Translational regulation by oncogenic proteins may be a rapid and efficient mechanism to modulate gene expression. We report here the identification of the CEBPB gene as a target of translational regulation in myeloid precursor cells transformed by the BCR/ABL oncogene. Expression of CEBPB was repressed in 32D-BCR/ABL cells and reinduced by imatinib (STI571) via a mechanism that appears to depend on expression of the CUG-repeat RNA-binding protein CUGBP1 and the integrity of the CUG-rich intercistronic region of c/ebpβ mRNA. Constitutive expression or conditional activation of wild-type CEBPB induced differentiation and inhibited proliferation of 32D-BCR/ABL cells in vitro and in mice, but a DNA binding-deficient CEBPB mutant had no effect. The proliferation-inhibitory effect of CEBPB was, in part, mediated by the CEBPB-induced GADD45A gene. Because expression of CEBPB (and CEBPA) is low in the blast crisis (BC) stage of chronic myelogenous leukemia (CML) and is inversely correlated with BCR/ABL tyrosine kinase levels, these findings point to the therapeutic potential of restoring C/EBP activity in CML-BC and, perhaps, other types of acute leukemia.
Introduction
Chronic myelogenous leukemia (CML) is a clonal myeloproliferative disorder arising from neoplastic transformation of the hematopoietic stem cell.1,2 Its clinical course involves progression from a protracted chronic phase (CML-CP), characterized by accumulation of apparently normal neutrophils, to a rapidly fatal blast crisis (CML-BC), characterized by clonal expansion of differentiation-arrested myeloid or lymphoid blasts. CML is consistently associated with a reciprocal translocation of the long arms of chromosomes 9 and 22,3,4 which generates the BCR/ABL fusion gene, in turn translated in the p210BCR/ABL oncoprotein in almost all patients.5 Expression activity of p210BCR/ABL is necessary and sufficient for hematopoietic cell transformation and disease maintenance as demonstrated by in vitro assays, leukemogenesis in mice, and the antileukemia effects of imatinib, a specific BCR/ABL tyrosine kinase inhibitor.6-9
BCR/ABL-dependent transformation of hematopoietic cells involves the assembly of multiprotein complexes and the phosphorylation of various substrates, which is essential to generate proliferative and antiapoptotic signals10-12 and is often accompanied by transcriptional and posttranscriptional changes in gene expression. In regard to the latter mechanism, BCR/ABL can regulate both positively and negatively mRNA translation and protein stability.13-15
We recently identified translation-regulatory mechanisms involving increased expression of RNA-binding proteins that modulate MDM2 and CEBPA levels in BCR/ABL-expressing cells.16,17 Enhanced expression of MDM2 by increased expression of the RNA-binding protein La reduces the susceptibility of BCR/ABL-expressing cells to apoptosis induced by DNA-damaging agents.16 Suppression of CEBPA expression by increased expression of the RNA-binding protein hnRNPE2 is important for the differentiation arrest of BCR/ABL-expressing leukemic cells as indicated by the rapid induction of differentiation on reactivation of CEBPA expression or activity.17,18
Because translational regulation by BCR/ABL might not be limited to MDM2 and C/EBPA, we sought to identify other translation-regulated genes by probing oligonucleotide microarrays with polysomal RNA of untreated and STI571-treated BCR/ABL-expressing myeloid precursor 32Dcl3 cells. Because polysomal mRNA is efficiently translated, some of the mRNAs more abundant in STI571-treated cells may be translationally repressed by BCR/ABL. The identification of these genes may be important for understanding the mechanisms underlying the antileukemia effects of STI571 and may reveal molecules able to bypass resistance to STI571, which develops in some CML-CP patients and in the majority of CML-BC patients.19 We report here the identification of CEBPB as one such STI571 target and show that restoring its activity in BCR/ABL-transformed cells inhibits proliferation and induces differentiation.
Materials and methods
Plasmids
WT-C/EBPβ-HA. The full-length CEBPB (including 37 nucleotides from the 5′UTR) was amplified by reverse transcription-polymerase chain reaction (RT-PCR) from 32Dcl3 total RNA using a sense (5′-GGACGCAGCGGAGCCCGC-3′), and an antisense (5′-CTCGGCGGGCCACTGCTAG-3′) primer. This PCR product was reamplified with a primer containing a 5′-flapping XhoI site flanked by the 5′UTR, and a primer containing a 5′-flapping EcoRI site flanked by the HAtag and a mutated CEBPB STOP codon and subcloned into the XhoI/EcoRI-digested MigRI vector.
ΔuORF-C/EBPβ-HA. This plasmid was obtained by PCR from pWT-C/EBPβ with a primer containing a 5′-flapping XhoI site (sense 5′-ATGGAAGTGGCCAACTTCTACTA-3′), and an antisense primer containing a 5′-flapping EcoRI site flanked by the HA tag and a mutated CEBPB STOP codon; the PCR product was subcloned into the XhoI/EcoRI-digested MigRI vector.
Δ(231-242) C/EBPβ-HA. This plasmid was generated by PCR with QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) from pΔuORF-C/EBPβ-HA.
ΔuORF-C/EBPβ-ERTAM and Δ(231-242) C/EBPβ-ERTAM. These plasmids were generated by PCR as follows. (1) The ligand-binding domain of the murine estrogen receptor (ER) was amplified by RT-PCR from 32Dcl3 RNA and point-mutated (Gly 525; Arg). (2) The PCR product was then reamplified with an oligomer containing a 5′-flapping BamHI site and a primer containing a 5′-flapping EcoRI site. (3) ΔuORF-C/EBPβ and Δ(231-242) CEBPB were amplified by PCR from the respective plasmids with a primer containing a 5′-flapping XhoI site and a primer containing a 3′-flapping BamHI site after a mutated CEBPB STOP codon. ΔuORF-C/EBPβ orΔ(231-242) CEBPB and ERTAM PCR products were subcloned into the XhoI/EcoRI-digested MigRI vector.
CUGBP1-Flag. The full-length CUGBP1 was generated by RT-PCR from 32Dcl3 RNA using a sense (5′-ATGAACGGCACCCTGGACCA-3′) and an antisense (5′-TCAGTAGGGCTTACTACTATTCT-3′) primer. The cDNA product was reamplified with a Flag-tagged 5′-flapping XhoI site oligomer, cloned into the TA cloning vector pCR2.1 (Invitrogen, Grand Island, NY), and reinserted into the XhoI/EcoRI-digested MigRI or MSCV vector.
GADD45α-HA. This plasmid was generated by RT-PCR from total RNA of 4-HT–treated C/EBPβ-ERTAM-expressing 32D BCR-ABL cells with a sense (5′-ATGACTTTGGAGGAATTCTCGG-3′) and an antisense (5′-ATCACCGTTCCGGGAGATTAAT-3′) primer. The cDNA product was PCR-amplified with a primer containing a 5′-flapping XhoI site preceding theATG starting site and 19 downstream nucleotides, and a primer containing a 5′-flapping EcoRI site after the HA tag and a mutated GADD45A STOP codon and subcloned into the XhoI/EcoRI-digested MigRI vector.
GADD45A shRNAs. Forward and reverse oligonucleotides targeting 3 different 19 bases (Gi1 nt747-765, Gi2 nt280-299, Gi3 nt92-110) of GADD45A mRNA were synthesized according to Brummelkamp et al20 ; a 19 mer sequence with a similar GC/AT composition but without homology to other mouse mRNAs was used as control. Each oligonucleotide pair was annealed and cloned into the BamHI/XhoI-digested pSRP vector (kindly provided by Stephen Lessnick, Dana Farber Cancer Center, Boston, MA). Each plasmid was sequenced to verify that it contained the designed hairpin sequence.
Cells, retroviral infections, proliferation, and differentiation assays
CD34+ normal bone marrow (NBM) cells from different healthy donors were purchased from AllCells (LLC, Berkley, CA) and their use was approved by the Ohio State University Institutional Review Board (IRB). Mononuclear hematopoietic cells from bone marrow of unidentifiable patients with CML were Ficoll-separated and used to isolate CD34+ cells (CD34 MultiSort Kit; Miltenyi Biotec, Auburn, CA). NBM, CML-CP, and CML-BC CD34+ cells were kept overnight in Iscove modified Dulbecco medium (IMDM) supplemented with 50% FBS, 2 mM glutamine, and human recombinant IL-3 (20 ng/mL), IL-6 (20 ng/mL), Flt-3 ligand (100 ng/mL), and KL (100 ng/mL) from StemCell Technologies (Vancouver, BC, Canada) and processed for Western blotting (see “Western blotting analysis and immunoprecipitation of UV–cross-linked cytoplasmic extracts”). Specimens from patients with CML, obtained from the Ohio State University Leukemia Tissue Bank (Columbus, OH), were used with approval from the Ohio State University IRB.
Murine IL-3–dependent 32Dcl3 cells, 32D BCR-ABL–expressing cells, and derivative cell lines were maintained in culture in IMDM supplemented with 10% FBS, 2 mM l-glutamine, and 10% of WEHI-conditioned medium as source of IL-3. Philadelphia1 K562 cells were maintained in culture in IMDM supplemented with 10% FBS and 2 mM l-glutamine. Retroviral infections of BCR/ABL-expressing 32Dcl3 cells and Philadelphia1 K562 cells were carried out as described21 ; 24 hours after infection, cells were sorted (EPICS Profile Analyzer; Coulter, Hialeah, FL) for green fluorescent protein (GFP) expression, or cultured in the presence of puromycin (2.5 μg/mL); thereafter, cells were kept in culture as described.16
For differentiation assays, cells were washed in phosphate-buffered saline (PBS) and plated in IMDM supplemented with 10% FBS, 2 mM l-glutamine, 25 ng/mL recombinant G-CSF (Calbiochem, San Diego, CA) with or without 4-hydroxytamoxifen (4-HT; Sigma, St Louis, MO) to activate C/EBPβ-ERTAM or Δ(231-242) C/EBPβ-ERTAM. Differentiation was evaluated by morphology (light microscopy of May-Grünwald-Giemsa–stained cytospins) or flow cytometry detection of the differentiation marker Gr-1 with a specific phycoerythrin (PE)–conjugated mouse monoclonal antibody (PharMingen, San Diego, CA). Inhibition of cell proliferation was assessed by methylcellulose colony assays or in liquid cultures, with or without 4-HT treatment.
Western blotting analysis and immunoprecipitation of UV–cross-linked cytoplasmic extracts
Cells were lysed in Laemmli buffer (2 × 105 cells/20 μL) and used for Western blot analysis. Proteins of interest were detected with anti-CEBPB (Δ198) polyclonal antibody, anti-CUGBP1 (3B1) monoclonal antibody, anti-G-CSFR (M-20) polyclonal antibody, anti-GADD45a (H-165) polyclonal antibody (all from Santa Cruz Biotechnology, Santa Cruz, CA), and anti-GRB2 monoclonal antibody (610112, BD Transduction Laboratories, Lexington, KY). Exogenous proteins were detected with monoclonal anti-HA.11 epitope antibody (MMS-101-P Covance) or with the anti-Flag M2 HRP-conjugated epitope monoclonal antibody (A8592; Sigma). Cytoplasmic extracts for UV–cross-linking and immunoprecipitation (IP) were obtained from 32D BCR/ABL cells expressing or not CUGBP1-Flag as described.15 Lysates were incubated (20 minutes at 23°C) with 2 × 105 cpm of 32P-labeled oligoribonucleotide βLAPu, which contains the sequence of mouse CEBPB mRNA from the first AUG to the STOP codon of the upstream ORF (5′-AUG CAC CGC CUG CUG GCC UGG GAC GCA GCA UGC CUC CCG CCG CCG CCC GCC GCC UUU AG-3′) or with 2 × 105 cpm of the 32P-labeled oligoribonucleotide sORFβ, which contains the sequence of the short uORF from the second AUG to the STOP codon (5′-AUG CCU CCC GCC GCC GCC CGC CGC CUU UAG-3′). Reactions were exposed to UV light for 30 minutes and immunoprecipitated with an anti-Flag M2-conjugated agarose resin; cytoplasmic extracts and IPs were resolved on a 4% to 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gradient, transferred onto a nitrocellulose membrane, immunoblotted with an anti-Flag HRP-conjugated antibody and exposed to an x-ray film for detection of the radioactive signal (overnight, –80°C).
Isolation of total/polysome-associated RNA and Northern blot analysis
Polysome-associated RNA was prepared from 108 untreated or STI571-treated (2 μM, 12 hours) 32D-BCR/ABL cells as described.16,22 Polysomal RNA was prepared by phenol/chloroform extraction and isopropanol precipitation, washed with ethanol, and resuspended in 30 μL DEPC-treated water. Total RNA was extracted using Tri-Reagent (Sigma, St. Louis, MO). For Northern blot analysis, total or polysomal RNA (10 μg) was fractionated onto denaturing 1% agarose/6.6% formaldehyde gels, transferred onto Hybond-nylon membrane (Amersham Pharmacia Biotech, Piscataway, NJ), and hybridized to a 32P-labeled murine CEBPB cDNA 500-bp fragment. For Northern blot analysis of 4-HT–treated, retrovirally transduced 32D-BCR/ABL cells, total RNA was extracted at the indicated times using Tri-Reagent, fractionated (10 μg) onto denaturing 1% agarose/6.6% formaldehyde gels and transferred onto Hybond-nylon membrane. The membrane was hybridized to the following 32P-labeled probes: a PstI 470-bp fragment of murine myeloperoxidase (MPO) 23 ; a 357-bp fragment of murine CD33 generated by RT-PCR from RNA of 32Dcl3 cells; the full-length cDNA of murine GADD45A from pGADD45α-HA. GAPDH mRNA levels were monitored as control for equal loading.
Microarray experiments
For identification of STI571-regulated mRNAs, 5 μg polysomal and total RNA were used to generate cRNA as described.24 Fragmented RNA (10 μg) was hybridized onto the Affymetrix MG-U74Av2 array interrogating the expression of about 6000 genes and about 6000 EST clusters. After hybridization, raw expression data were rescaled as described to account for chip intensity differences.25 Data were preprocessed using a minimum value of 20 and a maximum of 16 000 with a maximum/minimum filter of 2. After preprocessing, genes were ranked based on degree of fold changes.
EMSA and luciferase assay
Nuclear extracts were obtained from 293T cells transfected with the indicated plasmids by calcium-phosphate DNA precipitation and electrophoresis mobility shift assay (EMSA) was performed by incubating 10 μg of nuclear extracts with 50 000 cpm of double-stranded 32P-labeled oligonucleotide (5′-AGGTGTT-GCAATC-CCCAGC-3′, which includes the underlined C/EBP-binding site) as described.26 Unlabeled oligonucleotide was used as competitor at a 100-fold molar excess.
For luciferase assay, 293T cells were cotransfected with the reporter plasmid pTK-G-CSFR-luciferase (kind gift of D. G. Tenen, Harvard Institutes of Medicine) and the indicated CEBPB plasmids by calcium-phosphate DNA precipitation. Twenty-four hours after transfection, firefly luciferase activity was recorded on a luminometer using the Luciferase Assay System (Promega, Madison, WI). Results (counts of luciferase activity) are representative of 3 experiments. Expression of CEBPB was assessed by Western blotting.
Survival of C3H/Hej leukemic mice conditionally expressing a functional CEBPB
Four- to 6-week-old C3H/Hej mice (8/group) were sublethally irradiated (500 Gy) and, 24 hours later, injected intravenously with ΔuORF-C/EBPβ-ERTAM-expressing 32D-BCR/ABL cells (5 × 105/mouse). Two days later, mice were injected intraperitoneally with 4-HT (1 mg/d/15 d), dissolved in 100 μL sterile sunflower seed oil (Sigma), or treated with oil alone. Colony formation assays were performed 18 days after leukemic-cell injection using peripheral blood withdrawn retro-orbitally from 3 mice for each group. Blood was treated with a red-cell lysing solution (StemCell Technologies) and mononuclear cells used for morphologic examination (May-Grünwald-Giemsa staining of cytospins) or for methylcellulose plating in absence of cytokines. Images were photographed with an Olympus microscope with a 40 ×/0.65 NA objective lens and an Olympus SC35 type 12 camera (Hitech Instruments, Newtown Square, PA). JPEG images were viewed with Adobe Photoshop (Adobe Systems, San Jose, CA), and contrast adjustments were made.
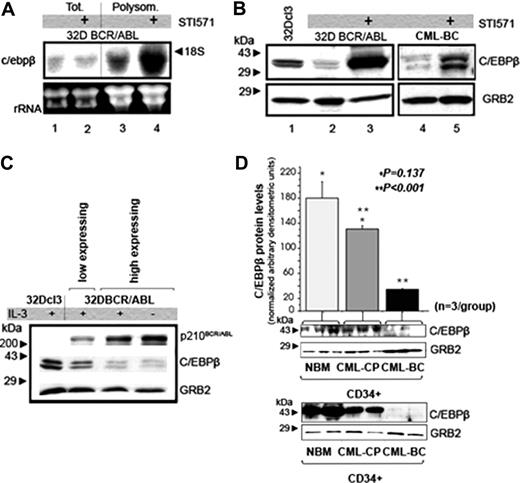
CEBPB mRNA and protein levels in STI571-treated 32D-BCR/ABL cells. (A) Northern blot analysis on total (lanes 1 and 2) or polysome-associated (lanes 3-4) RNA from untreated (lanes 1 and 3) or STI571-treated (2 μM, 12 hours) 32D BCR/ABL cells (lanes 2 and 4). A 500-base segment of the c/ebpβ mRNA 3′UTR was 32P-radiolabeled and used as probe; 28S and 18S are shown as a control of equal RNA loading. (B) Western blot analysis of CEBPB expression in total extracts from 32Dcl3 cells, untreated and STI571-treated (1 μM, 24 hours), 32D-BCR/ABL cells, untreated and STI571-treated (2 μM, 24 hours) mononuclear cells from a CML-BC sample. Western blotting with an anti-GRB2 antibody was used as control of equal loading. (C) Western blot shows CEBPB and tyrosine-phosphorylated BCR/ABL levels in 32Dcl3 cells transduced with MigRI-BCR/ABL and sorted by low and high GFP expression. Anti-GRB2 Western blotting was used as control of equal loading. (D) Histogram (top) and Western blot (bottom, first row) show CEBPB protein levels (expressed as mean ± SE of densitometric units after normalization with GRB2 levels) in the CD34+ fraction from bone marrow of healthy donors (NBM; n = 3), CML-CP (n = 3), and CML-BC (n = 3) patients (NBM versus CML-CP, *P = .137; CML-CP versus CML-BC, **P < .001; t test). The Western blot in the second row shows CEBPB levels in 2 of the 3 NBM, CML-CP, and CML-BC samples still available for a second analysis.
CEBPB mRNA and protein levels in STI571-treated 32D-BCR/ABL cells. (A) Northern blot analysis on total (lanes 1 and 2) or polysome-associated (lanes 3-4) RNA from untreated (lanes 1 and 3) or STI571-treated (2 μM, 12 hours) 32D BCR/ABL cells (lanes 2 and 4). A 500-base segment of the c/ebpβ mRNA 3′UTR was 32P-radiolabeled and used as probe; 28S and 18S are shown as a control of equal RNA loading. (B) Western blot analysis of CEBPB expression in total extracts from 32Dcl3 cells, untreated and STI571-treated (1 μM, 24 hours), 32D-BCR/ABL cells, untreated and STI571-treated (2 μM, 24 hours) mononuclear cells from a CML-BC sample. Western blotting with an anti-GRB2 antibody was used as control of equal loading. (C) Western blot shows CEBPB and tyrosine-phosphorylated BCR/ABL levels in 32Dcl3 cells transduced with MigRI-BCR/ABL and sorted by low and high GFP expression. Anti-GRB2 Western blotting was used as control of equal loading. (D) Histogram (top) and Western blot (bottom, first row) show CEBPB protein levels (expressed as mean ± SE of densitometric units after normalization with GRB2 levels) in the CD34+ fraction from bone marrow of healthy donors (NBM; n = 3), CML-CP (n = 3), and CML-BC (n = 3) patients (NBM versus CML-CP, *P = .137; CML-CP versus CML-BC, **P < .001; t test). The Western blot in the second row shows CEBPB levels in 2 of the 3 NBM, CML-CP, and CML-BC samples still available for a second analysis.
Results
Expression of CEBPB is translationally regulated in 32D-BCR/ABL cells
To identify genes translationally regulated by BCR/ABL, we probed an Affymetrix array with polysome- and monosome-associated RNA from untreated and STI571-treated 32D-BCR/ABL cells. In 32D-BCR/ABL cells treated with STI571 for 24 hours, approximately 700 polysome-associated mRNAs showed at least a 2-fold down-regulation, whereas 80 mRNAs were up-regulated more than 4-fold, suggesting that some of these mRNAs are efficiently transcribed and translated after inhibition of BCR/ABL kinase activity or that enhanced polysome loading reflects only translational regulation. Because none of the polysome-associated mRNAs of STI571-treated cells were more abundant in the monosome fractions of untreated 32D-BCR/ABL cells, as might be expected if poorly translated mRNAs become efficiently translated, we tested polysomal RNA and protein levels of selected genes. Expression of CEBPB, a member of the CCAAT/enhancer-binding family of transcription factors, was increased approximately 4-fold in polysomal RNA of STI571-treated cells.
By Northern blot hybridization, c/ebpβ mRNA levels were similar in total RNA of IL-3–cultured untreated and STI571-treated 32D-BCR/ABL cells (Figure 1A lanes 1 and 2), but showed a 4- to 5-fold increase in polysomal RNA of STI571-treated 32D-BCR/ABL cells (Figure 1A lanes 3 and 4). To assess whether c/ebpβ polysomal mRNA levels correlated with increased protein expression, Western blot analysis was performed with an anti-CEBPB antibody; after 24 hours of STI571 treatment, 32D-BCR/ABL cells showed a marked increase in CEBPB expression (compare lanes 2 and 3 in figure 1B). STI571 treatment also induced an increase in CEBPB levels in mononuclear cells from a CML-CP patient (not shown) and in mononuclear cells from a CML-BC patient with a wild-type (WT) BCR/ABL kinase domain (compare lanes 4 and 5 in Figure 1B).
To assess whether CEBPB and BCR/ABL expression were inversely correlated, 32Dcl3 cells were retrovirally transduced with MigRI-BCR/ABL and low- and high-expressing cells selected by sorting low- and high-expressing GFP+ cells. Western blots of lysate of GFP-sorted cells showed lower CEBPB expression in samples with higher levels of tyrosine-phosphorylated BCR/ABL (Figure 1C).
CEBPB protein levels were also compared in purified CD34+ cells from healthy marrow donors (NBM; n = 3), CML-CP (n = 3), and CML-BC (n = 3) bone marrow samples (Figure 1D). In 2 different Western blot experiments, one with cell lysates of all 3 NBM, CML-CP, and CML-BC samples and one with 2 of the 3, CEBPB expression was similar in NBM and CML-CP samples, but markedly reduced in CML-BC samples (Figure 1D). CEBPB expression was inversely correlated with BCR/ABL levels, which were higher in the CML-BC (∼75% increase) than in the CML-CP samples (not shown).
The CEBPB gene generates 2 functional isoforms (known as LAP1 and LAP2) and an N-terminal truncated form (known as LIP) whose levels are, in part, regulated translationally by an intercistronic segment that includes a short upstream open reading frame (uORF) and a CUG-rich region.27-29 Two CEBPB isoforms, one carrying the full-length CEBPB coding sequence (including the out-of-frame uORF) tagged to the HA epitope at the 3′ (WT-C/EBPβ) and one lacking the first initiation codon and the uORF-containing intercistronic region of c/ebpβ mRNA (ΔuORF C/EBPβ), were cloned in the MigRI vector (Figure 2A) and transduced in 32D-BCR/ABL cells and HA-tagged CEBPB levels assessed by anti-HA Western blotting in GFP-sorted cells before and after treatment with STI571. Treatment with STI571 induced increased expression of HA-tagged CEBPB and HA-tagged LIP in cells transduced with the WT-C/EBPβ retrovirus (compare lanes 3 and 4 in Figure 2B), but not in cells transduced with the ΔuORF C/EBPβ retrovirus (compare lanes 5 and 6 in Figure 2B). Because the c/ebpβ-HA transcript levels were identical in untreated and treated cells (not shown), these data suggest c/ebpβ mRNA translation is inhibited in BCR/ABL-expressing cells.
The CUG repeat binding protein CUGBP1 regulates CEBPB expression
A region between the first and the second AUG starting codon of c/ebpβ mRNA is reported to interact with a CUG repeat binding protein, CUGBP1.30 This interaction appears to be important for the translational regulation of c/ebpβ mRNA in hepatocytes and in mammary epithelial cells.30,31
To assess whether CUGBP1 regulates the increased CEBPB expression in STI571-treated 32D-BCR/ABL cells, levels of CUGBP1 and CEBPB were evaluated by Western blotting in parental and in untreated or STI571-treated 32D-BCR/ABL cells (Figure 3A). CUGBP1 expression is higher in parental than in 32D-BCR/ABL cells; after a 24-hour treatment with STI571, 32D-BCR/ABL cells showed an increase in CUGBP1 expression comparable to that of untreated parental cells. The induction of CUGBP1 expression correlated well with that of CEBPB, suggesting a functional link.
The relationship between CUGBP1 and CEBPB expression was also tested in CD34+ cells from the CML-CP and CML-BC samples used to measure CEBPB levels (Figure 1D). CUGBP1 levels were more abundant in CML-CP than in CML-BC samples (Figure 3B), in excellent correlation with those of CEBPB (Figure 1D).
To assess whether CUGBP1 regulates CEBPB protein levels, the murine CUGBP1 cDNA fused to a Flag tag was cloned in the MigRI vector and transduced into 32D-BCR/ABL cells. Anti-C/EBPβ Western blotting in sorted GFP+ cells overexpressing CUGBP1-Flag revealed markedly increased CEBPB levels compared to those of MigRI-transduced cells (Figure 3C lanes 1 and 2). Of interest, levels of LIP were also increased, consistent with a previous study.31 Levels of CEBPB were clearly more abundant also in CUGBP1-expressing Philadelphia1 K562 cells (Figure 3C lanes 3 and 4), but induction of LIP was not consistently observed (not shown).
To determine whether the functional relationship between CEBPB and CUGBP1 can be explained by a physical interaction between c/ebpβ mRNA and CUGBP1, the radiolabeled βLAPu or sORFβ oligo RNAs were UV–cross-linked to cytoplasmic extracts from MigRI-transduced or CUGBP1-Flag–overexpressing 32D-BCR/ABL cells and immunoprecipitated with an anti-Flag antibody. IPs and total extracts were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with an anti-Flag antibody (Figure 3D upper panel); after the decay of the peroxidase signal, the membrane was also exposed to an x-ray film to detect any radioactive labeling (Figure 3D lower panel). A shifted and radiolabeled complex of the same size was present in the immunoprecipitate of 32D-BCR/ABL CUGBP1-Flag cells with the βLAPu, but not with the sORFβ oligo-RNA (Figure 3D lower panel), consistent with a physical interaction between c/ebpβ mRNA and CUGBP1.
Role of the c/ebpβ mRNA intercistronic region in the STI571-dependent CEBPB expression in 32D-BCR/ABL cells. (A) Schematic representation of the expression vectors MigRI GFP WT-C/EBPβ (top) and MigRI GFP ΔuORF-C/EBPβ (bottom), in which the CEBPB-coding sequence is fused in-frame to the HA epitope at 3′. ATG1, 3, and 4 correspond to the initiation codon for LAP1, LAP2, and LIP, respectively. (B) Western blot analysis of HA-tagged CEBPB protein. Total lysate from 293T cells transfected with MigRI GFP WT-C/EBPβ-HA, used as positive control (lane 1), GFP-sorted 32D-BCR/ABL cells carrying the MigRI GFP empty vector (lane 2), GFP-sorted MigRI GFP WT-C/EBPβ cells (lanes 3-4), or MigRI GFP ΔuORF C/EBPβ cells (lanes 5-6) untreated (lanes 3 and 5) or treated (lanes 4 and 6) for 24 hours with 1 μM STI571 were probed with an antibody against the HA epitope. Equal loading was assessed by probing the filter with an anti-GRB2 antibody.
Role of the c/ebpβ mRNA intercistronic region in the STI571-dependent CEBPB expression in 32D-BCR/ABL cells. (A) Schematic representation of the expression vectors MigRI GFP WT-C/EBPβ (top) and MigRI GFP ΔuORF-C/EBPβ (bottom), in which the CEBPB-coding sequence is fused in-frame to the HA epitope at 3′. ATG1, 3, and 4 correspond to the initiation codon for LAP1, LAP2, and LIP, respectively. (B) Western blot analysis of HA-tagged CEBPB protein. Total lysate from 293T cells transfected with MigRI GFP WT-C/EBPβ-HA, used as positive control (lane 1), GFP-sorted 32D-BCR/ABL cells carrying the MigRI GFP empty vector (lane 2), GFP-sorted MigRI GFP WT-C/EBPβ cells (lanes 3-4), or MigRI GFP ΔuORF C/EBPβ cells (lanes 5-6) untreated (lanes 3 and 5) or treated (lanes 4 and 6) for 24 hours with 1 μM STI571 were probed with an antibody against the HA epitope. Equal loading was assessed by probing the filter with an anti-GRB2 antibody.
Role of CUGBP1 in imatinib-induced CEBPB expression. (A) Levels of expression of CUGBP1 and CEBPB proteins assessed by Western blotting in parental (lane 1) or 32D-BCR/ABL-expressing cells untreated or treated with 1 μM STI571 (lanes 2-4). Specific antibodies against C/EBPβ or CUGBP1 were used to probe the nitrocellulose filter. (B) Histogram (top) and Western blot (bottom) show CUGBP1 levels (expressed in the graph as mean ± SE of densitometric units after normalization with GRB2 levels) in the CD34+ fraction from NBM (n = 3), CML-CP (n = 3), and CML-BC (n = 3) marrow samples (NBM versus CML-CP, *P = .272; CML-CP versus CML-BC, **P = .003; t test). GRB2 levels were measured as loading control. (C) Effects of CUGBP1 on C/EBPβ expression in GFP-sorted 32D-BCR/ABL cells or puromycin-selected K562 cells. Western blot shows CEBPB levels in empty vector-transduced cells (lanes 1 and 3) or in cells ectopically expressing CUGBP1 (lanes 2 and 4). Exogenous CUGBP1 was detected by anti-Flag Western blotting. (D) Immunoprecipitation with anti-Flag–conjugated agarose beads: cytoplasmic extracts (lanes 1 and 4) from GFP-sorted cells carrying the empty vector (lane 4), or expressing CUGBP1-Flag (lane 1) were UV–cross-linked and immunoprecipitated after incubation with 32P-labeled oligo-RNAs βLAPu (lanes 3 and 5) or sORFβ (lane 2). Cytoplasmic extracts (lanes 1 and 4) and immunoprecipitates (lanes 2, 3, and 5) were resolved by SDS-PAGE and then probed in Western blotting by an anti-Flag epitope antibody (top panel); the filter was then exposed to an x-ray film to detect the radioactive signal of the RNA-protein complex (bottom panel).
Role of CUGBP1 in imatinib-induced CEBPB expression. (A) Levels of expression of CUGBP1 and CEBPB proteins assessed by Western blotting in parental (lane 1) or 32D-BCR/ABL-expressing cells untreated or treated with 1 μM STI571 (lanes 2-4). Specific antibodies against C/EBPβ or CUGBP1 were used to probe the nitrocellulose filter. (B) Histogram (top) and Western blot (bottom) show CUGBP1 levels (expressed in the graph as mean ± SE of densitometric units after normalization with GRB2 levels) in the CD34+ fraction from NBM (n = 3), CML-CP (n = 3), and CML-BC (n = 3) marrow samples (NBM versus CML-CP, *P = .272; CML-CP versus CML-BC, **P = .003; t test). GRB2 levels were measured as loading control. (C) Effects of CUGBP1 on C/EBPβ expression in GFP-sorted 32D-BCR/ABL cells or puromycin-selected K562 cells. Western blot shows CEBPB levels in empty vector-transduced cells (lanes 1 and 3) or in cells ectopically expressing CUGBP1 (lanes 2 and 4). Exogenous CUGBP1 was detected by anti-Flag Western blotting. (D) Immunoprecipitation with anti-Flag–conjugated agarose beads: cytoplasmic extracts (lanes 1 and 4) from GFP-sorted cells carrying the empty vector (lane 4), or expressing CUGBP1-Flag (lane 1) were UV–cross-linked and immunoprecipitated after incubation with 32P-labeled oligo-RNAs βLAPu (lanes 3 and 5) or sORFβ (lane 2). Cytoplasmic extracts (lanes 1 and 4) and immunoprecipitates (lanes 2, 3, and 5) were resolved by SDS-PAGE and then probed in Western blotting by an anti-Flag epitope antibody (top panel); the filter was then exposed to an x-ray film to detect the radioactive signal of the RNA-protein complex (bottom panel).
Ectopic expression of CEBPB inhibits proliferation and induces differentiation of 32D-BCR/ABL cells
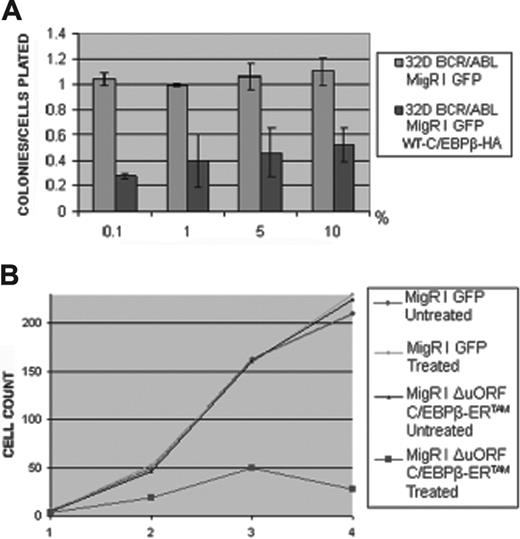
The biologic effects induced by restoring CEBPB expression in 32D-BCR/ABL cells were first tested by comparing colony formation of GFP-sorted control and CEBPB-transduced cells. Expression of CEBPB caused a 75% to 80% inhibition of colonies growing in the presence of decreasing concentrations of WEHI-conditioned medium (Figure 4A). Cell proliferation was also tested in liquid culture assays using cells expressing the C/EBPβ-ERTAM chimeric protein. On treatment with 4-HT, proliferation of ΔuORF C/EBPβ-ERTAM cells is markedly suppressed as indicated by an approximately 75% inhibition after 4 days in culture (Figure 4B). Similar data were obtained using individual clones (not shown).
The ability of CEBPB to induce granulocytic differentiation of 32D-BCR/ABL cells was tested by morphologic examination of cytospins from cultures of 4-HT–treated cells and by measuring the expression of granulocytic differentiation markers such as the Gr-1 and CD33 antigens, the granulocyte-colony stimulating factor receptor (G-CSFR) and myeloperoxidase (MPO).
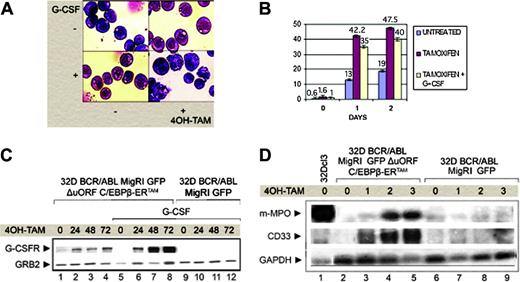
As indicated by May-Grünwald-Giemsa staining (Figure 5A), 4-HT treatment of ΔuORF C/EBPβ-ERTAM 32D-BCR/ABL cells induced morphologic features (reduction of the nucleus-cytoplasm ratio, nuclear indentation and segmentation) typical of differentiating cells, although end cells with trilobate or multilobate nuclei were infrequent.
By flow cytometry, there was a marked increase in the number of Gr-1+ cells, 1 and 2 days after 4-HT treatment (Figure 5B). Levels of G-CSFR, monitored by Western blotting, also showed an increase; it was modest at 24 hours, but more marked at 48 and 72 hours, especially in cells treated with G-CSF (Figure 5C). MPO and CD33 mRNA levels were also increased on CEBPB activation (Figure 5D).
The biologic effects of CEBPB in 32D-BCR/ABL cells require the DNA-binding activity
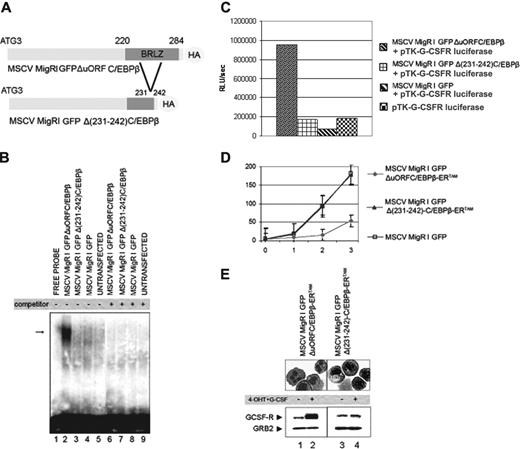
To test whether the effects of CEBPB in 32D-BCR/ABL cells require its DNA-binding activity, we generated a deletion mutant of the CEBPB basic region (Figure 6A) and tested its ability to interact with the C/EBP-binding site of the G-CSFR promoter and to activate C/EBP-dependent luciferase activity. Nuclear extracts from 293T cells ectopically expressing WT-C/EBPβ bound the C/EBP-binding site, whereas no binding was detected using nuclear extracts of Δ(231-242) C/EBPβ-transfected cells (Figure 6B). In transient transfection assays, Δ(231-242) C/EBPβ did not enhance luciferase activity driven by multiple C/EBP-binding sites of the G-CSFR promoter (Figure 6C).
Effect of CEBPB expression on 32D-BCR/ABL cell proliferation. (A) Methylcellulose colonies (as ratio on plated cells, on the y-axis) from GFP-sorted 32D-BCR/ABL cells transduced with MigRI GFP or MigRI GFP WT-C/EBPβ-HA and plated in the presence of increasing concentration of WEHI-conditioned medium as source of IL-3 (on the x-axis). Values are the mean plus SD of 3 different experiments. (B) Proliferation in liquid culture of untreated or 4-HT–treated (250 nM) cells carrying the MigRI GFP or the MigRI GFP ΔuORF C/EBPβ-ERTAM retrovirus; cell number (× 103) is on the y-axis, day of treatment is on the x-axis.
Effect of CEBPB expression on 32D-BCR/ABL cell proliferation. (A) Methylcellulose colonies (as ratio on plated cells, on the y-axis) from GFP-sorted 32D-BCR/ABL cells transduced with MigRI GFP or MigRI GFP WT-C/EBPβ-HA and plated in the presence of increasing concentration of WEHI-conditioned medium as source of IL-3 (on the x-axis). Values are the mean plus SD of 3 different experiments. (B) Proliferation in liquid culture of untreated or 4-HT–treated (250 nM) cells carrying the MigRI GFP or the MigRI GFP ΔuORF C/EBPβ-ERTAM retrovirus; cell number (× 103) is on the y-axis, day of treatment is on the x-axis.
CEBPB expression induces granulocytic differentiation of 32D-BCR/ABL cells. (A) May-Grünwald-Giemsa staining of cytospin from 32D-BCR/ABL transduced with the MigRI GFP ΔuORF C/EBPβ-ERTAM retrovirus, induced or not with 4-HT and untreated or treated with G-CSF. (B) Gr-1 expression in MigRI GFP C/EBPβ-ERTAM-transduced 32D-BCR/ABL cells. The percentage of Gr-1+ cells (assessed by immunostaining with a PE-conjugated anti–Gr-1 antibody) is expressed on the y-axis, whereas x values indicate length of treatment (in days). Values shown are the mean plus SD of 3 different experiments. (C) Western blot analysis of G-CSFR expression in transduced 32D-BCR/ABL cells. Cells were treated with 4-HT alone (lanes 1-4) or in combination with G-CSF (lanes 5-12). Equal loading was monitored by anti-GRB2 Western blotting. (D) MPO and CD33 antigen mRNA levels on total RNA from G-CSF–treated (3 days) 32Dcl3 cells as positive control (lane 1), MigRI GFP ΔuORF C/EBPβ-ERTAM (lanes 2-5), or MigRI GFP-transduced (lanes 6-9) 32D-BCR/ABL cells before (lanes 2 and 6) and after (lanes 3-5 and 7-9) 4-HT treatment.
CEBPB expression induces granulocytic differentiation of 32D-BCR/ABL cells. (A) May-Grünwald-Giemsa staining of cytospin from 32D-BCR/ABL transduced with the MigRI GFP ΔuORF C/EBPβ-ERTAM retrovirus, induced or not with 4-HT and untreated or treated with G-CSF. (B) Gr-1 expression in MigRI GFP C/EBPβ-ERTAM-transduced 32D-BCR/ABL cells. The percentage of Gr-1+ cells (assessed by immunostaining with a PE-conjugated anti–Gr-1 antibody) is expressed on the y-axis, whereas x values indicate length of treatment (in days). Values shown are the mean plus SD of 3 different experiments. (C) Western blot analysis of G-CSFR expression in transduced 32D-BCR/ABL cells. Cells were treated with 4-HT alone (lanes 1-4) or in combination with G-CSF (lanes 5-12). Equal loading was monitored by anti-GRB2 Western blotting. (D) MPO and CD33 antigen mRNA levels on total RNA from G-CSF–treated (3 days) 32Dcl3 cells as positive control (lane 1), MigRI GFP ΔuORF C/EBPβ-ERTAM (lanes 2-5), or MigRI GFP-transduced (lanes 6-9) 32D-BCR/ABL cells before (lanes 2 and 6) and after (lanes 3-5 and 7-9) 4-HT treatment.
We also generated a plasmid in which the Δ(231-242) CEBPB mutant was fused in-frame with the 4-HT–responsive ER ligand-binding domain. 4-HT treatment induced a marked decrease in the proliferation of ΔuORF C/EBPβ-ERTAM-expressing cells, most evident after 2 and 3 days (Figure 6D), but had no effect on Δ(231-242) C/EBPβ-ERTAM-expressing cells (Figure 6D).
CEBPB effects on proliferation and differentiation require its DNA-binding activity. (A) Schematic representation of MigRI GFP ΔuORF C/EBPβ (top) and MigRI GFP ΔP231-242) CEBPB (bottom) plasmids. (B) DNA-binding activity of WT and mutant CEBPB assessed by EMSA using a 32P-labeled double-stranded oligonucleotide corresponding to the G-CSFR C/EBP-binding site and lysates from untransfected 293T cells (lane 9) or cells transfected with MigRI GFP (lanes 4 and 8), MigRI GFP ΔuORF C/EBPβ (lanes 2 and 6), or MigRI GFP Δ(231-242) CEBPB (lanes 3 and 7). Binding specificity was assessed using the G-CSFR C/EBP-binding site oligonucleotide as cold competitor (lanes 6-9). Arrow indicates the DNA-protein complex. (C) Luciferase assay (representative of 3 different experiments) on cell lysate from 293T transfected with a reporter gene in which luciferase activity is driven by 4 C/EBP-binding sites (G-CSFR pTK G-CSFR-luciferase) alone, or in the presence of the MigRI GFP empty vector, or ΔuORF or Δ(231-242) CEBPB. Levels of HA-C/EBPβ expression (bottom) were tested by Western blotting with an anti-HA antibody; equal loading was demonstrated by monitoring GRB2 levels. (D) Proliferation of 4-HT–treated (250 nM) cells carrying the MigRI GFP (□), the MigRI GFP ΔuORF C/EBPβ-ERTAM (♦), or the MigRI GFP Δ(231-242) C/EBPβ-ERTAM (▴) retrovirus; cell number (× 103) is on the y-axis, day of treatment is on the x-axis. Values are the mean plus SD of 3 different experiments. (E) May-Grünwald-Giemsa staining of cytospin from MigRI GFP ΔuORF C/EBPβ-ERTAM (left) or MigRI GFP Δ(231-242) C/EBPβ-ERTAM (right) 32D-BCR/ABL cells 3 days after treatment with 4-HT (250 nM) and G-CSF (25 ng/mL); G-CSFR levels are shown at the bottom.
CEBPB effects on proliferation and differentiation require its DNA-binding activity. (A) Schematic representation of MigRI GFP ΔuORF C/EBPβ (top) and MigRI GFP ΔP231-242) CEBPB (bottom) plasmids. (B) DNA-binding activity of WT and mutant CEBPB assessed by EMSA using a 32P-labeled double-stranded oligonucleotide corresponding to the G-CSFR C/EBP-binding site and lysates from untransfected 293T cells (lane 9) or cells transfected with MigRI GFP (lanes 4 and 8), MigRI GFP ΔuORF C/EBPβ (lanes 2 and 6), or MigRI GFP Δ(231-242) CEBPB (lanes 3 and 7). Binding specificity was assessed using the G-CSFR C/EBP-binding site oligonucleotide as cold competitor (lanes 6-9). Arrow indicates the DNA-protein complex. (C) Luciferase assay (representative of 3 different experiments) on cell lysate from 293T transfected with a reporter gene in which luciferase activity is driven by 4 C/EBP-binding sites (G-CSFR pTK G-CSFR-luciferase) alone, or in the presence of the MigRI GFP empty vector, or ΔuORF or Δ(231-242) CEBPB. Levels of HA-C/EBPβ expression (bottom) were tested by Western blotting with an anti-HA antibody; equal loading was demonstrated by monitoring GRB2 levels. (D) Proliferation of 4-HT–treated (250 nM) cells carrying the MigRI GFP (□), the MigRI GFP ΔuORF C/EBPβ-ERTAM (♦), or the MigRI GFP Δ(231-242) C/EBPβ-ERTAM (▴) retrovirus; cell number (× 103) is on the y-axis, day of treatment is on the x-axis. Values are the mean plus SD of 3 different experiments. (E) May-Grünwald-Giemsa staining of cytospin from MigRI GFP ΔuORF C/EBPβ-ERTAM (left) or MigRI GFP Δ(231-242) C/EBPβ-ERTAM (right) 32D-BCR/ABL cells 3 days after treatment with 4-HT (250 nM) and G-CSF (25 ng/mL); G-CSFR levels are shown at the bottom.
Role of GADD45A expression in CEBPB-dependent proliferation inhibition of 32D-BCR/ABL cells. (A) Left: Northern blot analysis on total RNA from 32D-BCR/ABL cells transduced with MigRI GFP ΔuORF-C/EBPβ-ERTAM (lanes 1-5) or with MigRI GFP (lanes 6-9) before (lanes 1 and 6) or after (lanes 2-5 and 7-9) 4-HT treatment (250 nM). The full-length coding sequence of GADD45A was used as probe; the GAPDH probe was used to control equal loading. Right: Western blot analysis of GADD45A expression in STI571-treated CML-BC mononuclear cells. (B) Left: methylcellulose colony formation (as ratio on plated cells) from 32D-BCR/ABL cells transduced with the MigRI GFP or the MigRI GFP GADD45α-HA retrovirus. Right: proliferation of MigRI GFP or MigRI GFP GADD45α-HA-transduced 32D-BCR/ABL cells; values reported in both panels correspond to the mean plus SD of 3 different experiments. (C) Left: GADD45A expression in ΔuORF C/EBPβ-ERTAM 32D-BCR/ABL cells transduced with GADD45A or a control shRNA retrovirus before (lanes 1, 3, 5, 7, 9) and after (lanes 2, 4, 6, 8, 10) 4-HT treatment (250 nM). Right: proliferation of 4-HT treated ΔuORF C/EBPβ-ERTAM 32D-BCR/ABL cells (untransduced, transduced with pSRP GADD45ARNAi2 and pSRP GADD45α-RNAi3 together, or with a control shRNA). Representative of 3 different experiments; percentage of viable over untreated cells (y-axis) after 1, 2, or 3 days of 4-HT treatment (x-axis).
Role of GADD45A expression in CEBPB-dependent proliferation inhibition of 32D-BCR/ABL cells. (A) Left: Northern blot analysis on total RNA from 32D-BCR/ABL cells transduced with MigRI GFP ΔuORF-C/EBPβ-ERTAM (lanes 1-5) or with MigRI GFP (lanes 6-9) before (lanes 1 and 6) or after (lanes 2-5 and 7-9) 4-HT treatment (250 nM). The full-length coding sequence of GADD45A was used as probe; the GAPDH probe was used to control equal loading. Right: Western blot analysis of GADD45A expression in STI571-treated CML-BC mononuclear cells. (B) Left: methylcellulose colony formation (as ratio on plated cells) from 32D-BCR/ABL cells transduced with the MigRI GFP or the MigRI GFP GADD45α-HA retrovirus. Right: proliferation of MigRI GFP or MigRI GFP GADD45α-HA-transduced 32D-BCR/ABL cells; values reported in both panels correspond to the mean plus SD of 3 different experiments. (C) Left: GADD45A expression in ΔuORF C/EBPβ-ERTAM 32D-BCR/ABL cells transduced with GADD45A or a control shRNA retrovirus before (lanes 1, 3, 5, 7, 9) and after (lanes 2, 4, 6, 8, 10) 4-HT treatment (250 nM). Right: proliferation of 4-HT treated ΔuORF C/EBPβ-ERTAM 32D-BCR/ABL cells (untransduced, transduced with pSRP GADD45ARNAi2 and pSRP GADD45α-RNAi3 together, or with a control shRNA). Representative of 3 different experiments; percentage of viable over untreated cells (y-axis) after 1, 2, or 3 days of 4-HT treatment (x-axis).
Inhibition of proliferation induced by CEBPB depends, in part, on enhanced GADD45A expression
To identify CEBPB-regulated genes, total RNA from untreated and 4-HT–treated (12 hours) cells was used to probe an oligonucleotide microarray including 20 000 mRNAs. The mRNA of growth arrest and DNA damage-inducible gene (GADD45A), a p53-regulated gene involved in the regulation of the cell cycle, apoptosis, and genomic stability,32-34 was up-regulated approximately 6-fold after a 12-hour 4-HT treatment.
GADD45A induction was confirmed by Northern blotting of total RNA from 4-HT–treated ΔuORF C/EBPβ-ERTAM-expressing 32D-BCR/ABL cells (Figure 7A, left panel). Expression of GADD45A mRNA is barely detectable in untreated cells, but was readily evident on 4-HT treatment of ΔuORF C/EBPβ-ERTAM-transduced cells; in the control line, no expression is detectable up to 48 hours, but a moderate increase was visible at 72 hours, probably reflecting a stress response to the high density reached at 72 hours by the cultured cells.
Because the expression of CEBPB is up-regulated by STI571 and GADD45A levels are enhanced by CEBPB, we reasoned that STI571 treatment of BCR/ABL-expressing cells may also lead to increased GADD45A levels. As shown in Figure 7A (right panel), GADD45A levels were higher in STI571-treated CML-BC mononuclear cells (same sample as in Figure 1), although it cannot be excluded that the effect of STI571 is, at least in part, CEBPB dependent.
32D-BCR/ABL cells ectopically expressing GADD45A were less clonogenic (Figure 7B left) and proliferated more slowly (Figure 7B right) than parental cells. To assess whether the proliferation inhibition induced by CEBPB requires GADD45A expression, 3 different pSRP retroviruses expressing shRNAs targeting GADD45A mRNA and a retrovirus carrying a control sequence were transduced in ΔuORF C/EBPβ-ERTAM 32D-BCR/ABL cells and the ability of the shRNAs to down-regulate GADD45A expression was tested by Western blotting with an anti-GADD45A antibody after 4-HT treatment (Figure 7C left). Expression of GADD45A shRNA 2 and 3 prevented the CEBPB-induced up-regulation of GADD45A, whereas the control sequence had no effect (Figure 7C left). In cells transduced with ΔuORF C/EBPβ-ERTAM only, 4-HT treatment readily induced GADD45A expression (Figure 7C left). We then tested whether the inhibition of cell proliferation induced by CEBPB activation in 32D-BCR/ABL cells would be rescued by expression of GADD45 shRNAs 2 and 3. Each day after 4-HT treatment, cells transduced with GADD45A shRNAs were more numerous than control cells (Figure 7D right), consistent with a role of GADD45A expression in the inhibition of cell proliferation induced by CEBPB.
Conditional activation of CEBPB in vivo prolongs survival of leukemic mice
To test whether restoration of CEBPB expression in BCR/ABL-transformed cells suppresses leukemogenesis in mice, 4- to 6-week-old C3H/Hej mice were sublethally irradiated (500 Gy) and 24 hours later injected intravenously with ΔuORF C/EBPβ-ERTAM-expressing 32D-BCR/ABL cells (3 × 105/mouse). Two days after cell injection, mice of the treated group received a daily dose of 4-HT (1 mg/d for 15 days, intraperitoneally) dissolved in sunflower seed oil, whereas the control group was injected with oil alone, for 15 days or until death occurred.
Conditional activation of CEBPB in vivo prolongs survival of leukemic mice. (A) Methylcellulose colonies from peripheral-blood cells (5 × 102) of untreated and 4-HT–treated C3H/Hej mice injected with 32D-BCR/ABL MigRI GFP ΔuORF C/EBPβ-ERTAM cells; values indicate mean (plus SD) colony number from cells of 3 different mice for each group. (B) May-Grünwald-Giemsa staining of white blood cells from untreated (top panel) and 4-HT–treated (bottom panel) C3H/Hej mice injected with 32D-BCR/ABL MigRI ΔuORF GFP C/EBPβ-ERTAM cells. (C) Survival of untreated (▴) and 4-HT–treated (•) C3H/Hej mice injected with 32D-BCR/ABL MigRI GFP ΔuORF C/EBPβ-ERTAM cells. The y-axis represents survival percentage of untreated and treated mice; x-axis values indicate time of survival after injection (days).
Conditional activation of CEBPB in vivo prolongs survival of leukemic mice. (A) Methylcellulose colonies from peripheral-blood cells (5 × 102) of untreated and 4-HT–treated C3H/Hej mice injected with 32D-BCR/ABL MigRI GFP ΔuORF C/EBPβ-ERTAM cells; values indicate mean (plus SD) colony number from cells of 3 different mice for each group. (B) May-Grünwald-Giemsa staining of white blood cells from untreated (top panel) and 4-HT–treated (bottom panel) C3H/Hej mice injected with 32D-BCR/ABL MigRI ΔuORF GFP C/EBPβ-ERTAM cells. (C) Survival of untreated (▴) and 4-HT–treated (•) C3H/Hej mice injected with 32D-BCR/ABL MigRI GFP ΔuORF C/EBPβ-ERTAM cells. The y-axis represents survival percentage of untreated and treated mice; x-axis values indicate time of survival after injection (days).
Eighteen days after injection of leukemic cells, we evaluated the leukemic process by assessing morphology and colony-forming potential of peripheral-blood leukocytes of 3 mice for each group. Peripheral-blood cells from untreated mice retained a high colony-forming potential in the absence of cytokines, whereas cells from treated mice formed approximately one tenth of the colonies (Figure 8A). Most peripheral-blood cells from treated mice showed multilobated nuclei typical of terminally differentiated cells (reflecting recovery of normal granulopoiesis), whereas cytospins of peripheral-blood cells from untreated mice revealed high numbers of immature blasts (reflecting circulating transformed cells; Figure 8B). These morphologic features are consistent with the observation that, when the analyses were performed, the number of peripheral-blood GFP+ cells in untreated mice ranged from 30% to 50%, whereas it was less than 5% in 4-HT–treated mice (not shown).
To assess whether restoration of CEBPB activity was sufficient to prolong the survival of leukemic mice, we monitored the deaths of the animals in the 2 groups (Figure 8C). Treated mice survived longer than untreated controls (median survival of 29.1 versus 18.5 days), suggesting that continuous expression of a functional CEBPB can inhibit BCR/ABL-induced leukemogenesis in vivo.
Discussion
Translational regulation has emerged as a mechanism by which BCR/ABL can modulate gene expression.15 In this study, we attempted to identify translationally regulated genes by probing microarrays with efficiently (polysome-associated) and poorly translated (monosome-associated) mRNAs. We also reasoned that treatment of BCR/ABL-transformed cells with STI571 may promote changes in mRNA translation detectable by microarray hybridization.
Approximately 80 mRNAs were more abundant in the polysomal fraction of STI571-treated 32D-BCR/ABL than in that of untreated cells, suggesting that translational-dependent perturbation of protein synthesis may be important for the phenotype (inhibition of proliferation and enhanced apoptosis susceptibility) of drug-treated cells. Of the 80 mRNAs identified, CEBPB attracted our attention because: (1) it is a member of a family of transcriptional regulators that control differentiation and proliferation in many cell types35 ; (2) it is a functional equivalent of CEBPA based on its ability to restore granulocytic differentiation in CEBPA knockout mice36 ; and (3) it is induced by all-trans-retinoic acid (ATRA) in acute promyelocytic leukemia (APL) cells and is required for the ATRA-dependent differentiation of these cells.37
Translational regulation of c/ebpβ mRNA in BCR/ABL-expressing cells
CEBPB mRNA generates 3 protein isoforms: p38 CEBPB (LAP1, liver-enriched activated protein) from the first AUG, p34 CEBPB (LAP2) from the third, and p20 LIP (liver-enriched inhibiting protein) from the fourth.27-29 The LIP isoform lacks the N-terminal transactivation domain and inhibits the high-molecular-weight isoforms by forming nonfunctional heterodimers.38 An additional out-of-frame AUG, located between the first and the third AUG, has the potential to generate a small uORF that ends before the p34 CEBPB starting site. This small sequence can form a stem-loop secondary structure considered critical for translational regulation of CEBPB isoform expression by favoring translational reinitiation from a downstream AUG.28
By assessing the ability of STI571 to modulate the expression of HA-tagged CEBPB from 2 c/ebpβ mRNAs, one of which includes the first AUG and the intercistronic region, we identified an STI571-responsive sequence lying between the first and the third AUG. This sequence, which contains a CUG-repeat region potentially bound by members of the CUG-binding proteins (CUGBPs), was essential for the increased levels of LAP2 and LIP. The effects of STI571 were mimicked by overexpression of CUGBP1, which led to enhanced levels of the p34 and, to a lesser degree, the p38 CEBPB isoform. Expression of LIP was slightly induced in 32D-BCR/ABL cells, but not in K562 cells, suggesting that the relative induction of LAP2 and LIP may depend on the differential effect of CUGBP1 concentration in promoting translation initiation at AUG 3 and 4 and, perhaps, on cell-type–specific mechanisms. Based on the ratio between LAP2 and LIP in STI571-treated and CUGBP1-expressing cells (Figures 2 and 3), we favor the interpretation that STI571 and CUGBP1 primarily enhance LAP2 translation. Such an increase correlated with the physical interaction of CUGBP1 with the CUG-rich c/ebpβ oligoribonucleotide, suggesting that the enhanced expression of CEBPB in STI571-treated BCR/ABL cells depends on increased association of CUGBP1 with the CUG-rich region of c/ebpβ mRNA and enhanced translation of c/ebpβ mRNA.
Another RNA-binding protein implicated in regulation of c/ebpβ mRNA translation is calreticulin.39 Calreticulin mRNA levels were increased after STI571 treatment of 32D-BCR/ABL cells, but 32D-BCR/ABL cells ectopically expressing calreticulin showed no changes in CEBPB levels (not shown), suggesting that in these cells calreticulin does not regulate CEBPβ expression.
Biologic effects of CEBPB in BCR/ABL-transformed cells
C/EBP proteins are crucial regulators of the balance between differentiation and proliferation in various tissues35 ; in particular, CEBPA and C/EBPϵ are necessary for early and late normal granulopoiesis, respectively, as indicated by the phenotype of the corresponding knockout mice.40,41 CEBPB is a regulator of several biologic processes in the liver and adipose tissue,42,43 but recent findings have also implicated CEBPB in the control of granulopoiesis.36,37
Consistent with the observation that CEBPB expression is induced by and is required for ATRA-dependent differentiation of APL cells,37 we found that ectopic expression or inducible activation of CEBPB promotes morphologic differentiation and the expression of molecular markers of the granulocytic lineage in differentiation-arrested murine 32D-BCR/ABL cells and Philadelphia1 K562 cells (not shown).
Although enhanced activity of CEBPB can promote differentiation of normal44 and malignant myeloid precursors (this study), no alterations in normal granulocytic differentiation were described in the CEBPB knockout mice.45,46 This is not surprising because CEBPB knockout mice are likely to express normal levels of CEBPA, as CEBPA expression precedes that of CEBPB during normal myelopoiesis.47 However, expression of CEBPB from the c/ebpα gene locus is sufficient to restore normal granulopoiesis in CEBPA knockout mice,36 suggesting that CEBPα and CEBPB have redundant but not overlapping activities. In myeloid precursor cells transformed by BCR/ABL, the expression of both CEBPA17,48 and CEBPB (this study) is repressed, which may explain why either one can induce differentiation when activated. Nevertheless, CEBPA is a more potent inducer of granulocytic differentiation in 32D-BCR/ABL cells (data not shown), consistent with microarray screening data that indicate modulation of overlapping but also distinct genes (not shown). Like CEBPA,17 the expression of CEBPB is often repressed in blast cells of CML-BC patients (this study), suggesting that loss of CEBPA and CEBPB activity contributes to differentiation arrest and aggressive behavior of CML-BC cells.
CEBPB suppresses the leukemogenic potential of 32D-BCR/ABL cells by inducing granulocytic differentiation and by inhibiting cell proliferation. The relative abundance of LIP and LAP proteins has been considered important for the ability of certain cell types to proliferate or differentiate28 ; in regenerating liver of mice after partial hepatectomy, increased levels of LIP versus LAP CEBPB promote cell-cycle progression by induction of cell-cycle regulators such as cyclins, whereas enhanced LAP expression delays cell-cycle progression.49 In our study, ectopic expression of p34LAP or induction of C/EBPβ-ERTAM by 4-HT treatment was sufficient to inhibit proliferation of leukemic cells in murine 32D-BCR/ABL cells and in Philadelphia1 K562 cells (not shown).
Activation of CEBPB suppressed leukemogenesis in mice injected with C/EBPβ-ERTAM-expressing 32D-BCR/ABL cells as indicated by a reduced number of leukemic cells in the peripheral blood of 4-HT–-treated mice, the appearance of normal granulocytes, and the longer survival of 4-HT–treated mice. The antileukemia effects of CEBPB in vivo were, however, not striking, possibly reflecting the inability of CEBPB to induce terminal differentiation of the totality of BCR/ABL-expressing cells. Moreover, CEBPB activation did not induce apoptosis of 32D-BCR/ABL cells (not shown), suggesting that BCR/ABL-transformed cells not committed to terminal differentiation but proliferation arrested resumed their normal rate of proliferation when active CEBPB was no longer available. Alternatively, a cohort of 32D-BCR/ABL cells insensitive ab initio to CEBPB might have been responsible for the death of the mice.
The effects of CEBPB LAP on cell proliferation and differentiation appear to be dependent on its transcription activation function because a DNA binding-deficient mutant of CEBPB was unable to enhance C/EBP-dependent gene expression, to induce differentiation or to block proliferation.
Among the genes activated by CEBPB identified through microarray studies, GADD45A was of special interest because it appears to regulate cell-cycle progression by interacting with or modulating the activity of several cell-cycle regulators such as the proliferating-cell nuclear antigen (PCNA),50 p21,51,52 cdc2/Cdk1,53 and cyclin B1.54 In particular, GADD45A expression induces a G2 cell-cycle arrest32 by interacting with cdc2/Cdk1 and disrupting the formation of Cdk1-cyclin B1 complexes.54
As expected, ectopic expression of GADD45A in 32D-BCR/ABL cells markedly suppressed colony formation and proliferation. Of greater interest, inhibition of GADD45A expression by RNA interference reversed, in part, the proliferation-inhibitory effects of CEBPB, establishing that GADD45A is a novel effector of CEBPB.
In summary, we have identified CEBPB as a gene translationally regulated by STI571 in 32D-BCR/ABL cells. Activation of gene expression by this drug is consistent with mechanisms of cytotoxicity dependent on reactivation of pathways normally repressed by BCR/ABL. One of these pathways leads to suppression of p38 kinase activity,55 which may or may not be involved in regulating CEBPB expression. The effects of CEBPB in BCR/ABL-expressing cells suggest that enhanced CEBPB expression might contribute to the cytotoxic effects of STI571.
The transition from CML-CP to CML-BC is associated with the accumulation of genetic abnormalities (ie, p53 mutation) and with changes in gene expression (ie, down-modulation of CEBPA levels),56 which may explain the increased apoptosis resistance and differentiation arrest of CML-BC cells. Because CEBPB levels are also low in BCR/ABL-transformed cell lines and in CML-BC cells, a complete loss of C/EBP activity might be necessary to disrupt the differentiation potential of CML-BC progenitors. These data and those of CEBPA activation in BCR/ABL-expressing cells (G.F.-A., Karen Keeshon, Michelle Zattoni, C. G., George Iotti, Nick J. Donato, and B. C., submitted for publication) also point to the therapeutic potential of restoring C/EBP activity in CML-BC and, perhaps, in other types of acute myeloid leukemia.
Prepublished online as Blood First Edition Paper, January 17, 2006; DOI 10.1182/blood-2005-08-3181.
Supported, in part, by National Institutes of Health grants CA 95111 and PO1 78890 (B.C.) and CA 95512 (D.P.) and by the US Army, Chronic Myelogenous Leukemia Research Program grant DAMD 17-03-1-0184 (D.P.). C.G. was supported by a fellowship of the American-Italian Foundation for Cancer Research. G.F.-A. was supported, in part, by a fellowship of the A. Serra Foundation for Cancer Research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Cathy Tomastik for editorial assistance and Marea D. Pollard for mice irradiation.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal