Abstract
Exposure to HIV-1 does not necessarily result in infection and progression toward disease, thus suggesting that the control of viral infection may be achieved. Antibodies to CCR5 have been detected in HIV-exposed but uninfected subjects (ESNs); thus, these antibodies could be involved in HIV protection. To assess whether anti-CCR5 antibodies may also contribute to slow HIV disease progression, we searched for anti-CCR5 antibodies in 497 subjects, including 85 long-term nonprogressors (LTNPs), 70 progressors, 135 HIV+ patients treated with highly active antiretroviral therapy (HAART), and 207 seronegative donors. We found anti-CCR5 antibodies in a fraction of the LTNPs(23.5%) but not in the other populations studied (P < .001). These antibodies recognized a conformational epitope within the first extramembrane loop of CCR5, and they induced a stable and long-lasting downregulation of CCR5 on the surface of T lymphocytes, which inhibited HIV entry. In addition, CD4+ lymphocytes from LTNPs having anti-CCR5 antibodies are resistance to R5 strains of HIV-1. Follow-up studies showed that the loss of anti-CCR5 antibodies occurred in some subjects, and this loss was significantly associated with a progression toward disease, whereas subjects who retained anti-CCR5 Abs maintained their LTNP status. Induction of anti-CCR5 Abs could be relevant to vaccine design and therapeutics.
Introduction
HIV-1 infection is defined by different outcomes of disease progression over time. Rapid progressors (RPs) display a rapid decline in CD4+ T-lymphocyte count, developing AIDS in a few years and produce neutralizing antibodies poorly. Conversely, long-term nonprogressors (LTNPs),1,2 maintain CD4+ T-cell count (≥ 500 CD4+ T cell/mL) and healthy clinical parameters for 7 to 10 years, control viral replication without treatment, and mount cell-mediated and humoral response,3-6 and the production of neutralizing antibodies is usually robust.7-9 Multiple mechanisms have been invoked to explain the LTNP status,1,3 including specific HLA alleles (HLA-B27 and -B57) as well as mutations in human genes for cellular coreceptors CCR5 and CCR2 and in nonstructural HIV-1 genes, (eg, nef, vif, or vpr).10-15 Transmitted HIV-1 strains generally use the CCR5 coreceptor (R5 viruses).16-18 HIV-neutralizing anti-CCR5 antibodies have been found in HIV-exposed seronegative individuals (ESNs),19,20 where they have been hypothesized to play a protective role. We searched for such a humoral response in a diverse cohort of HIV-infected subjects.
Patients, materials, and methods
Study population
A total of 290 HIV-1–seropositive patients from 2 different cohorts were analyzed. The group included 85 LTNPs, 55 men and 30 women ranging in age between 14 and 65 years. Seventy-five LTNPs were in the Milan cohort of LTNPs,21 and 10 LTNPs were from prospective studies of HIV-1 infection in the United States: 5 LTNPs from the United States were enrolled in the Women's Interagency HIV Study (WIHS), an NIH multicenter study of the natural history of HIV-1 infection in women22 ; 5 United States LTNPs took part in an HIV-1 pathogenesis study performed at the Wadsworth Center of the New York State Department of Health (NYS-DOH). Most patients were examined at 6-month intervals, and at least 3 serial samples for each individual were collected. Every 6 months, all patients had T-lymphocyte analysis. Virus isolation, typing of virus coreceptor usage, and viral load were also assessed repeatedly in the course of follow-up of the Milan cohort. All patients were also screened for CCR5-Δ32, CCR2-64I; in the Milan cohort SDF1-3′A polymorphisms were also assayed.23-26 The inclusion criteria for the LTNP cohorts were (1) certified seroconversion at 7 or more years, (2) CD4+ T-cell counts of at least 500 cells/μL, (3) absence of antiretroviral therapy, and (4) asymptomatic HIV-1 infection and good health conditions.
Control subjects included 70 patients with AIDS (AIDS) selected because they exhibited one or more AIDS-defining diseases and CD4+ T-cell count of no more than 50 cells/μL at the time of enrollment regardless of pharmacotherapeutic regimen, and 135 chronic HIV-infected individuals receiving highly active antiretroviral therapy (HAART) (HIV+HAART), with CD4 counts at 500 cells/μL and no previous AIDS-defining diseases. All studied populations (LTNP, AIDS, and HIV+HAART) were matched for sex, age, and risk factors and were recruited at the Infectious Diseases Clinic of the San Raffaele Scientific Institute of Milan or selected from the WIHS and Wadsworth cohorts. Moreover, 202 healthy blood donors (HCs) from Milan and 5 HCs from the United States were also included.
The institutional review boards of each clinical site and the NYSDOH in Albany, NY, approved the investigations.
Serum and plasma collection
Peripheral venous blood was drawn and serum was separated, heat inactivated, and stored at –80°C until analysis. Samples from the United States consisted of plasma, which was separated as described.22
Cell lines
Glioma cell lines U87 and U87-transfected with the genes for different chemokine receptors and monoclonal antibodies (2D7 and 12G5) were obtained through the NIH AIDS Research and Reference Reagent Program.
Purification of total CD4+ T cells
Peripheral blood mononuclear cells (PBMCs) from healthy blood donors were stimulated for 3 days with PHA (3 μg/mL; Sigma-Aldrich, Steinheim, Germany) and rIL-2 (100 U/mL; Amersham, Buckinghamshire, United Kingdom). CD4+ T cells were purified by immune-adsorption to anti-CD4 antibody–coated magnetic beads (Oxoid, Hampshire, United Kingdom). Purified CD4+ T cells were stimulated for 3 days with IL-2 prior to testing for the CCL4/MIP1β-binding assay. Donors of PBMCs were characterized for the presence of the CCR5-Δ32 allele; only donors not carrying a CCR5-Δ32 allele were used as a source of PBMCs.
CCL4/MIP1β- and CCL2/MCP1-binding assay
The assay was performed according to published procedures.27 Briefly, 106 purified CD4+ T cells resuspended in 200 μL RPMI (Gibco-Life Technologies, Milan, Italy), containing 0.05 M NaN3, 1% BSA, and 25 mM HEPES, were incubated at 4°C with appropriate dilutions of sera and/or Ig-enriched fractions. After 45 minutes of incubation 125I-MIP1β (Dupont-NEN, Mechelem, Belgium) or 125I-MCP1 (Amersham) was added [final concentration 0.1 nM, 0.2 μCi (0.0074 MBq)], and incubation at 4°C was continued for an additional 2 hours. Unbound radioactivity was separated from cell-bound radioactivity by centrifugation in an Eppendorf centrifuge on a 2-step gradient28 in 0.3-mL tubes (Nunc, Roskilde, Denmark) where the lower layer consisted of fetal calf serum (FCS) containing 10% sucrose and the upper layer consisted of 80% silicone (Sigma-Aldrich) and 20% mineral oil (Sigma-Aldrich). The bound radioactivity in the cell pellets was measured in a gamma scintillator counter (Perkin Elmer, Turku, Finland). A specificity control consisting of a 100-fold excess of unlabeled MIP1β or MCP1 was included in all experiments. The binding of the 125I-MIP1β to activated CD4+ T cells ranged between 1000 and 6000 CPM. 125I-MCP1 was also tested on U87 and CCR2-transfected U87 cell line.
Immunoglobulin purification
To purify all classes of Ig, CNBr-activated Sepharose 4B (Pharmacia, Uppsala, Sweden) was coupled with rabbit anti–human Ig (2 mg/mL; Sigma Aldrich, Milan, Italy). Then, 96 μL of each sample (from all LTNPs and from 5 healthy blood donors) was incubated at room temperature for 15 minutes on columns containing 2400 μL Sepharose–anti-Ig. Columns were washed, and bound antibodies were eluted with 0.2 M glycine/HCl (pH 2). The pH of the eluate was neutralized with 300 μL of 1 M Tris buffer, and then it was dialyzed against PBS buffer.
Preparation of peptide/bead conjugates and affinity purification of antibodies
Peptides were synthesized by the solid-phase F-moc method29 using an Applied Biosystem model 433 A peptide synthesizer (Warrington, United Kingdom). After peptide assembly, resin-bound peptides were deprotected as previously described30 and purified to greater than 95% purity by semipreparative reverse-phase high-performance liquid chromatography (RP-HPLC). To obtain peptides in cyclized oxidized form, an extrasequence cysteine was added to peptides 1, 3, and 4. These peptides were treated overnight with a 5-fold excess of oxidized glutathione and purified by RP-HPLC.
Coupling of CCR5 or unrelated-peptides (Table 1) to tosyl-activated Dynabeads M280 (Dynal, Oslo, Norway) was performed according to manufacturer's instructions. To establish whether the region recognized by anti-CCR5 antibodies corresponded to a conformational epitope, the specific peptide/beads were incubated with 10 mM β mercapto-ethanol, and subsequently with 30 mM N-ethyl maleimide (final concentration, 30 mM) for 60 minutes prior to antibody binding.
Peptides covering CCR5 extracellular regions and Ig reactivity obtained with LTNP samples
. | CCR5 external domain sequence . | Ig reactivity . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide . | . | 4 . | 11 . | 20 . | 21 . | 22 . | 152 . | 110 . | 173 . | NY4 . | NY6 . | NY9 . | NY10 . | |||||||||||
| 1 | MDYQVSSPIYDINYYTSEPC | — | — | — | — | — | — | — | — | ND | ND | ND | ND | |||||||||||
| 3 | cYAAAQWDFGNTMCQ | 5.2 | 5.4 | 4 | 7.4 | 5.9 | 6.8 | 8 | 7.7 | + | + | + | + | |||||||||||
| 4 | CSSHFPYSQYQFWKNFQTLKc | — | — | — | — | — | — | — | — | ND | ND | ND | ND | |||||||||||
. | CCR5 external domain sequence . | Ig reactivity . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide . | . | 4 . | 11 . | 20 . | 21 . | 22 . | 152 . | 110 . | 173 . | NY4 . | NY6 . | NY9 . | NY10 . | |||||||||||
| 1 | MDYQVSSPIYDINYYTSEPC | — | — | — | — | — | — | — | — | ND | ND | ND | ND | |||||||||||
| 3 | cYAAAQWDFGNTMCQ | 5.2 | 5.4 | 4 | 7.4 | 5.9 | 6.8 | 8 | 7.7 | + | + | + | + | |||||||||||
| 4 | CSSHFPYSQYQFWKNFQTLKc | — | — | — | — | — | — | — | — | ND | ND | ND | ND | |||||||||||
A specific concentration of human Igs from the 12 LTNPs was obtained only when peptide 3 was used. CCR5 specific antibodies from some LTNP were quantified and expressed as percentage of total serum Igs. Ig reactivity was negative in all samples for peptide 2 with a CCR5 external domain sequence of YYTSEPCQKINVKQIAARLLP and for peptide 5 with a CCR5 external domain sequence of FQEFFGLNNCSSSNR. An unrelated peptide, with sequence of VQGEESNDK, was used as negative control.
ND indicates not done; —, Ig reactivity was negative; and +, that Ig reactivity was positive but not quantified.
Binding of anti–CCR5-specific Igs to peptide/beads was performed by incubating 9 μg Ig to 9 μg peptide/beads for 1 hour at 4°C. After washing Igs were eluted with 0.5 M acetic acid and dialyzed against RPMI medium.
Anti-CCR5 radiobinding assay
CCR5- and CXCR4-transfected U87 cells (1 × 105) were incubated with 200 ng anti–CCR5-specific Igs for 45 minutes on ice, then were processed as described for CCL4/MIP1β-binding assay. Total serum Igs from a pool of 5 HCs was used as negative control. Anti-CCR5 Abs from LTNPs bound to CCR5+ cells were revealed by 0.5 μCi/mL (0.0185 MBq/mL) 125I-sheep anti–human Ig F(ab′)2 Ab fragments (Amersham); monoclonal antibodies (2D7 and 12G5) were also used, and their binding was revealed by 0.5μCi/mL (0.0185 MBq/mL) 125I-sheep anti–mouse Ig F(ab′)2 Ab fragments (Amersham).
Quantification of serum Igs
Ninety-six microwell plates were coated with Ig fractions (up to 1:128 by 2-fold dilutions) in 50 mM carbonate buffer pH 9.5 for 1 hour at 37°C. A commercial preparation of human Ig (Sigma Aldrich) was used as standard. After saturation with PBS/1% milk powder (Humana 3, Herford, Germany), the plates were incubated with peroxidase-conjugated rabbit anti–human Ig (Dako, Glostrup, Denmark) for 30 minutes at 37°C. The enzymatic reaction was developed with TMB Microwell Peroxidase Substrate System (KPL, Gaithersburg, MD).
CCR5-internalization assay and flow cytometric analysis
Untransfected and CCR5-transfected U87 cell lines and CD4+ T lymphocytes were incubated with affinity-purified anti-CCR5 Ig (200 ng) at 37°C for 1, 12, 24, or 48 hours. Surface-expressed CCR5 was detected by using an anti-CCR5 mAb (2D7)31 and a secondary antimouse antibody conjugated with FITC according to a published protocol.32 RPMI medium containing 10% FCS and 200 ng of a pool of serum samples from 5 HCs was used as a negative control. Cells were also incubated with 50 nM RANTES for 1 hour and processed as described. In some assays, CCR5-U87 cells were incubated in the presence of hypertonic sucrose (0.45M), filipin (5 μg/mL), and/or nystatin (50 μg/mL) (Sigma Aldrich). Relative percentage of CCR5 surface expression was calculated by measuring mean channel fluorescence (MCF) according to the following expression: 100 × (MCF) (stimulated)–MCF (neg control)/MCF (medium)–MCF (neg control). Untransfected (U87) cells were used as the negative control.
Virus isolation and titration
Subtype B viruses were obtained as previously described.19,33 In brief, HIV-1 was isolated from the PBMCs of HIV-1–seropositive individuals by their cocultivation with PHA-stimulated PBMCs of 2 healthy donors; the cultures were maintained until increasing levels of HIV-1 p24 antigen (Ag) (AALTO Bio Reagents Ltd, Dublin, Ireland) were detected in 2 consecutive determinations. The infectivity (ID50) of each virus isolate was determined on PBMCs from 1 single donor as follows: 6 replicas (150 μL) of 5-fold serial dilutions (from 1:5 to 1:3125) of virus were added to 96 wells of a round-bottom Microtiter plate (Nunc) containing 105 resting PBMCs in 75 μL medium, incubated at 37°C for 2 hours. Medium was then removed by centrifugation, and fresh RPMI 1640 medium containing PHA (3 μg/mL) and rIL-2 (10 U/mL) was added. HIV-1 p24 Ag was titrated after 5 and 7 days of culture. ID50 titers were defined as the reciprocal of the virus dilution yielding 50% positive wells by Reed-Muench calculation.
Phenotypic characterization of viral coreceptor usage
Each virus isolate was used to infect U87 cell lines expressing one of the following chemokine receptors: CCR1, CCR2B, CCR3, CCR5, or CXCR4. Then, 1 mL virus-containing culture supernatant from the infections was incubated with 8 × 104 U87 cells in 12-well plates for 2 hours at 37°C. Culture supernatants, collected from each well 2, 5, and 7 days after infection, were analyzed for HIV-1 p24 Ag by standard enzyme-linked immunosorbent assay (ELISA; AALTO Bio Reagents Ltd).
Virus neutralization assays
The neutralizing activity was evaluated by 2 different methods. In the first method, 2 × 105 PBMCs, activated with PHA and IL-2 for 48 hours, was added to 75 μL serial dilutions of serum and/or purified Ig from LTNPs or HCs and incubated for 48 hours (required to obtain a complete CCR5 down-regulation), then 75 μL of a virus dilution corresponding to 20 TCID50 was added. After 2 hours the cultures were washed. Supernatant p24 levels were determined on postinfection days 5, 7, and 9, and the analysis was performed when the level of TCID50 ranging from 10 to 30 was achieved. Each assay correlated with negative (HC) and positive controls (a HIV-neutralizing Mab and a serum from HIV-seropositive subjects). Each dilution was tested in duplicate at virus concentrations ranging from 10 to 30 TCID50; each experiment was repeated by using fresh PBMCs from 3 different HCs. Each value obtained with a specific serum/Ig dilution was compared with the mean values from the 6 corresponding replicates without the addition of sera or Igs. Virus titration was repeated in each neutralization assay. All of the results were expressed as the sera or Ig concentrations leading to the 50% and 90% inhibition of viral replication (IC50 and IC90, respectively). In some assays, Abs were preincubated with 150 nmol/L peptide 3 for 30 minutes at 37°C prior to incubation to the cells.
In the second method, SOS pseudoviruses (kind gift of D. Burton and J. Binley) were used to infect CCR5-transfected U87 cell line as previously reported.34 Briefly, the plasmid pCAGGS was used to express membranebound Env of the R5 isolate JR-FL. Env proteins either as full-length gp160 or as a mutant truncated at residue 708, leaving 3 amino acids of the gp41 cytoplasmic tail, were expressed. Mutations were made to introduce cysteines at residues 501 and 605 (the SOS mutant). A mutation was also generated to replace the gp120/gp41 cleavage site REKR by the inefficiently cleaved GEKR. The plasmid pNL4-3.Luc.R-E-, expressing an HIV-1 genome fragment with frameshifts in Env and Vpr and a luciferase reporter gene in place of Nef34 was also used.
Pseudoviruses were produced by transfection of 293T cells with pNL4-3.Luc.R-E- and Env-expressing pCAGGS-based plasmids. As a negative control, VSV-G pseudovirus was used. U87 cells (2 × 104/well) were incubated with different concentrations of Abs for 48 hours; thus, SOS pseudoviruses (HIV-R5 and VSV-G) were incubated with U87 cells for 2 hours, after which the cultures were washed and treated with 5 mM DTT for 10 minutes, and the medium was replaced. Cells were then cultured for an additional 48 hours, and luciferase activity was measured by a previously reported method.35 The reaction was read with the use of a Top Count apparatus (Packard, Meriden, CT). Neutralizing activity was expressed as Ig concentrations leading to the 50% and 90% inhibition of viral replication (IC50 and IC90, respectively).
Statistical analysis
Chi-square test and Fisher exact test were used to compare variable frequencies between LTNPs and the other populations, including AIDS, HIV+HAART, and HC, as appropriate.
Individual regression curves were compared between LTNP subjects loosing or maintaining anti-CCR5 Abs by using the Mann-Whitney test.
Frequency of patients whose viremia increased during the follow-up period in contrast to those whose viremia failed to increase was compared in the 2 groups (ie, Ab to CCR5 lost versus Ab to CCR5 not lost) by using the Fisher exact test.
Individuals were classified in 2 groups according to an increasing level of viremia during follow-up (≥ 1 log increment). The distribution of time to change for anti-CCR5 Ab serostatus was analyzed by the Kaplan-Meier method. Comparison among survival curves between the 2 groups was accomplished by means of the log-rank test.
The association of basal viremia with change of anti-CCR5 Ab serostatus was investigated by means of the Cox proportional hazards regression model. Hazard ratio and 95% CI were reported.
Results
LTNP sera contain Abs recognizing a conformational epitope within the first loop of the extramembrane portion of CCR5
We analyzed sera from 85 LTNPs for the presence of anti-CCR5 antibodies using a MIP1β competitive radiobinding assay performed on CCR5+ CD4+ T lymphocytes.19 A pool of 6 sera from ESN individuals previously shown to be positive for anti-CCR5 Ab and 207 HIV-1–negative sera from healthy control subjects were included as positive and negative controls, respectively. Also serum samples from 70 AIDS patients and 135 HIV+HAART patients (chronic infected patients under HAART) were tested. The presence of LTNP/MIP1β-competing sera, in 20 of 85 LTNPs was highly significant as compared with AIDS, HIV+HAART, and healthy control subjects (P < .001 versus all of the 3 categories) (Figure 1A).
To verify that the MIP1β binding to CCR5 was specifically due to the presence of anti-CCR5 antibodies, samples from the 20 LTNP/MIP1β-competing sera and a pool of sera from 5 HCs were purified by affinity chromatography to obtain Ig-enriched and Ig-depleted fractions. As shown in Figure 1B, the ability of inhibiting MIP1β binding was retained by Ig but not by Ig-depleted fractions. Both fractions from the pool of 5 HCs presented no reactivity; thus, only Ig-enriched fractions from LTNP sera were able to interfere with MIP1β binding to CCR5, and the inhibition was concentration dependent as shown in Figure 1C.
Depending on the availability of samples, some of the 20 sera exhibiting MIP1β competition properties were evaluated further. To exclude the possibility that these Abs may compete nonspecifically to receptors expressed on CD4+ T-cell surface, a competition experiment was performed, comparing the binding to cells of either MCP1 or serum-purified Ig (20 μg/assay) of interest. Ig from LTNPs no. 20, no. 1, and no. 22 and HCs (a pool of purified Igs from 5 seronegative HCs) was tested on either CD4+ T lymphocytes or CCR2-transfected U87 cell lines. No competition of MCP1 binding to cells by human serum-purified Ig was seen, whereas an efficient inhibition by nonradioactive MCP1 was observed (Figure 1D).
To identify the CCR5 region recognized by anti-CCR5 Abs from LTNPs, the serum Igs of 12 individuals were tested on a panel of 5 synthetic peptides spanning the complete sequence of the extracellular portion of CCR5. An unrelated peptide was also used (Table 1). To titer the specific Ig fractions, all fractions were also assayed by ELISA. A specific concentration of human Igs from the 12 LTNPs was obtained only when peptide 3 was used. CCR5-specific Abs were then quantified. The mean of CCR5-specific Abs levels was 6.3% (range, 4%-8%) of total serum Ig, indicating that these Abs are elicited by a adequate antigenic stimulation (Table 1). Moreover, binding of the anti-CCR5 abs from LTNPs to peptide 3 was completely abolished by the addition of 2-mercaptoethanol and N-ethyl maleimide, which causes the reduction and alkylation of the cysteine loop (data not shown), suggesting that Ab recognition requires the maintenance of the epitope tertiary structure. The specificity of peptide 3–purified Abs was assessed by radiobinding to either CCR5- or CXCR4-transfected U87 cell line. All Igpurified fractions exhibited specific binding to CCR5but not to CXCR4-transfected cell lines (Figure 1E).
Competitive binding assays by human serum samples. (A) Inhibition of MIP1β binding to CD4+ T cells in sera samples of a pool of 6 HIV-exposed seronegative (ESN), 207 unexposed seronegative (HC), 85 HIV+ seropositive LTNP (20 competing and 65 not competing), 135 chronically HIV-infected (HIV+HAART), and 70 progressor(P) patients. The values are expressed as the percentage of inhibition of MIP1β binding and are representative of 3 experiments performed. Error bars represent SD of 5 replicates per each data point. The cutoff value was set at 12% (3 SDs above the mean value of the HC serum samples, as previously described36 ). • indicates mean value for each subject; ♦, mean value of each group. (B) Competitive binding assays comparing the 20 LTNP sera displaying competitive activity against MIP1β and CCR5. Abs were affinity purified on Sepharose column, then quantified by ELISA, and tested in MIP1β binding inhibition. Ig-enriched and Ig-depleted fractions from HCs were used as controls. The data are expressed as a percentage of MIP1β binding inhibition to CD4+ T cells and are representative of 3 experiments performed. Error bars represent SDs of 5 replicates per data point. (C) Dose-response curves of Ig-enriched fractions mediated binding inhibition of MIP1β binding to CD4+ T cells. Serum samples from 11 LTNP MIP1β competing and from a pool of 5 HCs were affinity purified by Sepharose column. Ig-enriched fractions were quantified by ELISA. The data are representative of 2 experiments performed. (D) MCP1 binding assay inhibition was carried out on CCR2-expressing transfected U87 cell line and CD4+ T lymphocytes from HCs. The data are representative of 3 experiments performed. Error bars represent SDs of 5 replicates per each data point. MCP1 cold and Igs from HCs were used as positive and negative controls, respectively. (E) Binding of pep 3 purified Igs to either CCR5- or CXCR4-expressing transfected U87 cell line. Peptide 3 corresponds to the second external domain of CCR5 as shown in Table 1. Total serum Abs were purified on Sepharose column and quantified by ELISA, and then Ig fractions were affinity purified on the relative CCR5 peptide as described in “Materials and methods.” A pool of serum Igs from the HCs was used as a negative control. A CCR5 specific mAb (2D7) and a CXCR4-specific mAb (12G5) were used at 0.3 mg/mL to bind to CCR5- and CXCR4-transfected cells, respectively. The data are representative of 3 experiments performed. Error bars represent SDs of 5 replicates per each data point.
Competitive binding assays by human serum samples. (A) Inhibition of MIP1β binding to CD4+ T cells in sera samples of a pool of 6 HIV-exposed seronegative (ESN), 207 unexposed seronegative (HC), 85 HIV+ seropositive LTNP (20 competing and 65 not competing), 135 chronically HIV-infected (HIV+HAART), and 70 progressor(P) patients. The values are expressed as the percentage of inhibition of MIP1β binding and are representative of 3 experiments performed. Error bars represent SD of 5 replicates per each data point. The cutoff value was set at 12% (3 SDs above the mean value of the HC serum samples, as previously described36 ). • indicates mean value for each subject; ♦, mean value of each group. (B) Competitive binding assays comparing the 20 LTNP sera displaying competitive activity against MIP1β and CCR5. Abs were affinity purified on Sepharose column, then quantified by ELISA, and tested in MIP1β binding inhibition. Ig-enriched and Ig-depleted fractions from HCs were used as controls. The data are expressed as a percentage of MIP1β binding inhibition to CD4+ T cells and are representative of 3 experiments performed. Error bars represent SDs of 5 replicates per data point. (C) Dose-response curves of Ig-enriched fractions mediated binding inhibition of MIP1β binding to CD4+ T cells. Serum samples from 11 LTNP MIP1β competing and from a pool of 5 HCs were affinity purified by Sepharose column. Ig-enriched fractions were quantified by ELISA. The data are representative of 2 experiments performed. (D) MCP1 binding assay inhibition was carried out on CCR2-expressing transfected U87 cell line and CD4+ T lymphocytes from HCs. The data are representative of 3 experiments performed. Error bars represent SDs of 5 replicates per each data point. MCP1 cold and Igs from HCs were used as positive and negative controls, respectively. (E) Binding of pep 3 purified Igs to either CCR5- or CXCR4-expressing transfected U87 cell line. Peptide 3 corresponds to the second external domain of CCR5 as shown in Table 1. Total serum Abs were purified on Sepharose column and quantified by ELISA, and then Ig fractions were affinity purified on the relative CCR5 peptide as described in “Materials and methods.” A pool of serum Igs from the HCs was used as a negative control. A CCR5 specific mAb (2D7) and a CXCR4-specific mAb (12G5) were used at 0.3 mg/mL to bind to CCR5- and CXCR4-transfected cells, respectively. The data are representative of 3 experiments performed. Error bars represent SDs of 5 replicates per each data point.
Anti-CCR5 Abs induce CCR5 internalization through a clathrin-dependent pathway
CD4+ T lymphocytes, CCR5-U87, and untransfected U87 cell lines were incubated with purified anti-CCR5 Ig from 3 LTNPs.37-39 The levels of surface CCR5 were determined by indirect staining with 2D7. As controls, similar assays were performed without adding Ig or with 200 ng of a pool of serum Ig from healthy controls. As a positive control, a CCR5-binding chemokine, RANTES (50 nM), was used. CCR5 down-regulation on target cells was achieved after 48 hours of treatment with CCR5-specific Igs (Figure 2A-B), whereas an intermediate value was obtained after 24 hours of incubation (Figure 2B). CCR5 modulation was not evident after 1 hour of LTNP-human anti-CCR5 antibody incubation, thus demonstrating that human Abs do not interfere with 2D7 binding on the cell surface of CD4+ cells. Moreover, the preincubation with 0.3 μg/mL 2D7 for 20 minutes on ice on CD4+ cells did not interfere with human antibody binding activity (data not shown).
Purified Ig from LTNPs induced a marked down-regulation on activated CD4+ T lymphocytes and CCR5-U87 cell line (Table 2). No evidence of CCR5 expression on the surface of CD4-U87 cell line was seen.
Anti-CCR5 antibody-mediated downregulation of surface CCR5 receptor
Cells and samples . | Mean surface expression of CCR5, % (range) . |
|---|---|
| CD4+ lymphocytes | |
| Fetal calf serum | 99 (98-100) |
| HC* | 96 (92-99) |
| LTNP no. 20 | 19 (14-24) |
| LTNP no. 21 | 32 (24-39) |
| LTNP no. 22 | 23 (15-37) |
| RANTES | 45 (41-49) |
| U87-CCR5 | |
| Fetal calf serum | 95 (91-100) |
| HC* | 89 (85-99) |
| LTNP no. 20 | 29 (24-34) |
| LTNP no. 21 | 28 (24-38) |
| LTNP no. 22 | 37 (30-45) |
| RANTES | 54 (52-58) |
Cells and samples . | Mean surface expression of CCR5, % (range) . |
|---|---|
| CD4+ lymphocytes | |
| Fetal calf serum | 99 (98-100) |
| HC* | 96 (92-99) |
| LTNP no. 20 | 19 (14-24) |
| LTNP no. 21 | 32 (24-39) |
| LTNP no. 22 | 23 (15-37) |
| RANTES | 45 (41-49) |
| U87-CCR5 | |
| Fetal calf serum | 95 (91-100) |
| HC* | 89 (85-99) |
| LTNP no. 20 | 29 (24-34) |
| LTNP no. 21 | 28 (24-38) |
| LTNP no. 22 | 37 (30-45) |
| RANTES | 54 (52-58) |
Relative percentage of surface expression of CCR5 is expressed as mean of 3 independent experiments with indication of range values.
A pool of total purified Ig from 5 healthy control subjects was used as negative control.
CCR5 modulation on surface of CD4+ lymphocytes. CCR5 expression was evaluated by 2D7 Mab indirect binding, and relative surface expression was evaluated as the percentage of CCR5 expression on CD4+ cells. (A) CCR5 down-regulation in response to presence of 200 ng Abs for 48 hours. Total serum Abs were purified on Sepharose column and quantified by ELISA. Ig fractions were then affinity-purified on the relative CCR5 peptide and incubated with CD4+ lymphocytes. From the top: cells were not incubated with Igs; total Igs from healthy control; RANTES (50 nM); CCR5/peptide 3-specific Ig from LTNPs no. 22, no. 21, and no. 20. The data are representative of 2 experiments performed. (B) Kinetic of CCR5 down-regulation by 200 ng CCR5/peptide 3–specific Ig from LTNP no. 20. Total serum Abs were affinity-purified on Sepharose column and quantified by ELISA. Ig fractions were then affinity-purified on the relative CCR5 peptide and incubated with CD4+ lymphocytes for 1, 12, 24, and 48 hours before analysis. As positive controls, cells were not incubated with Igs, and as negative control cells were incubated with Igs from HCs. The data are representative of 3 experiments performed. (C) CCR5 expression on CD4+ lymphocytes from some LTNP patients carrying (no. 22, no. 20, no. 152) or not carrying (L01, M02, and C03) anti-CCR5 Abs. CD4+ lymphocytes from LTNPs no. 22, no. 20, and no. 152 were cultured for a further 7 days in the absence of anti-CCR5 Abs. The data are representative of 2 experiments performed. Error bars represent SDs of 3 replicates per each data point. (D) Susceptibility of infection by either HIV no. 36 (R5) or HIV no. 45 (R5, X4, R3) on CD4+ lymphocytes from some LTNPs carrying (no. 22, no. 20, no. 152) or not carrying (L01, M02, and C03) anti-CCR5 Abs. The data are representative of 2 experiments performed. Error bars represent SDs of 3 replicates per each data point. (E) Abs to first loop of CCR5 induce receptor endocytosis by clathrin-dependent pathway. Serum Igs from 2 LTNPs carrying CCR5 specific Abs were affinity-purified on Sepharose column and quantified by ELISA. Ig fractions were then affinity-purified on the relative CCR5 peptide (peptide 3) and incubated on CCR5-transfected cells for 48 hours in the presence or not of specific chemicals. All experiments were repeated twice, and SDs are shown.
CCR5 modulation on surface of CD4+ lymphocytes. CCR5 expression was evaluated by 2D7 Mab indirect binding, and relative surface expression was evaluated as the percentage of CCR5 expression on CD4+ cells. (A) CCR5 down-regulation in response to presence of 200 ng Abs for 48 hours. Total serum Abs were purified on Sepharose column and quantified by ELISA. Ig fractions were then affinity-purified on the relative CCR5 peptide and incubated with CD4+ lymphocytes. From the top: cells were not incubated with Igs; total Igs from healthy control; RANTES (50 nM); CCR5/peptide 3-specific Ig from LTNPs no. 22, no. 21, and no. 20. The data are representative of 2 experiments performed. (B) Kinetic of CCR5 down-regulation by 200 ng CCR5/peptide 3–specific Ig from LTNP no. 20. Total serum Abs were affinity-purified on Sepharose column and quantified by ELISA. Ig fractions were then affinity-purified on the relative CCR5 peptide and incubated with CD4+ lymphocytes for 1, 12, 24, and 48 hours before analysis. As positive controls, cells were not incubated with Igs, and as negative control cells were incubated with Igs from HCs. The data are representative of 3 experiments performed. (C) CCR5 expression on CD4+ lymphocytes from some LTNP patients carrying (no. 22, no. 20, no. 152) or not carrying (L01, M02, and C03) anti-CCR5 Abs. CD4+ lymphocytes from LTNPs no. 22, no. 20, and no. 152 were cultured for a further 7 days in the absence of anti-CCR5 Abs. The data are representative of 2 experiments performed. Error bars represent SDs of 3 replicates per each data point. (D) Susceptibility of infection by either HIV no. 36 (R5) or HIV no. 45 (R5, X4, R3) on CD4+ lymphocytes from some LTNPs carrying (no. 22, no. 20, no. 152) or not carrying (L01, M02, and C03) anti-CCR5 Abs. The data are representative of 2 experiments performed. Error bars represent SDs of 3 replicates per each data point. (E) Abs to first loop of CCR5 induce receptor endocytosis by clathrin-dependent pathway. Serum Igs from 2 LTNPs carrying CCR5 specific Abs were affinity-purified on Sepharose column and quantified by ELISA. Ig fractions were then affinity-purified on the relative CCR5 peptide (peptide 3) and incubated on CCR5-transfected cells for 48 hours in the presence or not of specific chemicals. All experiments were repeated twice, and SDs are shown.
To investigate the mechanisms involved in the CCR5 internalization induced by human Abs, we treated CCR5-transfected cells with several chemicals, including sucrose (specific inhibitors of clathrin-coated pits pathway), filipin, or nystatin (inhibitors of caveolae) in the presence of peptide 3–purified Abs. As shown in Figure 2E, Ab/CCR5 internalization was specifically inhibited by sucrose but not by the other chemicals.
CCR5 down-regulation on the surface of CD4+ lymphocytes from LTNPs
To determine whether this internalization was occurring in vivo, the level of CCR5 expression on the surface of CD4+ T lymphocytes of LTNPs was compared between 3 Ab-positive and 3 Ab-negative subjects. Fluorescence-activated cell sorting (FACS) was performed with 2D7. As shown in Figure 2C, no binding could be detected on the cells from any of the 3 Ab-positive subjects, whereas high levels of CCR5 expression was found in Ab-negative subjects. When CD4+ T lymphocytes of LTNPs carrying anti-CCR5 Abs were cultured, CCR5 reappeared in 7 days (Figure 2C). Thereafter, cells were cultured in the presence of purified Igs (1 mg/mL) from LTNPs carrying CCR5 Abs, and after 48 hours CCR5 levels were significantly reduced (data not shown).
CD4+ lymphocytes of LTNPs positive for anti-CCR5 Abs are resistant to HIV infection
To correlate the CCR5 level and susceptibility to infection, CD4+ T lymphocytes from PBMCs of 3 LTNPs carrying anti-CCR5 and 3 LTNPs negative for anti-CCR5 Abs were infected with 2 viruses: HIV-36 (R5 strain) and -45 (R5, X4, R3 strain). As shown in Figure 2D, a low level of infection was obtained by HIV-36 on cells from all of the 3 subjects carrying anti-CCR5 antibodies and CCR5– phenotype, whereas efficient infection was found in subjects not carrying anti-CCR5 antibodies. HIV-45 infects CD4+ T lymphocytes from all of the 6 LTNPs (Figure 2D). CD4+ T lymphocytes were isolated from LTNPs with a viremia level below 1000 copies/mL.
CCR5-specific Abs prevent infection of HIV-1 from different subtypes
To investigate whether anti-CCR5 Abs block HIV infection, Ig-enriched fractions from 5 LTNPs with anti-CCR5 Abs were tested in neutralization assays by examining 2 subtype B-R5 tropic isolates (HIV-36 and HIV-40). As positive controls, we used a mAb against CD4 (SIM4) and Igs from a neutralizing serum of a seropositive subject (HIV+). As shown in Figure 3, HIV infectivity reduction was concentration dependent. Neutralization was demonstrated to be R5-specific because Igs from LTNPs failed in blocking the infectivity of X4-tropic HIV-26 and X4-, R5-, X3-tropic HIV-45 (data not shown). Further evidence that neutralization was associated with HIV-R5 viruses was obtained by using R5-viral molecular clone (SOS-pseudovirus). No blocking capacity was obtained with the unrelated virus SOS/VSV-G (vesicular stomatitis virus) (Table 3). Anti-CCR5 antibody-mediated neutralization of the HIV-1 isolates belonging to subtype B (HIV-36 and -40), C (no. 92BR025), and A (no. 92TH007) was observed when Abs eluted on peptide 3–coated beads were tested, as shown in Table 3. Furthermore, preincubation of purified Igs from LTNPs no. 11 and no. 20 with peptide 3 decreased the blocking capacity (IC50) from 2 and 0.8 μg/mL to 9 and 4 μg/mL, respectively (data not shown).
Viral subtype, coreceptor usage, and infectivity reduction by CCR5 specific Igs from LTNP
. | . | Coreceptor usage . | IC50/IC90, μg/mL . | . | . | . | . | . | . | . | . | . | . | . | . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Viruses . | Strain . | . | 4 . | 11 . | 20 . | 21 . | 22 . | 152 . | 110 . | 173 . | NY4 . | NY6 . | NY9 . | NY10 . | HC* . | ||||||||||||
| HIV-36 | B | R5 | 0.85/> 2 | 0.64/> 2 | 0.42/> 2 | 0.27/1.8 | 0.42/> 2 | ND | ND | ND | > 2/> 2 | > 2/> 2 | > 2/> 2 | > 2/> 2 | > 45/> 45 | ||||||||||||
| HIV-40 | B | R5 | 0.51/> 2 | 0.60/> 2 | 0.96/1.7 | 0.28/1.9 | 0.46/1.5 | ND | ND | ND | 0.89/> 2 | 0.88/> 2 | 0.56/> 2 | 1.4/> 2 | > 45/> 45 | ||||||||||||
| 92BR025 | C | R5 | ND | ND | 0.45/> 2 | 1.9/> 2 | 0.8/> 2 | ND | ND | ND | ND | ND | ND | ND | > 45/> 45 | ||||||||||||
| 92TH007 | A | R5 | ND | ND | 0.01/> 2 | 2/> 2 | 0.7/> 2 | ND | ND | ND | ND | ND | ND | ND | > 45/> 45 | ||||||||||||
| HIV-45 | B | X4, R5, R3 | > 2/> 2 | > 2/> 2 | > 2/> 2 | > 2/> 2 | > 2/> 2 | ND | ND | ND | > 2/> 2 | > 2/> 2 | > 2/> 2 | > 2/> 2 | > 45/> 45 | ||||||||||||
| HIV-26 | B | X4 | ND | ND | > 2/> 2 | > 2/> 2 | > 2/> 2 | ND | ND | ND | ND | ND | ND | ND | > 45/> 45 | ||||||||||||
| SOS/HIV | JRFL | R5 | 0.5/2.9 | 1.1/6.1 | 0.4/2.2 | 0.6/3.3 | 0.8/4.4 | 1.2/6.7 | 1.3/7.2 | 1/5.9 | ND | ND | ND | ND | > 45/> 45 | ||||||||||||
| SOS/VSV-G | NA | NA | > 10/> 10 | > 10/> 10 | > 10/> 10 | > 10/> 10 | > 10/> 10 | > 10/> 10 | > 10/> 10 | > 10/> 10 | ND | ND | ND | ND | > 45/> 45 | ||||||||||||
. | . | Coreceptor usage . | IC50/IC90, μg/mL . | . | . | . | . | . | . | . | . | . | . | . | . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Viruses . | Strain . | . | 4 . | 11 . | 20 . | 21 . | 22 . | 152 . | 110 . | 173 . | NY4 . | NY6 . | NY9 . | NY10 . | HC* . | ||||||||||||
| HIV-36 | B | R5 | 0.85/> 2 | 0.64/> 2 | 0.42/> 2 | 0.27/1.8 | 0.42/> 2 | ND | ND | ND | > 2/> 2 | > 2/> 2 | > 2/> 2 | > 2/> 2 | > 45/> 45 | ||||||||||||
| HIV-40 | B | R5 | 0.51/> 2 | 0.60/> 2 | 0.96/1.7 | 0.28/1.9 | 0.46/1.5 | ND | ND | ND | 0.89/> 2 | 0.88/> 2 | 0.56/> 2 | 1.4/> 2 | > 45/> 45 | ||||||||||||
| 92BR025 | C | R5 | ND | ND | 0.45/> 2 | 1.9/> 2 | 0.8/> 2 | ND | ND | ND | ND | ND | ND | ND | > 45/> 45 | ||||||||||||
| 92TH007 | A | R5 | ND | ND | 0.01/> 2 | 2/> 2 | 0.7/> 2 | ND | ND | ND | ND | ND | ND | ND | > 45/> 45 | ||||||||||||
| HIV-45 | B | X4, R5, R3 | > 2/> 2 | > 2/> 2 | > 2/> 2 | > 2/> 2 | > 2/> 2 | ND | ND | ND | > 2/> 2 | > 2/> 2 | > 2/> 2 | > 2/> 2 | > 45/> 45 | ||||||||||||
| HIV-26 | B | X4 | ND | ND | > 2/> 2 | > 2/> 2 | > 2/> 2 | ND | ND | ND | ND | ND | ND | ND | > 45/> 45 | ||||||||||||
| SOS/HIV | JRFL | R5 | 0.5/2.9 | 1.1/6.1 | 0.4/2.2 | 0.6/3.3 | 0.8/4.4 | 1.2/6.7 | 1.3/7.2 | 1/5.9 | ND | ND | ND | ND | > 45/> 45 | ||||||||||||
| SOS/VSV-G | NA | NA | > 10/> 10 | > 10/> 10 | > 10/> 10 | > 10/> 10 | > 10/> 10 | > 10/> 10 | > 10/> 10 | > 10/> 10 | ND | ND | ND | ND | > 45/> 45 | ||||||||||||
The values are expressed as Ig concentration (μg/mL) inhibiting 50% (IC50) and 90% (IC90) virus replication. The results are representative of 3 independent experiments. Six primary isolates were used to infect PBMCs from HCs, and 2 viral molecular clones (SOS/HIV/JRFL and an HIV-unrelated clone SOS/VSV-G) were used to infect CCR5-U87 cells.
ND indicates not done; NA, not applicable.
A pool of total serum Ig from 5 HCs was used as negative control (HC). The highest Ig concentration tested in neutralization assay was 45 and 10 μg/mL for HCs and LTNsP, respectively.
Virus neutralization assays carried out with Ig fractions from 5 LTNP sera displaying anti-CCR5 Abs. A neutralizing anti-CD4 mAb (SIM4) and a neutralizing serum from a patient with AIDS (tested at the same dilutions of the LTNP sera) were used as positive control. As negative control, a pool of sera from 5 HCs was also used. The data are expressed as the percentage of infectivity reduction and are representative of 3 experiments. (A) Mediated antibody infectivity reduction obtained with subtype B isolate HIV-36. (B) Mediated antibody infectivity reduction obtained with subtype B isolate HIV-40.
Virus neutralization assays carried out with Ig fractions from 5 LTNP sera displaying anti-CCR5 Abs. A neutralizing anti-CD4 mAb (SIM4) and a neutralizing serum from a patient with AIDS (tested at the same dilutions of the LTNP sera) were used as positive control. As negative control, a pool of sera from 5 HCs was also used. The data are expressed as the percentage of infectivity reduction and are representative of 3 experiments. (A) Mediated antibody infectivity reduction obtained with subtype B isolate HIV-36. (B) Mediated antibody infectivity reduction obtained with subtype B isolate HIV-40.
Longitudinal immunovirologic and clinical follow-up
To determine whether anti-CCR5 Abs might contribute to maintain LTNP status, anti-CCR5 Abs were tested over at least a 3-year follow-up in patients from the Milan LTNP cohort.
Mean ± SD CD4+ T-cell count and viral load varied between 718.59 ± 222.88 and 28 071.22 ± 66 998.20 (at enrollment) to 734.15 ± 400.80 and 22 377.0 ± 58 163.91 (at the end of the study), respectively.
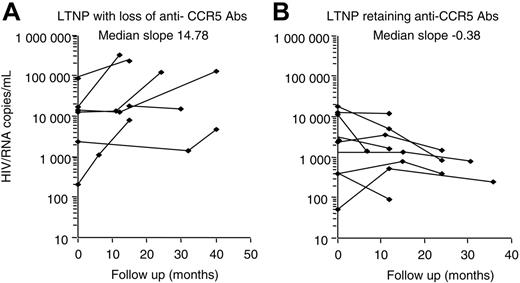
Nine individuals showed a loss of anti-CCR5 Abs during follow-up, at a timing corresponding to the loss of their LTNP status, and to the beginning of treatment. A linear regression test was used for each patient, and the resulting slopes for the 2 groups were compared by Mann-Whitney test to investigate the possible association between the variation of viremia and/or CD4 levels during the follow-up in those LTNPs who retained Abs to CCR5 and those who lost them. No statistical correlation was found with circulating CD4+ T cells, but a significant increase of viral load was observed only in subjects with loss of anti-CCR5 Abs, with a median slope value of 14.38 (Figure 4A). Interestingly, patients who maintained anti-CCR5Abs showed relatively low levels of viremia, stable over time, with a median slope value of –0.38 (Figure 4B). All LTNPs who lost anti-CCR5 Abs showed a progression toward diseases as shown in Table 4. In particular, 8 of 9 progressors were on therapy. Nevertheless, 3 patients developed AIDS (no. 131, no. 39, and no. 110). Strikingly, all of the 11 subjects who retained anti-CCR5 Abs maintained stable clinical and immunologic conditions without treatment during the study period (data not shown).
Immunologic and virologic parameters in LTNPs who lost anti-CCR5 Abs during the study period
Subject code and sample date . | CD4 count, cells/μL . | HIV-RNA, copies/mL . | Therapy . | CCR5 Abs . |
|---|---|---|---|---|
| 173 | ||||
| Oct 95 | 533 | 184 | No | Pos |
| Sep 03 | 429 | 483 000 | Yes | Neg |
| Oct 04 | 305 | 330 000 | Yes | Neg |
| 167 | ||||
| Jan 99 | 443 | 10 800 | No | Pos |
| Nov 03 | 511 | 12 960 | No | Neg |
| Apr 04 | 331 | 7 000 | No | Neg |
| 131 | ||||
| Oct 00 | 656 | 17 000 | No | Pos |
| Jul 02 | 964 | 82 000 | No | Pos |
| Nov 04 | 905 | < 50 | Yes | Neg |
| 39 | ||||
| Jan 02 | 551 | 94 000 | No | Pos |
| Jan 04 | 369 | 127 658 | No | Neg |
| Dec 04 | 664 | 71 | Yes | Neg |
| 110 | ||||
| Feb 96 | 625 | 14 000 | No | Pos |
| Feb 02 | 577 | 46 000 | Yes | Neg |
| Oct 04 | 238 | 199 603 | Yes | Neg |
| 4 | ||||
| Sep 95 | 454 | 13 550 | No | Pos |
| Sep 96 | 326 | 233 063 | Yes | Neg |
| Jun 04 | 525 | < 50 | Yes | Neg |
| 22 | ||||
| May 95 | 728 | 400 | No | Pos |
| Apr 96 | 579 | 1 102 | No | Pos |
| Jun 97 | 604 | 7 800 | Yes | Neg |
| 11 | ||||
| Apr 96 | 464 | 4 900 | No | Pos |
| Apr 98 | 806 | 39 000 | Yes | Neg |
| May 99 | 873 | 33 000 | Yes | Neg |
| 21 | ||||
| Jan 95 | 638 | 2 218 | No | Pos |
| Sep 96 | 468 | 1 827 | No | Pos |
| Nov 97 | 417 | 15 000 | Yes | Neg |
Subject code and sample date . | CD4 count, cells/μL . | HIV-RNA, copies/mL . | Therapy . | CCR5 Abs . |
|---|---|---|---|---|
| 173 | ||||
| Oct 95 | 533 | 184 | No | Pos |
| Sep 03 | 429 | 483 000 | Yes | Neg |
| Oct 04 | 305 | 330 000 | Yes | Neg |
| 167 | ||||
| Jan 99 | 443 | 10 800 | No | Pos |
| Nov 03 | 511 | 12 960 | No | Neg |
| Apr 04 | 331 | 7 000 | No | Neg |
| 131 | ||||
| Oct 00 | 656 | 17 000 | No | Pos |
| Jul 02 | 964 | 82 000 | No | Pos |
| Nov 04 | 905 | < 50 | Yes | Neg |
| 39 | ||||
| Jan 02 | 551 | 94 000 | No | Pos |
| Jan 04 | 369 | 127 658 | No | Neg |
| Dec 04 | 664 | 71 | Yes | Neg |
| 110 | ||||
| Feb 96 | 625 | 14 000 | No | Pos |
| Feb 02 | 577 | 46 000 | Yes | Neg |
| Oct 04 | 238 | 199 603 | Yes | Neg |
| 4 | ||||
| Sep 95 | 454 | 13 550 | No | Pos |
| Sep 96 | 326 | 233 063 | Yes | Neg |
| Jun 04 | 525 | < 50 | Yes | Neg |
| 22 | ||||
| May 95 | 728 | 400 | No | Pos |
| Apr 96 | 579 | 1 102 | No | Pos |
| Jun 97 | 604 | 7 800 | Yes | Neg |
| 11 | ||||
| Apr 96 | 464 | 4 900 | No | Pos |
| Apr 98 | 806 | 39 000 | Yes | Neg |
| May 99 | 873 | 33 000 | Yes | Neg |
| 21 | ||||
| Jan 95 | 638 | 2 218 | No | Pos |
| Sep 96 | 468 | 1 827 | No | Pos |
| Nov 97 | 417 | 15 000 | Yes | Neg |
To establish whether baseline viremia levels could be associated with changes in anti-CCR5 Ab serostatus, the Cox proportional hazard regression model was used. The hazard ratio was 1.8 (95% CI, 1.12-4.15), indicating that an increment of 10 000 viremia units is associated with the loss of anti-CCR5 Abs. Moreover, all of the viral isolates from LTNPs carrying CCR5 Abs were R5.
Lack of correlation between anti-CCR5 Abs and other factors, including genetic polymorphisms and alteration in immunoglobulin pattern
The presence of genetic polymorphisms was assessed. Two LTNPs were heterozygous for CCR5-Δ32 mutation (LTNPs no. 21 and no. 67). All other LTNPs were homozygous wild type for CCR5. Four LTNPs carried CCR2-64I polymorphism (LTNP no. 4, LP no. 11, LP no. 39, NY no. 4), which is in linkage dysequilibrium with a mutation in the CCR5 promoter gene.24 One individual was homozygous for SDF1-3′A mutation (LTNP no. 11), whereas the other 5 LTNPs were heterozygous for SDF1-3′A (LTNPs no. 4, no. 20, no. 21, no. 22, no. 39).
Viremia variability correlation between patients who retained and lost anti-CCR5 Abs during follow-up period. Individual regression curves were compared between LTNP subjects changing (A) or maintaining (B) anti-CCR5 Abs. Samples were evaluated at the time of entry and at least twice during 3 years of follow-up. Median slope values per each group of LTNPs are shown. Analysis was performed by the Mann-Whitney test.
Viremia variability correlation between patients who retained and lost anti-CCR5 Abs during follow-up period. Individual regression curves were compared between LTNP subjects changing (A) or maintaining (B) anti-CCR5 Abs. Samples were evaluated at the time of entry and at least twice during 3 years of follow-up. Median slope values per each group of LTNPs are shown. Analysis was performed by the Mann-Whitney test.
We evaluated whether the loss of anti-CCR5 antibodies was due to a general deterioration of the immune system. No alteration of Ig pattern (total-IgG, -IgA, -IgM, and -IgE, tetanus- and measles specific Ig) was found in LTNPs analyzed at 2 time points corresponding to dates when they were positive for anti-CCR5 Ig and after the loss of these antibodies (data not shown).
To verify whether the presence of anti-CCR5 Abs could be associated to autoimmune diseases, all samples with anti-CCR5 Abs were analyzed for systemic and organ-specific autoimmunity, including Abs to DNA, extractable nuclear antigens, mitochondria, and thyroglobulin. No sign of autoimmune diseases was found in these subjects (data not shown).
Furthermore, no significant correlations were found between the presence of anti-CCR5 Abs and age, sex, and HIV risk factor; although a prevalence of men versus women was found (16 men versus 4 women).
Discussion
Viruses that use the CCR5 coreceptor molecule are the strains primarily transmitted in vivo.18 Consequently, anti-CCR5 antibodies, previously found in some ESN subjects may play a protective role against HIV-1 infection. HIV-1 resistance linked to immune responses, such as the production of antibodies to HLA class I, CD4, and other surface receptors, has been previously reported.33,36,40 These findings suggest a paradoxical protective role of autoantibodies, which may act by preventing HIV-1 infection, likely delaying disease progression.23-25,41,42
In this study, we detected anti-CCR5 antibodies in 23.5% of sera from LTNP individuals. No anti-CCR5 Abs were found in sera obtained from progressing HIV-1–seropositive individuals with AIDS, HAART-treated chronic infected patients without AIDS (HIV+HAART), or from healthy control subjects. Thus, these finding suggest a possible association between anti-CCR5 antibodies and the LTNP condition.
Anti-CCR5 antibodies recognized the cyclic peptide 3 (13-mer; YAAAQWDFGNTMCQ, aa 89-102) corresponds to the first loop of CCR5.43 Affinity-purified Abs on peptide 3 were able to induce CCR5 internalization, both in CD4+ T lymphocytes and in CCR5-transfected U87 cells (Figure 2A-B,E; Table 2) and to block infection with R5 strains of HIV (Table 3).
The signaling activity of CCR5 is partially controlled by internalization, recycling, and/or degradation, and it is not yet fully understood. CCR5 internalization may follow different pathways, including a clathrin-independent32,44 or -dependent pathway.45,46 We found that human Abs in LTNPs are able to induce internalization through the clathrin rather than the caveolae pathway. There are several differences between this study and previous studies that explain our different findings. The major difference is that we analyzed antibody-CCR5 internalization through binding to the first loop of CCR5, whereas previous works described the activity of CCR5-agonist–mediated endocytosis, which specifically bind the second external loop of CCR5. We cannot exclude different properties of each CCR5 extramembrane region, as revealed by the use of monoclonal antibodies to the N-terminus and to the second loop of CCR5, which differentially modulated and influenced receptor activity.47 For the first time, we found that a region not involved in HIV entry48 efficiently blocks HIV infection through an internalization of CCR5 that remains stable for several days (Figure 2C).
Natural and/or modified CCR5 ligands neutralize HIV by blocking the HIV-binding site and/or inducing signaling (internalization and interference with receptor recycling) following short-time kinetics,49 whereas antibodies to the first loop of CCR5-mediated HIV neutralization appears to result from the combination of 2 mechanisms time related. The shorter one, probably due to steric hindrance, includes a low level of HIV entry inhibition, reduction of chemotaxis MIP1β dependent, as we previously reported in ESN.19 In the second one, more importantly, these Abs induce a long-lasting CCR5 down-regulation. Thus, the 2 mechanisms suggest that a complete block of HIV replication is achieved by anti-CCR5 antibodies.
Moreover, in an experimental BalB/c mouse model, we induced and reproduced the specificities found in ESN and in LTNP. Mouse Abs to first loop of CCR5 in vivo induce CCR5 internalization and reexpression, following a very slow kinetics that was gradually recovered 4 weeks after immunization.50 This finding is potentially relevant for the design of appropriate microbicide candidates that may prevent HIV infection for several days after specific application of a compound.
We can hypothesize that either (1) anti-CCR5 antibodies are elicited by low levels of HIV-specific stimulation, thus explaining why these Abs have been found in ESNs and LTNPs but not in subjects who did not experienced HIV exposure or (2) anti-CCR5 Abs are elicited by some HIV-unrelated antigenic stimulation. The autoantibodies produced during the course of other viral infections have been attributed to virus-induced self-alterations that become autoimmunogenic.51 Host factors, including endogenous retroviruses (ERVs), could induce perturbations in cell membranes, through changes in surface receptors, that lead to the exposition of non-self epitopes. Thus, the acquisition of unusual, highly immunogenic conformations of CCR5 could be due to atypical viral exposure or local factors that could induce autoimmune antibodies, able to block HIV replication by long-lasting down-regulation of CCR5, rather than to interfere with gp120 binding on the cell surface. In this last situation, these antibodies may be present in some rare subjects (< 1/412, which is the sum of the 3 control populations: HC, AIDS, and HIV+HAART). (3) Alternatively, some individuals could have undergone a previous natural/immune priming because of retroviral proteins that share a partial homology with env-proteins; this early event could generate a pool of B-memory cells able to trigger cross-reactive immune responses. Once exposed to HIV, these subjects can readily mount a more HIV-specific and protective response, addressed to antigens that mimic viral particles.
During the course of follow-up, 9 of 20 LTNPs lost anti-CCR5 Abs and experienced a statistically significant increase in viremia (Figure 4), requiring the assumption of therapy (Table 4), thus becoming progressors. Strikingly subjects who retained anti-CCR5 Abs maintained stable conditions without any treatment. This finding suggests that the loss of anti-CCR5 Abs is associated to progression toward the disease, and it is strongly supported by the development of AIDS despite antiretroviral therapy in some subjects (Table 4). The patients were also classified into 2 groups according to whether they experienced an increasing level of viremia during follow-up (at least 1 log increment). The distribution of time interval to change in anti-CCR5 serostatus was analyzed by the Kaplan-Meier method. Comparison among survival curves between the 2 groups was made by means of log-rank test. A longer survival was observed in LTNPs whose viremia levels showed less than 1 log of increment (data not shown).
Moreover, the viral phenotype in LTNPs carrying anti-CCR5 antibodies does not shift in the presence of such antibodies, thus confirming that the selective pressure of CCR5 inhibitors does not induce a change of viral phenotype, as already reported in a monkey model.52 In addition, anti-CCR5 antibodies do not induce any apparent alterations in immune function, as demonstrated by the continued health of subjects who retained anti-CCR5 antibodies. Taken together, the results reported here are relevant for vaccine design strategies and also provide argument against theoretical concerns about using drugs that may block CCR5, such as CCR5 agonists and antagonists.
Prepublished online as Blood First Edition Paper, March 7, 2006; DOI 10.1182/blood-2005-06-2463.
Supported by Istituito Superiore di Sanità (grant 40D50; L.L.), (grant 30D63; G.P.), and National Institutes of Health (grants R01-AI-42555 and U01-AI-35004; H.B. and B.W.).
C.P., C.B., and S.G. performed immunologic and virologic analyses of Italian subjects and contributed to writing the paper; B.W., H.B., and K.K. performed genetic and immunologic analyses of subjects from the United States; C.U.-F.,A.L., and G.P. helped design the research; R.L. synthesized peptides and helped analyze the data; G.C. performed the statistical analysis; and L.L. designed the research, analyzed the data, and wrote the paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank D. Montefiori, B. Chackerian, and S. Shapiro for helpful discussion and critical reading of the manuscript. We also thankE. Faioni and J. Wethers for the samples from HIV-1–seronegative individuals; Sean Philpott for determining the CCR5 and CCR2 genotypes of the US subjects; D. Burton and J. Binley for providing SOS pseudoviruses; G. Morsica, G. Gallotta, F. Canducci, andF. Lillo for management of clinical data; and S. Russo and J. Li for editorial help.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal