cells was observed (Table 2, patient 10). Third, in one patient with early-stage disease, we observed that the circulating CD158kdim subset increased with disease progression. Patient 1 initially had erythematous and squamous plaques and then developed erythroderma. Interestingly, a significant increase in the circulating CD158kdim population was evidenced in this patient, from 16% at the beginning of the disease to 30% 1 year later; her condition did not improve despite treatment with topical mechlorethamine and chlorambucil (Table 2, patient 1). Fourth, with the use of immunohistochemistry, we identified a CD158k population within the predominantly CD3+ and CD4+ cutaneous infiltrates in erythrodermic skin of the 3 patients studied (Table 3; patients 1, 2, 3). Representative tissue sections from patient 3 are shown in Figure 1C. Interestingly, in patient 1, the proportion of CD158k+ cells also seemed to increase in the skin in accordance with the increasing CD158k+ circulating population. Indeed, 4-fold more CD158k+ cells were found in the epidermis 1 year after the onset of erythroderma (Table 3, patient 1). Findings from RT-PCR, performed with mRNA prepared from the skin samples used for immunohistochemistry, were positive for KIR3DL2/CD158k but negative for KIR3DL1, indicating that the AZ158 mAb identified CD158k+ cells (data not shown). It should be noted that anti-SC5 mAb could not be used for immunohistochemistry studies because it does not distinguish outer from inner cell membrane VIM localization. Altogether, these findings further support that CD158k receptor expression allows the separation of malignant from normal lymphocytes because it delineates malignant Sézary cells, whereas cell-surface expression of VIM is detected on tumor and nontumor cells.
Phenotypic analyses of circulating lymphocytes
. | . | . | Flow cytometry, % . | . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | Sézary cells, % . | Immunoscopy: Vβ family of T-cell clones . | Expanded TCR (Vβ) . | TCRαβ . | CD4 . | CD158k . | SC5 . | TCR Vβ and CD158k . | TCR Vβ and SC5 . | ||||||
| 1 | 30 | 17 | 38 (Vβ17) | 85 | 80 | 16 | 25 | 16 | 8 | ||||||
| 2 | 70 | 2 | 95 (Vβ2) | 90 | 90 | 80 | ND | 80 | ND | ||||||
| 3 | 70 | 3, 4, 8 | 85 (Vβ8) | 95 | 90 | 75 | 10 | 70 | 10 | ||||||
| 4 | 80 | 7 | 95 (Vβ7) | 90 | 95 | 85 | 55 | 85 | 53 | ||||||
| 5 | 80 | 3, 13, 16 | 20 (Vβ13.1) | 45 | 60 | 60 | 20 | 13 | 2 | ||||||
| 6 | 50 | 11 | 16 (Vβ11) | 56 | 60 | 18 | 70 | 15 | 11 | ||||||
| 7 | 95 | 8 | 92 (Vβ8) | 98 | 95 | 30 | 10 | 30 | 10 | ||||||
| 8 | 90 | 5, 17, 18 | 10 (Vβ17) | 90 | 95 | 65 | 35 | 0 | 5 | ||||||
| 9 | 10 | 14, 6 | 20 (Vβ14) | 65 | 47 | 8 | 36 | 0 | 10 | ||||||
| 10 | 80 | ND | ND | 95 | 95 | 70 | 23 | ND | ND | ||||||
| 10* | 0 | 14, 17, 20, 21 | 37 (Vβ14) | 95 | ND | 0 | 95 | 0 | 50 | ||||||
| 11 | 35 | 22 | 5 (Vβ22) | 45 | 65 | 25 | 15 | 0 | 5 | ||||||
| 11† | 0 | 22 | 3 (Vβ22) | 50 | 30 | 3 | 40 | ND | ND | ||||||
| 12 | 50 | 5, 15, 23 | None | 95 | 92 | 50 | 25 | ND | ND | ||||||
| 13 | 60 | 3, 15, 17, 23 | None | 80 | 80 | 15 | 34 | ND | ND | ||||||
. | . | . | Flow cytometry, % . | . | . | . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | Sézary cells, % . | Immunoscopy: Vβ family of T-cell clones . | Expanded TCR (Vβ) . | TCRαβ . | CD4 . | CD158k . | SC5 . | TCR Vβ and CD158k . | TCR Vβ and SC5 . | ||||||
| 1 | 30 | 17 | 38 (Vβ17) | 85 | 80 | 16 | 25 | 16 | 8 | ||||||
| 2 | 70 | 2 | 95 (Vβ2) | 90 | 90 | 80 | ND | 80 | ND | ||||||
| 3 | 70 | 3, 4, 8 | 85 (Vβ8) | 95 | 90 | 75 | 10 | 70 | 10 | ||||||
| 4 | 80 | 7 | 95 (Vβ7) | 90 | 95 | 85 | 55 | 85 | 53 | ||||||
| 5 | 80 | 3, 13, 16 | 20 (Vβ13.1) | 45 | 60 | 60 | 20 | 13 | 2 | ||||||
| 6 | 50 | 11 | 16 (Vβ11) | 56 | 60 | 18 | 70 | 15 | 11 | ||||||
| 7 | 95 | 8 | 92 (Vβ8) | 98 | 95 | 30 | 10 | 30 | 10 | ||||||
| 8 | 90 | 5, 17, 18 | 10 (Vβ17) | 90 | 95 | 65 | 35 | 0 | 5 | ||||||
| 9 | 10 | 14, 6 | 20 (Vβ14) | 65 | 47 | 8 | 36 | 0 | 10 | ||||||
| 10 | 80 | ND | ND | 95 | 95 | 70 | 23 | ND | ND | ||||||
| 10* | 0 | 14, 17, 20, 21 | 37 (Vβ14) | 95 | ND | 0 | 95 | 0 | 50 | ||||||
| 11 | 35 | 22 | 5 (Vβ22) | 45 | 65 | 25 | 15 | 0 | 5 | ||||||
| 11† | 0 | 22 | 3 (Vβ22) | 50 | 30 | 3 | 40 | ND | ND | ||||||
| 12 | 50 | 5, 15, 23 | None | 95 | 92 | 50 | 25 | ND | ND | ||||||
| 13 | 60 | 3, 15, 17, 23 | None | 80 | 80 | 15 | 34 | ND | ND | ||||||
The population of viable lymphocytes was gated according to morphologic criteria of forward and side scatter.
ND indicates not determined.
Data during the course of a treatment with anti-CD52 monoclonal antibodies (alemtuzumab).
Data at the time of clinical remission under treatment with extracorporeal photochemotherapy.
Molecular studies and immunohistochemistry in cutaneous samples from patients 1, 2, and 3
. | Molecular studies . | . | . | Immunohistochemistry . | . | . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | Epidermis, no. positive cells/mm . | . | . | . | Dermis, ratio category . | . | . | ||||||||
| Patient . | Immunoscopy: Vβ family of T-cell clones . | CD158k* . | CD158e† . | CD3 . | CD4 . | CD8 . | CD158k‡ . | CD4/CD3 . | CD8/CD3 . | CD158k‡/CD3 . | ||||||||
| 1 | 17, 6, 16 | + | - | 68 | 59 | 2 | 2 | 4 | 1 | 1 | ||||||||
| 1§ | 17, 15, 16 | + | - | 54 | 44 | 12 | 9 | 4 | 2 | 2 | ||||||||
| 2 | 2 | + | - | 6 | 3 | 0 | 3 | 4 | 0 | 2 | ||||||||
| 3 | 8 | + | - | 32 | 30 | 0 | 14 | 4 | 0 | 3 | ||||||||
. | Molecular studies . | . | . | Immunohistochemistry . | . | . | . | . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | Epidermis, no. positive cells/mm . | . | . | . | Dermis, ratio category . | . | . | ||||||||
| Patient . | Immunoscopy: Vβ family of T-cell clones . | CD158k* . | CD158e† . | CD3 . | CD4 . | CD8 . | CD158k‡ . | CD4/CD3 . | CD8/CD3 . | CD158k‡/CD3 . | ||||||||
| 1 | 17, 6, 16 | + | - | 68 | 59 | 2 | 2 | 4 | 1 | 1 | ||||||||
| 1§ | 17, 15, 16 | + | - | 54 | 44 | 12 | 9 | 4 | 2 | 2 | ||||||||
| 2 | 2 | + | - | 6 | 3 | 0 | 3 | 4 | 0 | 2 | ||||||||
| 3 | 8 | + | - | 32 | 30 | 0 | 14 | 4 | 0 | 3 | ||||||||
With the use of immunohistochemistry, the proportion of labeled cells was evaluated separately in the epidermis and in the dermis. In the epidermis, the numbers of CD3+, CD4+, CD8+, and AZ158+ lymphocytes per millimeter were evaluated on the total epidermal length. In the dermis, semiquantitative analysis was performed in which CD4/CD3, CD8/CD3, and AZ158/CD3 ratios were categorized into 4 groups: group 1 (0%-25%), group 2 (greater than 25%-50%), group 3 (greater than 50%-75%), and group 4 (greater 75%-100%).
Molecular study of CD158k in the skin consisted of RT-PCR with consensus primers applied to total RNA extracted from the skin samples used for immunohistochemistry.
Molecular study of CD158e in the skin consisted of RT-PCR with consensus primers applied to total RNA extracted from the skin samples used for immunohistochemistry.
Immunohistochemistry analysis for the CD158k+ population was performed using the AZ158 mAb, which recognizes KIR3DL1/CD158e and KIR3DL2/CD158k.
Data after 1 year of disease progression.
Expression of CD158k and VIM by circulating and cutaneous lymphocytes. (A) Flow cytometry analysis in patient 11 showed CD158kdim and cell-surface VIM positivity before treatment (top row). After treatment with extracorporeal photochemotherapy, there was a dramatic decrease in the CD4+ population associated with a loss of CD158k expression and a significant increase in VIM (bottom row). (B) RT-PCR for CD158k was positive for PBMCs in patients 1, 2, 3, and 4 and for the Sézary cell line Pno, whereas it was negative for CD4+ lymphocytes in a healthy donor. (C) Immunohistochemistry analysis performed in a skin sample from patient 3 showed a predominantly CD3+CD4+ infiltrate along with CD158k+ cells in the dermis and the epidermis. Intraepidermal nests of lymphocytes, or Pautrier microabscesses, can be seen (arrows). Few CD8+ lymphocytes were present in the dermis (arrowheads), whereas only CD4+ T cells are seen in the epidermis. Slides are shown at 200 × original magnification (20 ×/0.50 NA objective), and images were captured with an Axioskop2 microscope (Zeiss, Oberkochen, Germany). Pictures were scanned in with a DP70 camera and software (Olympus, Hamburg, Germany). Hematoxylin and eosin staining is shown on the left, whereas a biotin/avidin system conjugated to alkaline phosphatase was used for CD3, CD4, CD8, and CD158k labeling.
Expression of CD158k and VIM by circulating and cutaneous lymphocytes. (A) Flow cytometry analysis in patient 11 showed CD158kdim and cell-surface VIM positivity before treatment (top row). After treatment with extracorporeal photochemotherapy, there was a dramatic decrease in the CD4+ population associated with a loss of CD158k expression and a significant increase in VIM (bottom row). (B) RT-PCR for CD158k was positive for PBMCs in patients 1, 2, 3, and 4 and for the Sézary cell line Pno, whereas it was negative for CD4+ lymphocytes in a healthy donor. (C) Immunohistochemistry analysis performed in a skin sample from patient 3 showed a predominantly CD3+CD4+ infiltrate along with CD158k+ cells in the dermis and the epidermis. Intraepidermal nests of lymphocytes, or Pautrier microabscesses, can be seen (arrows). Few CD8+ lymphocytes were present in the dermis (arrowheads), whereas only CD4+ T cells are seen in the epidermis. Slides are shown at 200 × original magnification (20 ×/0.50 NA objective), and images were captured with an Axioskop2 microscope (Zeiss, Oberkochen, Germany). Pictures were scanned in with a DP70 camera and software (Olympus, Hamburg, Germany). Hematoxylin and eosin staining is shown on the left, whereas a biotin/avidin system conjugated to alkaline phosphatase was used for CD3, CD4, CD8, and CD158k labeling.
Identification of single or multiple T-cell clones in the blood and in erythrodermic skin
T-cell populations were analyzed using immunoscopy in the blood of all 13 patients and in the skin of patients 1, 2, and 3 (Tables 2, 3). In the blood, 28 circulating T-cell clones were identified among all patients. Immunoscopy identified a single circulating T-cell clone in 6 patients (patients 1, 2, 4, 6, 7, 11) and multiple clones in the remaining 7 patients. Up to 4 circulating T-cell clones could be found in the same patient (Table 2; patients 10, 13). In addition to T-cell clonal patterns, abnormalities of nonclonal TCR Vβ families were found in the PBMCs of all patients, as in patient 2 (Figure 2A). They consisted of apparent loss of TCR Vβ families or restricted spectratype patterns.
Immunoscopy applied to PBMCs and a skin specimen of patient 2: TCR Vβ spectratyping. (A) GeneScan Analysis of PBMCs in patient 2 revealed a monoclonal signature for Vβ2 (shown to be malignant by 2-color flow cytometry analysis), apparent loss of several families (*) and many contracted profiles (**). (B) Gene-Scan Analysis of a skin sample from the same patient revealed a complete loss of all Vβ families, with a single monoclonal peak of a size identical to that of the malignant T-cell clone identified in the blood, indicating the presence of the malignant T-cell clone in the skin and the complete loss of other T cells.
Immunoscopy applied to PBMCs and a skin specimen of patient 2: TCR Vβ spectratyping. (A) GeneScan Analysis of PBMCs in patient 2 revealed a monoclonal signature for Vβ2 (shown to be malignant by 2-color flow cytometry analysis), apparent loss of several families (*) and many contracted profiles (**). (B) Gene-Scan Analysis of a skin sample from the same patient revealed a complete loss of all Vβ families, with a single monoclonal peak of a size identical to that of the malignant T-cell clone identified in the blood, indicating the presence of the malignant T-cell clone in the skin and the complete loss of other T cells.
Immunoscopy applied to the erythrodermic skin of patients 1, 2, and 3 identified 3 distinct cutaneous T-cell clones each time in patient 1 (TCR Vβ6, 16, 17 and then TCR Vβ15, 16, 17 a year later; Table 3) and unique cutaneous T-cell clones in patients 2 (TCR Vβ2; Figure 2B; Table 3) and 3 (TCR Vβ8; Table 3). Interestingly, in these 3 patients, a unique T-cell clone was found in the skin and blood that appeared to correspond to the malignant CD158kdim lymphocytes (TCR Vβ17 in patient 1, TCR Vβ2 in patient 2, and TCR Vβ8 in patient 3; Table 2). Patients 2 and 3, in whom the malignant T-cell clone was single in the skin, both died of the disease (Table 1).
Phenotypic studies distinguished between malignant and nonmalignant circulating T-cell clones
Results from the phenotypic analyses are summarized in Table 2. Flow cytometry studies using anti–TCR Vβ mAb were performed according to the results of immunoscopy. Among the 28 circulating T-cell clones, 21 could be stained using an mAb specific to the corresponding TCR Vβ family. Eleven (52%) T-cell clones corresponded to a significant subset of gated lymphocytes, each in a different patient, delineating a clonotypic circulating T-cell population. According to TCR Vβ expression, the proportion of the expanded T-cell clones ranged from 5% to 95%. In 2 patients (patients 12 and 13), flow cytometry analyses supplied negative results for all TCR Vβ families in whom a T-cell clone was identified by immunoscopy. However, each displayed an abnormally high T-cell population (95% and 80% CD4+ T lymphocytes, respectively) and a high percentage of Sézary cells (determined by cytomorphology), strongly suggesting the presence of an expanded malignant T-cell clone that could be labeled dimly with anti-CD158k (Table 2) mAb. Two-color flow cytometry using anti–TCR Vβ and anti-CD158k or -SC5 mAb was performed for the 11 patients displaying an expanded TCR Vβ population. In 7 patients (patients 1-7), the clonotypic T-cell population was found to express CD158kdim, as shown by representative patient results (Figure 3A; patient 4). Similarly, in patient 3, a TCR Vβ8+ CD158k+ clonal population was identified, that was expanded to 70% to 80% in the peripheral blood. Interestingly, as previously reported for other patients,28,33 a long-term cultured Sézary cell line (TCR Vβ8+/CD158k+) could be established from the PBMCs of patient 3 peripheral blood. The identity of this cell line was confirmed by TCR Vβ analysis showing a unique TCR Vβ8-Jβ2.5 chain with a CDR3 sequence identical to that of the circulating malignant T-cell clone (D.H., N.O., Sabine Le Gouvello, Anne Tessier-Marteau, François Berrehar, A.M.-C., M.B., and A.B., manuscript in preparation). In addition to the malignant T cells, additional T-cell clones were found in patients 3 and 5. In patient 3, although additional TCR Vβ3 and TCR Vβ4 T-cell clones were identified by immunoscopy, only the anti–TCR Vβ8 mAb labeled 85% of gated lymphocytes. In patient 5, flow cytometry with TCR Vβ3 and TCR Vβ16 mAb supplied negative results, possibly reflecting that the rearranged gene products were not expressed at the cell surface, that these T-cell clones were not expanded, or that these T-cell clones did not react with the specific mAbs. In these patients, SC5 mAb stained the malignant clonal population, identified by the appropriate anti–TCR Vβ mAb, and the remaining gated reactive lymphocytes. In contrast, in 4 patients (patients 8-11), we identified one expanded T-cell clone with the anti–TCR Vβ mAb that did not express CD158k, as shown by representative patient results (Figure 3B; patient 8). In this patient, 10% of gated lymphocytes were stained with anti–TCR Vβ17 mAb and failed to react with anti-CD158k mAb, which labeled 65% of the gated lymphocyte population corresponding to the malignant cell population. Patients 10 and 11 especially drew our attention. In patient 10, no CD158k+ population was present at the time of treatment with anti-CD52 mAb, resulting in clinical remission with the clearance of all Sézary cells. However, several T-cell clones were identified by immunoscopy, including an expanded TCR Vβ14+CD158k– population, showing a strong cell-surface vimentin expression (anti-SC5 mAb reactivity; Figure 3C). In patient 11, immunoscopy identified a single TCR Vβ22 T-cell clone that appeared to lack CD158k expression. This T-cell clone corresponded to 5% of the gated peripheral blood lymphocytes and remained detectable by immunoscopy and cytometry study at the time of clinical remission. It is noteworthy that in all patients, SC5 mAb positivity was demonstrated not only within the clonal subset but also in the remaining gated lymphocytes. Thus, these results indicate that among the circulating T-cell clones that could be identified by a specific anti–TCR Vβ mAb, only the ones that expressed CD158k corresponded to the malignant Sézary cells.
Some T-cell clones displayed heterogeneous levels of cell surface expression of CD3/TCRαβ, CD158k, and VIM
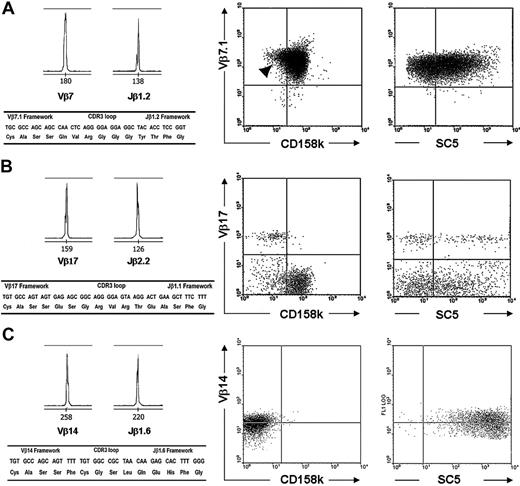
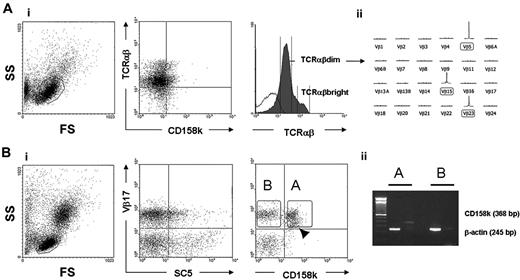
In agreement with findings of a previous report,6 we found in some patients that circulating lymphocytes expressed at their cell surfaces a diminished amount of CD3/TCRαβ molecules. Among these patients, one (patient 13) displayed clearly distinct TCRαβbright and TCRαβdim populations. Two-color flow cytometry analysis showed similarly low expression of CD158k in bright and dim TCRαβ subsets (Figure 4Ai), indicating the presence of malignant cells in both populations. This was further demonstrated through immunoscopy, which was applied to these separated cells. The results revealed that the TCRαβdim population consisted only of monoclonal TCR Vβ5, TCR Vβ15, and TCR Vβ23 (Figure 4Aii), whereas the TCRαβbright subset not only contained all TCR Vβ families, with the same profile seen in PBMCs, it included the monoclonal TCR Vβ5, TCR Vβ15, and TCR Vβ23 (data not shown). TCRαβbright and TCRαβdim lymphocyte populations displayed heterogeneous expression of cell surface VIM (data not shown). Heterogeneous expression of VIM was actually demonstrated in most T-cell clones. Importantly, we also noticed heterogeneous expression of CD158k by some malignant T-cell clones, as seen with patient 1 (Figure 4Bi), that expressed a unique TCR Vβ17+ clone in early-stage disease (Table 2). Flow cytometry cell sorting of TCR Vβ17+ CD158k+ and TCR Vβ17+ CD158k– subsets was performed on PBMCs from this patient. We found that CD158k mRNA was detectable in only the former subset (Figure 4Bii). We believe that a subset of malignant lymphocytes lacking CD158k expression was present in other patients, but it was more difficult to distinguish because of the low level of CD158k expression. For instance, in patient 4, a minor CD158k– population could be distinguished within the TCR Vβ7 malignant clonal population (Figure 3A, arrowhead). These latter results indicate that although CD158k is a reliable marker for CD4+ Sézary syndrome cells, the malignant clonal population can be heterogeneous for its expression at the transcript and the protein product levels.
Identification of malignant and nonmalignant T-cell clones in patients 4, 8, and 10 using TCR Vβ, CD158k, and cell-surface VIM phenotyping. Molecular characteristics of the T-cell clones are shown on the left, and the results of 2-color flow cytometry analyses are shown on the right. (A) A strongly expanded TCR Vβ7-Jβ1.2 T-cell clone was identified in patient 4, expressing CD158k and displaying heterogeneous labeling with SC5 mAb. A minor CD158k– population could be recognized within the Vβ7+ population (arrowhead). (B) A nonmalignant TCR Vβ17-Jβ2.2 T-cell clone was identified in patient 8, accounting for 10% of gated lymphocytes. Two-color flow cytometry showed that the TCR Vβ17+ population displayed heterogeneous expression of cell-surface vimentin (SC5) but was negative for CD158k. (C) A nonmalignant TCR Vβ14-Jβ1.6 T-cell clone was identified in patient 10 during treatment with anti–CD52 mAb (alemtuzumab) that was heavily expanded and did not express CD158k, as seen on the right based on the results of 2-color flow cytometry.
Identification of malignant and nonmalignant T-cell clones in patients 4, 8, and 10 using TCR Vβ, CD158k, and cell-surface VIM phenotyping. Molecular characteristics of the T-cell clones are shown on the left, and the results of 2-color flow cytometry analyses are shown on the right. (A) A strongly expanded TCR Vβ7-Jβ1.2 T-cell clone was identified in patient 4, expressing CD158k and displaying heterogeneous labeling with SC5 mAb. A minor CD158k– population could be recognized within the Vβ7+ population (arrowhead). (B) A nonmalignant TCR Vβ17-Jβ2.2 T-cell clone was identified in patient 8, accounting for 10% of gated lymphocytes. Two-color flow cytometry showed that the TCR Vβ17+ population displayed heterogeneous expression of cell-surface vimentin (SC5) but was negative for CD158k. (C) A nonmalignant TCR Vβ14-Jβ1.6 T-cell clone was identified in patient 10 during treatment with anti–CD52 mAb (alemtuzumab) that was heavily expanded and did not express CD158k, as seen on the right based on the results of 2-color flow cytometry.
Intraclonal heterogeneity for CD158k and TCR Vβ expression in patients 1 and 13 and cell sorting analysis. (Ai) Patient 13 showed 2 distinct TCRαβdim and TCRαβbright circulating T-cell populations. Two-color flow cytometry showed that CD158k+ cells were present within both T-cell populations. (Aii) GeneScan Analysis applied to sorted populations revealed that the TCRαβdim population only disclosed clonal TCR Vβ5, TCR Vβ15, and TCR Vβ23. In contrast, the TCRαβbright population had a profile similar to that of whole PBMCs, including clonal TCR Vβ5, Vβ23, and Vβ15, suggesting that malignant and nonmalignant T-cell clones were present in the TCRαβbright and TCRαβdim subsets. (Bi) Two-color flow cytometry revealed heterogeneity for CD158k expression within the malignant TCR Vβ17 population in patient 1. A proportion of Vβ17+ cells did not express CD158k. A minor Vβ17dim subpopulation could be recognized within the TCR Vβ17+ CD158k+ population (arrowhead). (Bii) Through RT-PCR, CD158k mRNA was detected at significant levels in the TCR Vβ17+ CD158k+ population; negative results were obtained in the TCR Vβ17+ CD158k– population.
Intraclonal heterogeneity for CD158k and TCR Vβ expression in patients 1 and 13 and cell sorting analysis. (Ai) Patient 13 showed 2 distinct TCRαβdim and TCRαβbright circulating T-cell populations. Two-color flow cytometry showed that CD158k+ cells were present within both T-cell populations. (Aii) GeneScan Analysis applied to sorted populations revealed that the TCRαβdim population only disclosed clonal TCR Vβ5, TCR Vβ15, and TCR Vβ23. In contrast, the TCRαβbright population had a profile similar to that of whole PBMCs, including clonal TCR Vβ5, Vβ23, and Vβ15, suggesting that malignant and nonmalignant T-cell clones were present in the TCRαβbright and TCRαβdim subsets. (Bi) Two-color flow cytometry revealed heterogeneity for CD158k expression within the malignant TCR Vβ17 population in patient 1. A proportion of Vβ17+ cells did not express CD158k. A minor Vβ17dim subpopulation could be recognized within the TCR Vβ17+ CD158k+ population (arrowhead). (Bii) Through RT-PCR, CD158k mRNA was detected at significant levels in the TCR Vβ17+ CD158k+ population; negative results were obtained in the TCR Vβ17+ CD158k– population.
Discussion
Sézary syndrome is an aggressive, leukemic variant of CTCL that clinically manifests as erythroderma with diffuse adenopathies. SS is characterized by a malignant memory CD4+CD45RO+ T-cell clone that can be roughly identified in the blood and skin by morphologic analysis. In addition to the lack of CD263-5 and CD7,7 new phenotypic markers of Sézary cells have been characterized—KIR3DL2/CD158k10-12 and cell-surface VIM.14,15 The diagnosis of SS relies on the circulating Sézary cell count, as determined by cytomorphology, and T-cell clonality, as determined by molecular approaches.16,22-25 However, the malignancy to T-cell clones has never been determined in patients with SS, and it is unknown whether molecular studies would detect nonmalignant expanded populations.
Through immunoscopy, we identified 28 circulating T-cell clones in 13 patients with SS. Six patients had unique T-cell clones, and the 7 remaining had several. In addition to monoclonal profiles, GeneScan Analysis (Applied Biosystems) revealed abnormal profiles across multiple TCR Vβ families, with lost or restricted patterns, in agreement with findings of a recent study.20 These abnormal profiles, reminiscent of those seen in patients with advanced-stage HIV, are thought to reflect a global loss of T-cell populations in SS.20 Phenotypic studies with anti–TCR Vβ monoclonal antibodies, together with immunoscopy, allowed us to identify an expanded circulating T-cell clone population in 11 patients. For 7 of them, we demonstrated that the anti–TCR Vβ mAb–reactive clonal population coexpressed CD158kdim and VIM and corresponded to malignant Sézary cells. In contrast, with 4 patients, we evidenced an expanded circulating T-cell clone that exhibited heterogeneous expression of cell-surface VIM and failed to express detectable CD158k. We found that the phenotypically identified T-cell clone in these patients corresponded to a nonmalignant population. Finally, for 2 patients, commercially available anti–TCR Vβ mAbs failed to react with any of the T-cell clones identified by immunoscopy. The inability of TCR Vβ–specific antibodies to detect certain T-cell clones is well known,4,16,17,20 and, as an explanation for this phenomenon, it was suggested that differences in the TCR Jβ-Cβ junction, or in CDR3 within a given TCR Vβ family, might influence the reactivity of the monoclonal antibodies.
SS is characterized by cutaneous erythroderma, which is thought to reflect the presence of the malignant T-cell clone in the skin. Our study further confirms that malignant Sézary cells are present in the skin. Indeed, the malignant T-cell clone identified in the blood was evidenced in the skin by GeneScan Analysis (Applied Biosystems) in the 3 patients studied. Interestingly, in addition to the malignant clone, T-cell clones found in the blood and skin seemed to be restricted to one compartment. It is already known that in patients with SS, some T-cell clones may be found in the skin without involvement of the blood.33 This finding is in agreement with the increased diagnostic value of T-cell clonality studies when a similar T-cell clone is found in the skin and the blood.27 Immunoscopy in erythrodermic skin revealed a unique T-cell clone in 2 patients who died of SS, forcing into question the prognostic significance of the restriction of the T-cell repertoire in the skin of patients with SS.
This study shows that 2 distinct types of clonal T-cell expansion, malignant and nonmalignant, can occur in the blood and skin of patients with SS. Therefore, malignant cells in patients with SS cannot be identified by T-cell clonality studies or with the use of molecular analysis of the TCR or flow cytometry with anti–TCR Vβ mAbs. Although the finding of a similar T-cell clone in the blood and skin may provide a defense for the diagnosis of SS,27 we show that only CD158k is a reliable marker for malignant Sézary cells, suitable for diagnosis and follow-up of the disease. The origin of malignant and nonmalignant T-cell clones, their relationship to each other, and the nature of the antigens that may be involved in promoting their emergence or proliferation are interesting issues. Malignant Sézary cells are mature, transformed T cells that can be activated upon TCR engagement. In this study, we present further evidence that malignant and nonmalignant T-cell clones in patients with SS undergo activation in vivo. Indeed they variably express cell-surface VIM, which we have shown to be an early activation marker of T cells.12,13 However, the nature of the antigenic stimulation involved in triggering malignant and nonmalignant cell proliferation in SS is unknown. The finding of expanded nonmalignant T-cell clones in the blood of SS patients is not an unexpected finding. Cytotoxic T-cell populations indeed can be found in the skin and in the blood of patients with CTCL,34-37 and their ability to react specifically to tumor cells has been shown.37 Whether expanded nonmalignant T-cell clones in SS are mostly antitumor cytotoxic expanded T-cell clones is a tempting hypothesis. Interestingly, in one patient receiving anti-CD52 mAb, we identified an expanded nonmalignant TCR Vβ14 T-cell clone that had not been found before the initiation of treatment. Thus, we may also ask whether treatment in itself may generate T-cell clones through the exertion of negative selection on the entire T-cell repertoire. The role of alemtuzumab in generating such a clonal expansion in our patient remains speculative, but, in agreement with this hypothesis, it has been shown that alemtuzumab cross-linking may result in T-cell activation in vitro38,39 and that alemtuzumab may promote the expansion of CD52– T-cell clones in vivo.40-42 On the other hand, the expansion of nonmalignant T-cell clones may be unrelated to the disease or the treatment because expanded T-cell clones can be detected by molecular studies in the blood of patients, especially elderly patients, with benign dermatoses or non–CTCL malignant diseases.27
Identifying one or several common antigen(s) specific to malignant Sézary cells, which may trigger or promote the proliferation of the malignant clone, would represent significant progress in the understanding of the pathogenesis of SS. Studies focusing on the T-cell repertoire in CTCL have suggested that malignant T-cell clones in SS may initially expand in the skin as a result of superantigenic stimulation.43-46 In agreement with this hypothesis, it has been shown that malignant and nonmalignant T-cell clonal expansion in CTCL may occur within the same TCR Vβ family in the blood and the skin of a given patient.43-47 Moreover, in a large retrospective series of CTCLs, it was suggested that TCR Vβ use in CTCLs may be skewed toward TCR Vβ5.1, a finding thought to reflect either TCR Vβ5 expansion or TCR Vβ depletion among other families after superantigenic stimulation.48 The role of superantigens in the pathogenesis of CTCL remains controversial. In other studies, no bias was found in the TCR Vβ use of the expanded population.49 In this series, we found a slightly predominant use of TCR Vβ17, TCR Vβ8, and TCR Vβ22 (Table 4), whereas only one TCR Vβ5 T-cell clone was found with negative staining in the corresponding mAb using flow cytometry. This does not support the role of superantigens in the pathogenesis of T-cell clones in SS. In addition to superantigens, it is tempting to speculate that nominal antigen(s) may be involved in the pathogenesis of SS. We could not identify a common sequence when we compared the CDR3-deduced amino acid sequences of all nonmalignant and malignant T-cell clones characterized in this study (data not shown).
TCR Vβ use by T-cell clones found in the 13 patients studied and by Sézary cell lines
. | T-cell clones defined by immunoscopy . | . | Expanded T-cell clones phenotypically characterized . | . | . | |||
|---|---|---|---|---|---|---|---|---|
| TCR Vβ family . | PBMC, 13 patients . | Skin, 3 patients . | Malignant + SCLs . | Nonmalignant . | Total . | |||
| 1 | 0 | 0 | 0 | 0 | 0 | |||
| 2 | 1 | 1 | 1 | 0 | 1 | |||
| 3 | 3 | 0 | 0 | 0 | 0 | |||
| 4 | 1 | 0 | 0 | 0 | 0 | |||
| 5 | 2 | 0 | 0 | 0 | 0 | |||
| 6 | 1 | 0 | 0 | 0 | 0 | |||
| 7 | 1 | 0 | 1 | 0 | 1 | |||
| 8 | 2 | 1 | 2 | 0 | 2 | |||
| 9 | 0 | 0 | 0 | 0 | 0 | |||
| 11 | 1 | 0 | 1 | 0 | 1 | |||
| 12 | 0 | 0 | 0 | 0 | 0 | |||
| 13 | 1 | 0 | 1 | 0 | 1 | |||
| 14 | 2 | 0 | 0 | 2 | 2 | |||
| 15 | 2 | 1 | 0 | 0 | 0 | |||
| 16 | 1 | 1 | 0 | 0 | 0 | |||
| 17 | 4 | 1 | 1 | 1 | 2 | |||
| 18 | 1 | 0 | 0 | 0 | 0 | |||
| 20 | 1 | 0 | 0 | 0 | 0 | |||
| 21 | 1 | 0 | 0 | 0 | 0 | |||
| 22 | 2 | 0 | 1 | 2 | 3 | |||
| 23 | 2 | 0 | 1 | 0 | 1 | |||
| 24 | 0 | 0 | 0 | 0 | 0 | |||
| Total | 28 | 5 | 9 | 5 | 14 | |||
. | T-cell clones defined by immunoscopy . | . | Expanded T-cell clones phenotypically characterized . | . | . | |||
|---|---|---|---|---|---|---|---|---|
| TCR Vβ family . | PBMC, 13 patients . | Skin, 3 patients . | Malignant + SCLs . | Nonmalignant . | Total . | |||
| 1 | 0 | 0 | 0 | 0 | 0 | |||
| 2 | 1 | 1 | 1 | 0 | 1 | |||
| 3 | 3 | 0 | 0 | 0 | 0 | |||
| 4 | 1 | 0 | 0 | 0 | 0 | |||
| 5 | 2 | 0 | 0 | 0 | 0 | |||
| 6 | 1 | 0 | 0 | 0 | 0 | |||
| 7 | 1 | 0 | 1 | 0 | 1 | |||
| 8 | 2 | 1 | 2 | 0 | 2 | |||
| 9 | 0 | 0 | 0 | 0 | 0 | |||
| 11 | 1 | 0 | 1 | 0 | 1 | |||
| 12 | 0 | 0 | 0 | 0 | 0 | |||
| 13 | 1 | 0 | 1 | 0 | 1 | |||
| 14 | 2 | 0 | 0 | 2 | 2 | |||
| 15 | 2 | 1 | 0 | 0 | 0 | |||
| 16 | 1 | 1 | 0 | 0 | 0 | |||
| 17 | 4 | 1 | 1 | 1 | 2 | |||
| 18 | 1 | 0 | 0 | 0 | 0 | |||
| 20 | 1 | 0 | 0 | 0 | 0 | |||
| 21 | 1 | 0 | 0 | 0 | 0 | |||
| 22 | 2 | 0 | 1 | 2 | 3 | |||
| 23 | 2 | 0 | 1 | 0 | 1 | |||
| 24 | 0 | 0 | 0 | 0 | 0 | |||
| Total | 28 | 5 | 9 | 5 | 14 | |||
Phenotypic characterization of expanded T-cell clones relies on TCR Vβ, CD 158k, and cell-surface VIM expression. For PBMCs, n = 13 patients; for skin, n = 3.
SCLs indicates Sézary cell lines (HUT78 and Pno).
Interestingly, heterogeneous expression of TCRαβ, CD158k, and cell-surface VIM was demonstrated within the circulating T-cell clones. The presence of both TCRαβdim and TCRαβbright subsets in the blood of patients with SS may suggest that the CD3/TCRαβ complex is recruited at the cell surface in a subset of circulating lymphocytes undergoing activation. Interestingly, the ability of a malignant Sézary cell line to be activated upon TCRαβ stimulation was previously shown.28 Nevertheless, some of our findings indicate that malignant and nonmalignant T-cell clones might have a diminished CD3/TCRαβ expression. Therefore, in contrast to findings of another study,6 we believe that this feature cannot be used as a marker for Sézary cells. In addition, we observed that malignant and nonmalignant T-cell clones in patients with SS displayed heterogeneous expression of cell-surface VIM, recently shown to be a marker of early activation.14,15 Altogether these findings suggest that subsets of malignant and nonmalignant clones are activated in the blood of patients with SS. Different hypotheses have been proposed to explain why malignant Sézary cells display heterogeneous expression of CD158k. One is that malignant Sézary cells may initially lack CD158k and may differentiate toward a CD158k+ phenotype. Another is that CD158k may be recruited at the cell surface by its ligands, resulting in a transient modulation of its cell-surface expression. The finding of clearly distinct CD158k– and CD158k+ subsets within the malignant T-cell clones in patient 1 with early disease and the absence of detectable levels of CD158k mRNA in the CD158k– subset support the first hypothesis. Recent studies indicate that KIR expression is stable in NK cells and T cells, in contrast to NKG2A, whose expression is increased by cytokines. However, several lines of evidence indicate that KIR expression in normal T cells is acquired at a mature stage, after rearrangement of the TCR. In patients with rheumatoid arthritis, it has been shown that CD158b+ and CD158b– subsets of a unique memory T-cell clone may coexist in the synovium50 and that the KIR repertoire may be diverse within each circulating CD4+CD28null T-cell clone.51 Our findings present further evidence that KIR expression may be acquired by mature T cells and by transformed malignant T cells.
In conclusion, this study demonstrates that CD158k delineates most malignant Sézary cells and that nonmalignant, activated T-cell clones may be present in the blood of patients with SS. Thus, malignant cells in patients with SS cannot be definitively identified by molecular T-cell clonality or by flow cytometry studies using anti–TCR Vβ mAbs. Rather, CD158k appears to be the tool of choice for the diagnosis of SS and the follow-up of patients, and it can be used in routine practice. However, the significance of the heterogeneous expression of CD158k by malignant Sézary cells remains to be explained. It is especially important to determine whether this heterogeneous expression reflects different steps in the progression toward malignancy and whether CD158k engagement by its ligand may play a role in controlling the proliferation or apoptosis of malignant cells.
Prepublished online as Blood First Edition Paper, January 17, 2006; DOI 10.1182/blood-2005-10-4239.
Supported by grants from the INSERM, Université Paris XII, Société Française de Dermatologie, Société de Recherche Dermatologique, and l'Association pour la Recherche sur le Cancer (M.B., A.B).
M.B., P.M., and A.B. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Hervé Bachelez (Department of Dermatology, Hôpital Saint-Louis, Paris, France) for providing clinical information about patient 10, Marie-Hélène Delfau-Larue (Department of Immunobiology, Hôpital Henri Mondor, Créteil France) for T-cell clonality studies using PCR-DGGE, Nadine Martin-Garcia (Department of Pathology, Hôpital Henri Mondor) for technical assistance with the immunostaining procedure, and Valérie Velayoudame-Ortonne (IM3, Hôpital Henri Mondor) for spectratyping and sequencing techniques and the use of GeneScan and ABI Prism 3100 sequencer software.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal