Abstract
Macrophages seed all tissues in which they have the ability, in specific and rare instances, to fuse with themselves and to differentiate into osteoclasts in bone or into giant cells in chronic inflammatory reactions. Although these cells play a central role in osteoporosis and in foreign body rejection, respectively, the molecular mechanism used by macrophages to fuse remains poorly understood. Macrophages might also fuse with somatic and tumor cells to promote tissue repair and metastasis, respectively. We reported that CD44 expression is highly induced in macrophages at the onset of fusion in which it plays a role. We report now that the intracellular domain of CD44 (CD44ICD) is cleaved in macrophages undergoing fusion and that presenilin inhibitors prevent the release of CD44ICD and fusion. We also show that CD44ICD promotes the fusion of tissue macrophages and bone marrow-derived macrophages. Finally, we report that CD44ICD is localized in the nucleus of macrophages in which it promotes the activation of NF-κB. These observations open avenues to study the role of CD44ICD in blood cells and tumors. (Blood. 2006;107: 796-805)
Introduction
Macrophages seed all tissues and have the capacity, in specific and rare instances, to undergo homotypic fusion and to differentiate into multinucleate osteoclasts or giant cells, in bone or in chronic inflammatory reactions, respectively. Multinucleation endows macrophages with the ability to resorb components located extracellularly such as bone and foreign bodies, respectively.1,2 In as much as osteoclasts are essential for the remodeling of bone, they are chiefly responsible for the loss of bone that leads to osteoporosis. Osteoclasts are also responsible for the local loss of bone that results from inflammation, such as in rheumatoid arthritis and in periodontal disease. Giant cells differentiate around foreign bodies such as pathogens, implants, and transplants that they resorb.
Increasing evidence suggests that macrophages possess the ability to fuse also with somatic cells to repair tissues and organs and with tumor cells to trigger the metastatic process.2 So macrophages, which are ubiquitously present in tissues where they perform a wide array of functions, might be endowed with a larger repertoire of biologic activities than originally anticipated.3
The molecular mechanisms used by macrophages to adhere to, and to fuse with each other, and possibly with other cells, is an essential step that remains to be characterized. Indeed, cell-to-cell fusion itself, whether it concerns that of sperm cells with oocytes or myoblasts with myoblasts, leading to fertilization and muscle development, respectively, remains unclear.4
To gain insight into the fusion mechanism of macrophages, we had subjected fusing rat alveolar macrophages to genome-wide oligonucleotide microarray, an approach that revealed the transiently induced expression of presenilin 2 (PS2), a protein associated with Alzheimer disease (for a review, see Hutton and Hardy5 ). The fact that the deletion of the ps1 gene in mice leads to severe skeletal defects,6 and that the deletion of the ps2 gene enhances the embryonic lethal phenotype of ps1 deletion,7 triggered our interest in PS.
Presenilins (PS1 and PS2) are transmembrane proteins identified originally through genetic linkage analysis of families with autosomal dominant forms of Alzheimer disease.5,8 PS1 and PS2 proteins are part of a large complex referred to as γ-secretase, share strong sequence homology, and are highly conserved and ubiquitously expressed. They contain 8 transmembrane domains and are rapidly cleaved on completion of synthesis into an approximate 30-kDa amino-terminus and an approximate 20-kDa carboxy-terminus fragment. PSs were originally thought of as targeting the γ-cleavage site of amyloid precursor protein (APP), which leads to the release of 40 to 42 amino acid peptides that accumulate intracellularly and form the so-called amyloid plaques. PSs are now known to cleave the intracellular domain (ICD) of at least 8 type I membrane proteins, including CD44,9,10 a receptor that we reported plays a role in the fusion of macrophages.11 Following the cleavage by membrane bound metalloproteases of the extracellular domain (ECD) of proteins that are a substrate for PS, intramembrane proteolysis occurs (for a review, see Huovila et al12 ). This event appears to be mediated by a γ-secretase protein complex that includes, in addition to PS1 and PS2, nicastrin, APH-1, and PEN-2 (for a review, see De Strooper13 ). In the case of CD44, which is a receptor for hyaluronic acid, its ECD is cleaved by MT1-MMP and ADAM-like proteases.14 The intracellular domain of CD44 (CD44ICD) is subsequently cleaved by γ-secretase9,15,16 and translocates to the nucleus in which it cooperates functionally with CBP/p300 cells to potentiate transcriptional activation in COS cells.16 We have reported that the expression of the hemopoietic form of CD44 is highly induced in macrophages at the onset of fusion.11 The similar increase in the abundance of PS2 transcripts that occurs at the onset of fusion in macrophages prompted us to ask whether PS plays a role in that event and, if so, whether it targets CD44.
We report now that the expression of PS2, like that of CD44, is induced at the onset of fusion, and that inhibitors of PS prevent the fusion of macrophages and the release of CD44ICD. We show that CD44ICD translocates to the nucleus and promotes the activation NF-κB, a transcription factor that is required for the differentiation of multinucleate osteoclasts.17,18 This work reveals the expression of PS1 and PS2 by macrophages and a new function for one of their targets, CD44, which is to promote fusion.
Materials and methods
Reagents
Recombinant mouse RANKL was purchased from RDI (Flanders, NJ), and macrophage colony-stimulating factor (M-CSF) from R&D Systems (Minneapolis, MN). DAPT19 and DupE20 were gifts from Dr Todd Golde (Mayo Clinic, Jacksonville, FL). JLK221 was a gift from Dr Frédéric Checler (UMR 6097, Valbonne, France). NBD peptides, wild type and mutant, were a gift from Shankar Ghosh (Yale School of Medicine, New Haven, CT).22 Helenalin, Bay 11-7082, CAPE, and HD were obtained from Biomol (Plymouth Meeting, PA). Unless otherwise stated, all chemicals were from Sigma (St Louis, MO). The monoclonal antibody (mAb) mouse anti-rat CD44 was purchased from Serotec (Raleigh, NC). This antibody recognizes the ECD of rat CD44 in its native conformation only, hence, not reduced. The mAb mouse anti-myc mAb 9E10 was obtained from the Hybridoma Bank at the University of Iowa. Rabbit anti-PS1, anti-PS2, and anti-NF-κB p65 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). mAb Y12 anti-Sm proteins were kindly provided by Dr Joan Steitz (Yale School of Medicine, New Haven, CT),23 and mouse mAb anti-MFR/SIRPα was previously published.24 Mouse anti-GAPDH was purchased from Ambion (Austin, Texas). Rabbit Ab anti-p38, mouse mAbs anti-I-κB and phospho-I-κB were obtained from Cell Signaling Technology (Beverly, MA); horseradish peroxidase-conjugated F(ab′)2 antimouse and antirabbit Abs were from Jackson ImmunoResearch (West Grove, PA).
Cells
Rat alveolar and peritoneal macrophages were obtained from 12-week-old Fisher rats (Charles River, Kingston, NY) by tracheobronchial and peritoneal lavage, respectively, and cultured in fusogenic conditions as previously described.25 The National Research Council's guide for the care and use of laboratory animals was followed. The study protocol was approved by the Yale Animal Care and Use Committee.
Bone marrow-derived macrophages were obtained from 6- to 12-week-old C57Bl/6J (Jackson Laboratories, Bar Harbor, ME) mouse tibiae and femurs by flushing with Dulbecco modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) as we previously described.26 Bone marrow cells (1 × 107) were cultured at an initial density of 1 × 106 cells/mL/10-cm dish (Becton Dickinson, Franklin Lakes, NJ) in α-minimum essential medium (α-MEM; Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS) in the presence of M-CSF (5 ng/mL) for 12 to 18 hours at 37°C in humidified 5% CO2. Nonadherent cells were harvested and cultured with M-CSF (30 ng/mL) in 10-cm dishes at the same density as before for an additional 48 hours. Floating cells were removed and attached cells that are tartrate resistant acid phosphatase (TRAP+)/macrophage-like were used as osteoclast precursors.27 To generate osteoclasts, adherent cells were cultured in the presence of RANKL (100 ng/mL) and 30% L929 cells supernatant in either 96-well dishes (2 × 104 cells/0.2 mL/well) or 60-mm dishes (2.5 × 106 cells/5 mL/dish). Adherent cells were cultured for the indicated times. Raw 264.7 cells were provided by Dr Bryant Darnay (MD Anderson Cancer Center, Houston, TX).
CD44 constructs
Full-length CD44 (CD44FL) constructs with C-terminal myc-epitope tag were generated by polymerase chain reaction (PCR) using murine CD44 cDNA (GenBank no. BC005676) and rat CD44 cDNA11 as templates, and the following primers: CD44FL forward, ggagatcttcatggacaagt/gtttggtggca (t/g corresponds to F in mice and V in rats, in the leader peptide), and CD44FL reverse, ggaattccCTACAGATCCTCTTCTGAGATGAGTTTTTGTTCcaccccaatcttcatgtcca (capital letters denote the C-terminal myc-epitope tag). The PCR products were subcloned into the BamHI/EcoRI sites of pcDNA3 (Invitrogen), and BglII/EcoRI sites of the retrovirus vector MigR1 (provided by Dr Warren Pear, University of Pennsylvania, Philadelphia, PA).28 In that vector, the target gene and GFP are separated by an internal ribosome entry site (IRES) so they can be expressed separately. The infection efficiency with that vector is very high, ranging between 60% and 90% routinely.
CD44 constructs lacking the ECD (CD44ΔE), with a signal peptide and a C-terminal myc-epitope tag were generated by PCR using the following primers: CD44ΔE forward, ggagatcttcATGGACAAGTTTTGGTGGCACACAGCTTGGGGACTTTGCCTCTTGCAGTTGAGCCTGGCAcaggacagtggagtgaccaca (capital letters denote the signal peptide), and CD44ΔE reverse, ccgctcgagcggCTACAGATCCTCTTCTGAGATGAGTTTTTGTTCcaccccaatcttcattgtcca (capital letters denote the C-terminal myc-epitope tag). The PCR product was subcloned into the BamHI/XhoI sites of pcDNA3 and BglII/XhoI sites of MigR1.
CD44ICD constructs with C-terminal myc-epitope tag were generated by PCR using the following primers: CD44ICD forward, ggagatcttatggcggtcaatagtaggagaagg, and CD44ICD reverse, ggaattccCTACAGATCCTCTTCTGAGATGAGTTTTTGTTCcaccccaatcttcatgtcca (capital letters denote the C-terminal myc-epitope tag). The PCR product was subcloned into the BamHI/EcoRI sites of pcDNA3, and BglII/EcoRI sites of the retrovirus vector MigR1.
CD44ΔE construct with c-terminal GFP was generated by releasing CD44ΔE from pcDNA3-CD44ΔE HindIII/KpnI sites and subcloning into HindIII/KpnI sites of pEGFP-N1vector (Clontech, Palo Alto, CA) to generate pN1-CD44ΔE-eGFP. CD44ΔE-eGFP was excised from HindIII/NotI sites and subcloned into HindIII/NotI sites of the LZRSpBMN-Z vector (provided by Dr Ira Mellman, Yale School of Medicine, New Haven, CT) to generate pBMN-CD44ΔE-eGFP.
The resulting constructs were sequenced for verification (W. M. Keck Biotechnology Resource Laboratory at Yale University).
Retrovirus production and infection. Packaging cells GPG293 (kindly provided by Dr Betty Lamothe, MD Anderson Cancer Center, Houston, TX) were cultured in DMEM supplemented with 10% heat-inactivated FBS, 50 U/mL penicillin/streptomycin (P/S), 2 mM l-glutamine, 1 μg/mL tetracycline, 2 μg/mL puromycin, and 0.3 μg/mL G418. GPG293 cells were split 1:5 into 10-cm dishes 2 days prior to transfection. Twelve μg DNA supplemented with Fugene 6 was added to each dish. Two days after transfection, the medium was changed to viral production medium (DMEM with 10% heat-inactivated FBS, 50 U/mL P/S). Cells were grown for 2 more days, viruses were then harvested by filtering through a 0.2-μm syringe filter, and assayed for viral titer by infection on NIH3T3 cells. Viral supernatants were supplemented with Polybrene (Clontech) to reach a final concentration of 10 μg/mL, and stored at 4°C.
Rat alveolar macrophages and mouse bone marrow macrophages were cultured at a density of 1 × 106 cells/mL in 10-cm dishes in MEM supplemented with 10% FCS and 30% L929 cell supernatant for 2 days, infected with virus-containing medium overnight, and then cultured in the same but fresh medium to recover for at least 24 hours. Raw 264.7 cells were infected with virus-containing medium overnight, then cultured in fresh medium to recover for at least 24 hours. Infection efficiency was monitored using an Olympus microscope (IMT-2) equipped with UV light and an eyepiece that contains a grid for quantification. Cultures with at least 45% to 50% GFP+ cells were selected for the experiments.
Subcellular fractionation: isolation of plasma membrane, nuclear and cytosolic fractions. Approximately 1 × 107 macrophages were used for each condition. Raw 264.7 cells infected with MigR1-CD44ICD were cultured for 2 days in the presence of L929 supernatant and RANKL. Cells were scraped in cold phosphate-buffered saline (PBS) supplemented with a cocktail of protease inhibitors (Roche Molecular Biochemicals, Mannheim, Germany), pelleted by low-speed centrifugation, washed in ice-cold PBS, and resuspended in 200 μL of buffer A (20 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, and protease inhibitors). The cells were then placed on ice and allowed to swell for 10 minutes before being lysed with Nonidet P-40 (NP-40) detergent. Following centrifugation, the nuclear pellet was washed once more in 100 μL buffer A and then resuspended in 50 μL buffer B (20 mM HEPES, pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM dithiothreitol, and protease inhibitors). The supernatant recovered from the NP-40 lysis solution was further centrifuged at 90 000g for 30 minutes at 4°C to separate the membrane from the cytosolic fraction. The nuclear pellet was shaken for 15 minutes at 4°C and then centrifuged at 10 000g to remove debris. Protein concentration was determined using a protein assay kit from Bio-Rad (Hercules, CA), and the nuclear, membrane and cytosolic fractions were subjected to Western blot analysis.
Western blotting. Cultured cells were lysed directly in Laemmli sample buffer supplemented with a cocktail of protease and phosphatase inhibitors, then subjected to Western blot analysis using enhanced chemiluminescence (ECL; Pierce, Rockford, IL). We have observed that macrophages are particularly rich in lysosomal enzymes that degrade proteins while in lysis buffer, even when supplemented with protease inhibitors and kept at 4°C. Unless proteins need to be purified using immunoaffinity, for instance, we lyse cells directly in sample buffer supplemented with protease inhibitors, then subject them to Western blot analysis to equilibrate the protein concentration among lysates. Tissue macrophages, such as alveolar macrophages, are postmitotic so we equilibrate their number at the time of plating.
Luciferase reporter assay. Raw 264.7 cells were plated at a density of 3 × 105 cells/well in 24-well plates 1 day before transfection. Cells from 6 wells were transfected using Fugene 6 (Roche Molecular Biochemicals, Indianapolis, IN) with 0.2 μg pBIIX-Luc, pAP-1-Luc (provided by Dr Jianjiang Ye, Yale University School of Medicine, New Haven, CT), or pNFAT-Luc (provided by Dr Gerald Crabtree, Stanford University School of Medicine, HHMI, Stanford, CA)29 and the indicated amounts of pcDNA3-CD44FL-myc, pcDNA3-CD44ΔE-myc, and pcDNA3-CD44ICD-myc. The total amount of transfected plasmid was equalized by supplementing the reaction mixture with the empty vector pcDNA3. Equivalent numbers of macrophages were harvested 48 hours after transfection, washed in PBS, lysed, and assayed for luciferase activity (Roche Molecular Biochemicals) and measured in a luminometer (Lumat LB9501, Wallac, Gaithersburg, MD) for 10 seconds (2 measures were made for each sample). The data were normalized to total protein concentration, for each sample, determined using a protein assay kit from Bio-Rad. Results are presented as percentage of luciferase activity of cells transfected with pcDNA3-CD44ICD-myc over that of cells transfected with empty vector pcDNA3. Some cells from parallel wells were treated with RANKL for 6 hours prior to lysis and used as a positive control. Other cells were lysed directly in Laemmli sample buffer and subjected to Western blotting analysis to monitor CD44ICD expression.
Nuclear extracts and electrophoretic mobility shift assays. Nuclear proteins were extracted from Raw cells transduced with MigR1 empty or encoding CD44ICD and treated or not with lipopolysaccharide (LPS; 100 μg/mL) or RANKL (100 ng/mL). NF-κB-binding reactions were carried out using 5 to 10 μg nuclear proteins and a 32P double-stranded oligonucleotide containing the NF-κB consensus sequence derived from the mouse κ intronic enhancer (GATCAGAGGGGACTTTCCGAG). Some cells were incubated with NBD peptides, mutant and wild type (50 μM each), for 4 hours prior to performing the binding reaction with 32P-NF-κB. Supershift analyses were performed by preincubating nuclear extracts with 1 μL/reaction of an anti-p65 or anti-myc mAb for 1 hour at 4°C prior to addition of the NF-κB-labeled probe. The samples were then electrophoresed under standard conditions, as previously described.30
Microscopy
Microscopy was performed using an IMT-2 Olympus microscope equipped with UV light and an OM-4 camera; images were captured using a × 10 lens, and the films were scanned and edited using Adobe Photoshop (Adobe, San Jose, CA). Histomorphometry was performed using a Nikon microscope coupled to IBM software and a digitalizing tablet. We used an Axiovert S100-TV (Carl Zeiss Microimaging, Oberkochen, Germany) coupled with a confocal system (MTC 1024, Bio-Rad Laboratories, Hercules, CA) to perform the confocal microscopy.
Statistical analysis
Data represent the mean ± 1 SD. Statistical differences among the experimental groups were evaluated based on the analysis of variances. The significance of the mean change was determined using unpaired Student 2-tailed t test and significance was considered to be P < .05.
Results
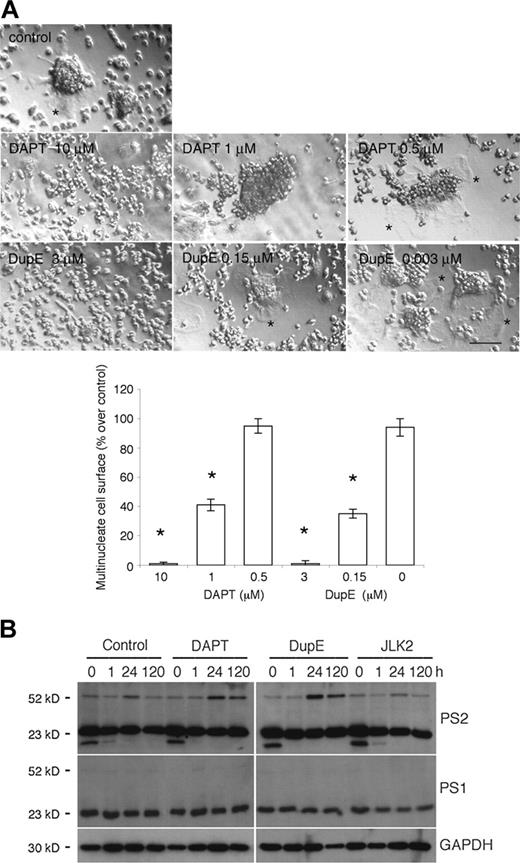
Genome-wide oligonucleotide microarray of fusing rat alveolar macrophages, using Affymetrix technology, revealed that transcripts coding for PS2 (accession no. X99267) were induced 5.1 ± 0.8-fold (SD; n = 3) 24 hours after plating, then decreased to 2.9 ± 0.6-fold at day 5 as compared with freshly isolated cells (Juan Zhang, Jun Li, Randy Barton, and A.V., unpublished data). These tissue macrophages offer an efficient and pure cell model system to study the fusion mechanism of macrophages9,25,31,32 because they spontaneously fuse in vitro when plated at cell-to-cell contact, and, hence, do not require the addition of cytokines. To initiate an investigation on the role of PS in fusion, we treated fusing rat alveolar macrophages with DAPT and DupE, 2 compounds that inhibit the cleavage of the ICD of APP and Notch19,20 by γ-secretase. Under these culture conditions, DAPT and DupE inhibited fusion in a dose-dependent manner (Figure 1A). This is best illustrated by the lack of spreading of the plasma membrane that characterizes multinucleated macrophages. In contrast, JLK2, a PS inhibitor that inhibits the cleavage of APP but not Notch,21 failed to inhibit the fusion of rat alveolar macrophages (data not shown). This confirmed that JLK2, unlike DAPT and DupE, affects γ-secretase activity differently, and suggested that it might target different substrates, or different domains of the same substrates within the γ-secretase complex. We surmised that perhaps these inhibitors altered fusion differently by affecting the expression of PS2. To test this possibility, we subjected lysates from fusing alveolar macrophages treated with DAPT, DupE, or JLK2 for increasing times, to Western blotting analysis. Under these conditions, PS1 and PS2 amino-terminal fragments migrated around 23 kDa and their abundance remained relatively constant over time and unaffected by PS inhibitors. In contrast, PS2 intact protein migrated around 50 kDa and its expression was transiently induced 24 hours after plating, hence, paralleling that of its transcripts (Figure 1B). PS1 intact protein could not be detected, possibly as a result of its low abundance or its rapid posttranslational cleavage or both. Of note, a lower molecular weight protein of about 15 kDa that reacted with the anti-PS2 antibody was detected in macrophages at time zero only. The significance of this PS2-immunoreactive protein remains unclear at this time. Together, these results revealed that macrophages express PS1 and PS2 and suggested that PS2, the expression of which is regulated during the multinucleation of macrophages, plays a role in fusion.
The PS inhibitors DAPT and DupE inhibit the fusion of macrophages and prolong the induced expression of PS2. (A) Rat alveolar macrophages were cultured in fusogenic conditions and treated with increasing concentrations of DAPT or DupE for 3 days. Whereas control cells fused efficiently into multinucleated macrophages, as reflected by the extensive plasma membrane of multinucleate macrophages (*), DAPT and DupE inhibited fusion of macrophages dose dependently, as illustrated by the lack of plasma membrane extension (bar represents 1 mm; *P < .001 versus control; SD; n = 6). (B) Rat alveolar macrophages were cultured in fusogenic conditions in 96-well dishes for the indicated times in the absence or presence of 10 μM DAPT, 3 μM DupE, or 50 nM JLK2. Cells were subjected to Western blotting analysis (0.2 × 106 cells/lane) using antibodies directed against PS1, PS2, or GAPDH. Note that PS1 and PS2 migrated as 23-kDa cleaved amino-terminal fragments, whereas intact PS2 ran as an intact protein of 50 kDa. Also, note the lower molecular weight band of about 15 kDa that is detected in all samples, but at time zero only.
The PS inhibitors DAPT and DupE inhibit the fusion of macrophages and prolong the induced expression of PS2. (A) Rat alveolar macrophages were cultured in fusogenic conditions and treated with increasing concentrations of DAPT or DupE for 3 days. Whereas control cells fused efficiently into multinucleated macrophages, as reflected by the extensive plasma membrane of multinucleate macrophages (*), DAPT and DupE inhibited fusion of macrophages dose dependently, as illustrated by the lack of plasma membrane extension (bar represents 1 mm; *P < .001 versus control; SD; n = 6). (B) Rat alveolar macrophages were cultured in fusogenic conditions in 96-well dishes for the indicated times in the absence or presence of 10 μM DAPT, 3 μM DupE, or 50 nM JLK2. Cells were subjected to Western blotting analysis (0.2 × 106 cells/lane) using antibodies directed against PS1, PS2, or GAPDH. Note that PS1 and PS2 migrated as 23-kDa cleaved amino-terminal fragments, whereas intact PS2 ran as an intact protein of 50 kDa. Also, note the lower molecular weight band of about 15 kDa that is detected in all samples, but at time zero only.
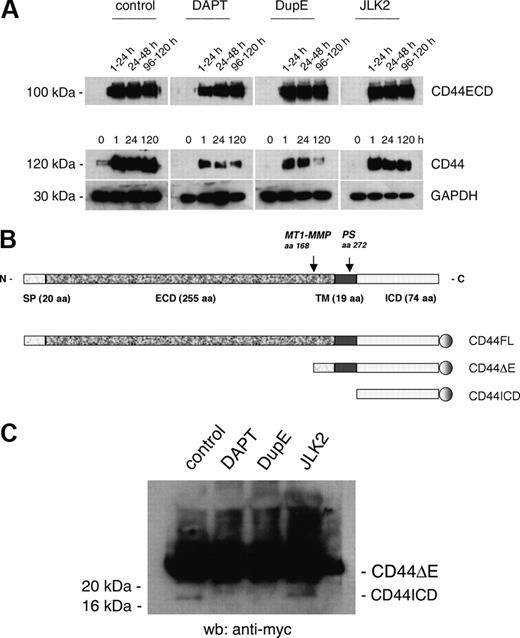
PS inhibitors DAPT and DupE, not JLK2, decrease the expression of CD44 and inhibit the release of CD44ICD. (A) Rat alveolar macrophages (5 × 106 cells/35-mm dish) were cultured for the indicated times, treated or not with DAPT (10 μM), DupE (0.5 μM), or JLK2 (20 μM), then lysed and subjected to Western blot analysis. In parallel wells, 24-hour cell supernatants from macrophages were collected and analyzed for CD44 content using Western blotting. (B) Schematic diagram of the CD44 constructs generated for this study. (C) Rat alveolar macrophages were transduced with the retroviral vector MigR1 empty (control) or encoding CD44ΔE, and then treated or not with DAPT (10 μM), DupE (0.5 μM), or JLK2 (20 μM) for 24 hours. The cell lysates were subjected to Western blotting analysis using an mAb directed against myc. The cleavage of CD44 ΔE into CD44ICD was inhibited by DAPT and DupE, but not by JLK2.
PS inhibitors DAPT and DupE, not JLK2, decrease the expression of CD44 and inhibit the release of CD44ICD. (A) Rat alveolar macrophages (5 × 106 cells/35-mm dish) were cultured for the indicated times, treated or not with DAPT (10 μM), DupE (0.5 μM), or JLK2 (20 μM), then lysed and subjected to Western blot analysis. In parallel wells, 24-hour cell supernatants from macrophages were collected and analyzed for CD44 content using Western blotting. (B) Schematic diagram of the CD44 constructs generated for this study. (C) Rat alveolar macrophages were transduced with the retroviral vector MigR1 empty (control) or encoding CD44ΔE, and then treated or not with DAPT (10 μM), DupE (0.5 μM), or JLK2 (20 μM) for 24 hours. The cell lysates were subjected to Western blotting analysis using an mAb directed against myc. The cleavage of CD44 ΔE into CD44ICD was inhibited by DAPT and DupE, but not by JLK2.
Because CD44 has been recently identified as a target for PS, and because its surface expression is transiently induced in macrophages at the onset of fusion,9 we asked whether PS alters the fusion of macrophages by cleaving the ICD of CD44. Because proteins targeted by PS must have their ECD cleaved first, we ensured that it is the case for CD44 in macrophages. We collected 24-hour supernatant from rat alveolar macrophages cultured in fusogenic conditions for 1 to 5 days. We used different cells/wells for each time point because changing the medium alters fusogenicity (A.V., unpublished observation). Macrophages cultured in parallel wells were lysed and analyzed, along with the supernatants, for CD44 expression using Western blotting. Of note, the mAb we used recognizes the native conformation of the ECD of rat CD44 and requires proteins to be electrophoresed in nondenaturing conditions. Under these conditions, CD44ECD migrated as a 100-kDa protein that was readily detected in supernatant from fusing macrophages at all time points examined (Figure 2A). Neither DAPT, DupE, nor JLK2 altered the abundance of CD44ECD. In contrast, the abundance of CD44 tended to be reduced by DAPT and DupE, but not JLK2 treatment. Together, these data revealed that CD44ECD sheds from fusing macrophages and is released into the supernatant as an intact protein independent of PS activity. Because CD44ICD increases the abundance of CD44 transcripts,16 these data suggested that DAPT and DupE might prevent the release of CD44ICD in fusing macrophages. To next investigate whether CD44ICD is cleaved by PS, we engineered a series of fusion proteins that contain the full length of CD44 (CD44FL), its transmembrane and intracellular domain (CD44ΔE), or its intracellular domain (CD44ICD), each tagged with myc at its carboxy terminus (Figure 2B). We inserted each one of these constructs into in the retroviral vector MigR1. We first transduced rat alveolar macrophages that had been plated at low density and cultured for 2 days in the presence of the growth factor, M-CSF, with the retroviral vector MigR1 either empty or encoding CD44ΔE. Macrophages were then further treated with M-CSF for 24 hours. Under these culture conditions, macrophages expressed CD44ΔE (Figure 2C). A lower molecular weight form that reacted with anti-myc antibody, hence was most likely CD44ICD, was detected in lysates from control and from JLK2-treated cells, but not from DAPT and DupE-treated cells (Figure 2C). These data indicated that PS cleaves the ICD of CD44 in rat alveolar macrophages.
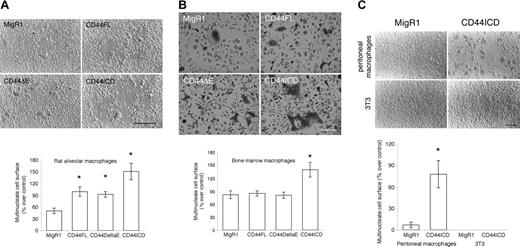
CD44ICD promotes the fusion of macrophages and the differentiation of TRAP+multinucleated osteoclasts. (A) Rat alveolar macrophages were transduced with the retroviral vector MigR1 empty or encoding CD44FL, CD44ΔE, or CD44ICD, plated in fusogenic conditions, and then cultured for only 2 days. Whereas macrophages transduced with empty vector, CD44FL, or CD44ΔE had barely initiated fusion by day 2, those transduced with MigR1 encoding CD44ICD were well fused (bar represents 3 mm; *P < .001 versus MigR1; SD; n = 5). (B) Mouse bone marrow cells were transduced with the retroviral vector MigR1 empty or encoding CD44FL, CD44ΔE, or CD44ICD, treated with M-CSF supplemented with RANKL for only 2 days to differentiate into osteoclasts, then reacted for TRAP, an osteoclast marker. Whereas very few macrophages transduced with empty vector or vector encoding CD44FL or CD44ΔE were fused by day 2, macrophages transduced with CD44ICD were highly differentiated into TRAP+ multinucleated osteoclasts (bar is 2 mm; *P < .001 versus MigR1; SD; n = 6). (C) CD44ICD promotes the fusion of peritoneal macrophages but does not induce the fusion of 3T3 cells. Rat peritoneal macrophages and 3T3 cells were cultured in fusogenic conditions for 18 hours and 48 hours, respectively, following transduction with MigR1 either empty or encoding CD44ICD. CD44ICD strongly promoted the fusion of rat peritoneal macrophages (bar represents 1 mm; *P < .001 versus MigR1; SD; n = 4).
CD44ICD promotes the fusion of macrophages and the differentiation of TRAP+multinucleated osteoclasts. (A) Rat alveolar macrophages were transduced with the retroviral vector MigR1 empty or encoding CD44FL, CD44ΔE, or CD44ICD, plated in fusogenic conditions, and then cultured for only 2 days. Whereas macrophages transduced with empty vector, CD44FL, or CD44ΔE had barely initiated fusion by day 2, those transduced with MigR1 encoding CD44ICD were well fused (bar represents 3 mm; *P < .001 versus MigR1; SD; n = 5). (B) Mouse bone marrow cells were transduced with the retroviral vector MigR1 empty or encoding CD44FL, CD44ΔE, or CD44ICD, treated with M-CSF supplemented with RANKL for only 2 days to differentiate into osteoclasts, then reacted for TRAP, an osteoclast marker. Whereas very few macrophages transduced with empty vector or vector encoding CD44FL or CD44ΔE were fused by day 2, macrophages transduced with CD44ICD were highly differentiated into TRAP+ multinucleated osteoclasts (bar is 2 mm; *P < .001 versus MigR1; SD; n = 6). (C) CD44ICD promotes the fusion of peritoneal macrophages but does not induce the fusion of 3T3 cells. Rat peritoneal macrophages and 3T3 cells were cultured in fusogenic conditions for 18 hours and 48 hours, respectively, following transduction with MigR1 either empty or encoding CD44ICD. CD44ICD strongly promoted the fusion of rat peritoneal macrophages (bar represents 1 mm; *P < .001 versus MigR1; SD; n = 4).
To initiate an investigation on the role of CD44ICD in macrophage fusion, we transduced rat alveolar macrophages treated with M-CSF with the retroviral vector MigR1 empty or encoding CD44FL, CD44ΔE, or CD44ICD. We then further cultured the cells for only 2 days in the presence of M-CSF. Under these culture conditions, macrophages transduced with MigR1 empty or encoding CD44FL and CD44ΔE had initiated fusion but remained for the most part mononucleated. In contrast, macrophages transduced with CD44ICD were multinucleated (Figure 3A). These results suggested that CD44ICD promotes the fusion of macrophages.
To ask next whether CD44ICD promotes also the fusion of bone marrow-derived macrophages, hence preosteoclasts, we cultured mouse bone marrow cells in the presence of M-CSF for 2 days to clonally expand macrophages, then with M-CSF supplemented with RANKL for another 2 days to induce the fusion of macrophages and initiate their differentiation into multinucleate osteoclast-like cells that requires 4 to 5 days.27 Again, macrophages were transduced with MigR1 empty (control) or encoding mouse CD44FL, CD44ΔE, or CD44ICD. Under these conditions, relatively few cells transduced with MigR1 alone or encoding CD44FL or CD44ΔE became multinucleated (Figure 3B). In contrast, bone marrow macrophages transduced with CD44ICD fused efficiently and differentiated into TRAP+ multinucleate cells, hence, osteoclasts. CD44ICD also promoted the fusion of peritoneal macrophages, which fuse at a lower rate than alveolar macrophages cultured here for 18 hours (Figure 3C).31 CD44ICD did not, however, induce the fusion of 3T3 cells that do not fuse spontaneously, even when plated in the same fusogenic conditions as tissue macrophages (Figure 3C). These data indicated that CD44ICD promotes but does not initiate cell-to-cell fusion.
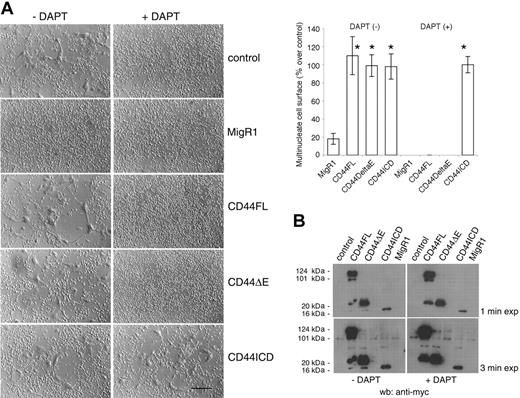
We next asked whether CD44ICD could rescue fusion of cells treated with a PS inhibitor. We infected rat alveolar macrophages in the same manner as described in Figure 3, and then treated or did not treat the macrophages with DAPT, but this time for 4 days. Under these conditions, DAPT blocked the fusion of macrophages transduced or not with CD44FL or CD44ΔE. However, macrophages transduced with CD44ICD fused even in the presence of DAPT, indicating that CD44ICD rescued fusion (Figure 4A). Of note, cells transduced with empty vector MigR1 fused poorly when compared with cells untreated, control cells, or cells transduced with MigR1 encoding CD44FL or CD44ΔE. The effects of MigR1 empty vector on macrophages remain unclear at this time.
To confirm that DAPT prevents the cleavage of CD44ΔE and the release of CD44ICD, we subjected the lysates from these cells to Western blotting analysis using an anti-myc antibody to reveal CD44ΔE and CD44ICD. CD44FL migrated as a diffuse band of about 120 kDa molecular weight, similar to that of endogenous CD44 in these cells (Figure 2A). CD44FL migrated also as proteins of 100, 35, and 20 kDa (Figure 4B). Because the lowest molecular weight form of CD44FL and CD44ΔE comigrated with CD44ICD, and was not detected in the presence of DAPT, it suggested that it was CD44ICD (Figure 4B).
To ask next whether CD44ICD translocates to the nucleus, as in COS cells,16 we engineered a protein that contains CD44ΔE tagged with green fluorescent protein (eGFP) at its C-terminus. We then inserted CD44ΔE-eGFP or eGFP alone in the retroviral vector MigR1. Raw macrophages transduced with eGFP exhibited an overall diffuse fluorescent signal. In contrast, the fluorescent signal of cells transduced with CD44ΔE-eGFP was restricted to the plasma membrane and the nucleus, suggesting that CD44ΔE is anchored in the plasma membrane and that its ICD is released and translocates to the nucleus in which it accumulates (Figure 5A). On treatment with DAPT, macrophages exhibited a “stellar” shape, and, in the case of CD44ΔE-eGFP, a “spiky” and “clustered” plasma membrane fluorescence associated with a weak nuclear signal (Figure 5A). In contrast, cells treated with JLK2 appeared similar to the controls. Together, these data revealed that in macrophages, CD44ICD is released and translocates to the nucleus. This translocation occurred despite the lack of a nuclear localization signal in CD44ICD (data not shown).
To confirm the nuclear localization of CD44ICD biochemically, we subjected fusing rat alveolar macrophages transduced with MigR1-CD44ΔE to subcellular fractionation followed by Western blotting analysis using an anti-myc antibody. We used also antibodies directed against proteins expressed in the plasma membrane (MFR), in the cytosol (p38 MAPK), and in the nucleus (Sm) to confirm the nature of the subcellular fractions. As expected, CD44ΔE and CD44ICD localized to the same fraction as MFR and Sm, respectively, hence, in the plasma membrane and in the nucleus (Figure 5B). The fraction of CD44ΔE detected in the cytosol was unexpected and might result from the overflow from the plasma membrane fraction in which it is very abundant. Indeed, CD44ICD was not detected in the cytosol, possibly as a consequence of its low abundance or of its rapid translocation to the nucleus.
CD44ICD rescues the fusion of macrophages treated with DAPT. (A) Rat alveolar macrophages were transduced with MigR1 empty or encoding CD44FL, CD44ΔE, or CD44ICD, and treated or not with DAPT (10 μM) for 4 days (bar represents 2 mm; *P < .01 versus MigR1; SD; n = 6). (B) Cells from panel A were subjected to Western blotting analysis using an mAb directed against myc. Overexposed blots revealed CD44ICD in CD44FL- and CD44ΔE-transduced cells, but not in the presence of DAPT.
CD44ICD rescues the fusion of macrophages treated with DAPT. (A) Rat alveolar macrophages were transduced with MigR1 empty or encoding CD44FL, CD44ΔE, or CD44ICD, and treated or not with DAPT (10 μM) for 4 days (bar represents 2 mm; *P < .01 versus MigR1; SD; n = 6). (B) Cells from panel A were subjected to Western blotting analysis using an mAb directed against myc. Overexposed blots revealed CD44ICD in CD44FL- and CD44ΔE-transduced cells, but not in the presence of DAPT.
CD44ICD localizes to the nucleus. (A) Raw 264.7 cells were transduced with pBMN-eGFP or pBMN-CD44ΔE-eGFP and treated or not with JLK2 (20 μM) or DAPT (10 μM) for 16 hours. Note that in DAPT-treated cells that express CD44ΔE-eGFP, not JLK2-treated cells, the nucleus does not fluoresce. Also, DAPT affects the shape of macrophages and induces the clustering of CD44ΔE-eGFP in the plasma membrane (bar is 7 μm). (B) Rat alveolar macrophages were transduced with the retroviral vector MigR1 encoding CD44ΔE and then subjected to subcellular fractionation. Fractions were analyzed by Western blotting using antibodies directed against myc, MFR (plasma membrane), p38 (cytosol), and Sm (nucleus).
CD44ICD localizes to the nucleus. (A) Raw 264.7 cells were transduced with pBMN-eGFP or pBMN-CD44ΔE-eGFP and treated or not with JLK2 (20 μM) or DAPT (10 μM) for 16 hours. Note that in DAPT-treated cells that express CD44ΔE-eGFP, not JLK2-treated cells, the nucleus does not fluoresce. Also, DAPT affects the shape of macrophages and induces the clustering of CD44ΔE-eGFP in the plasma membrane (bar is 7 μm). (B) Rat alveolar macrophages were transduced with the retroviral vector MigR1 encoding CD44ΔE and then subjected to subcellular fractionation. Fractions were analyzed by Western blotting using antibodies directed against myc, MFR (plasma membrane), p38 (cytosol), and Sm (nucleus).
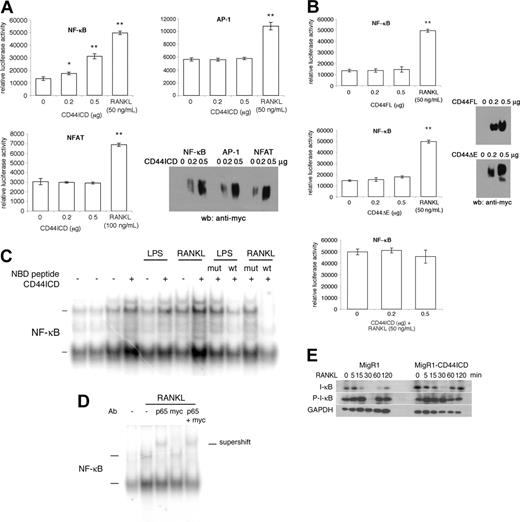
Because CD44ICD has been shown by Okamoto et al16 to potentiate transcriptional activation in COS-7 cells, we asked whether CD44ICD alters gene expression in macrophages. Because NF-κB is essential for osteoclastogenesis,17,18 we cotransfected mouse macrophages Raw 264.7 cells with CD44ICD and a luciferase reporter gene driven by an NF-κB promoter motif. We also cotransfected Raw 264.7 cells with luciferase reporter genes driven by AP-1 and NFAT promoter motifs. This is because c-Fos is a component of the AP-1 complex that is also required for osteoclastogenesis,33 and NFAT induces osteoclastogenesis.34,35 Indeed, NFAT1c rescues fusion of c-Fos deficient macrophages.34,35 Transfected cells were lysed and assayed for luciferase activity. We used receptor activator of NF-κB (RANKL) as a positive control for NF-κB activation. To confirm CD44ICD expression, we subjected cotransfected macrophages from parallel wells to Western blotting analysis using an anti-myc antibody. Under these conditions, CD44ICD dose dependently increased NF-κB transcriptional activity (Figure 6A). In contrast, AP-1 and NFAT-driven luciferase activity remained unchanged. This was despite the presence of CD44ICD (Figure 6A). Of interest, neither CD44FL nor CD44ΔE activated NF-κB transcriptional activity, possibly because the abundance of CD44ICD that is required to activate NF-κB was not reached during the transfection time (Figure 6B).
To initiate an investigation on the mechanism by which CD44ICD promotes activation of NF-κB, we asked whether CD44ICD altered the association of the NF-κB protein complex with DNA. We subjected nuclear extracts isolated from Raw cells transduced with MigR1 empty or encoding CD44ICD, and stimulated or not for 30 minutes with RANKL, which promotes the fusion of macrophages and osteoclastogenesis, or with LPS, which is a potent activator of macrophages and osteoclasts, to electrophoretic mobility shift assay. We used radiolabeled oligonucleotides that encode an NF-κB consensus sequence. In the absence of stimulation, nuclear protein complexes retarded modestly the gel migration of radiolabeled probes, independent of CD44ICD expression (Figure 6B). Of note, 2 sets of nuclear protein complexes from macrophages retarded the mobility of radiolabeled probes, as reported by Bouchon et al.36 As expected, both RANKL and LPS augmented the abundance of gel-retarded radiolabeled probes. Of interest, nuclear proteins extracted from cells transduced with CD44ICD augmented further the intensity of the signal from the gel-retarded radiolabeled probes. This suggested that CD44ICD promotes the binding of NF-κB protein complex to DNA. To ensure that the proteins bound to NF-κB DNA probes belonged to the NF-κB family of proteins, we preincubated RANKL- and LPS-stimulated cells that had been transduced with CD44ICD, with NEMO-binding peptides (NBD).22 NBD prevents the translocation of NF-κB to the nucleus, hence, the gel-retardation of NF-κB oligonucleotides. As a negative control, we used the mutant NBD peptide that poorly prevents the translocation of NF-κBtothe nucleus. As shown in Figure 6C, NBD wild type (wt) reduced the abundance of gel-retarded radiolabeled probes. The mutant peptide poorly reduced the abundance of bound radiolabeled probe. These data confirmed that the protein complex bound to an NF-κB reporter oligonucleotide is NF-κB.
CD44ICD activates NF-κB. (A) Raw 264.7 cells were cotransfected with pcDNA3-CD44ICD and NF-κB, AP-1 or NFAT luciferase reporter constructs, grown for 2 days, and then either assayed for luciferase activity (*P < .05 and **P < .001 versus 0 μg CD44ICD; SD; n = 3) or subjected to Western blot analysis using anti-myc antibody. RANKL was used as a positive control. (B) Raw 264.7 cells were cotransfected with pcDNA3-CD44FL or pcDNA3-CD44ΔE and NF-κB luciferase reporter construct, grown for 2 days, and either assayed for luciferase activity (**P < .001 versus 0 μg CD44FL or CD44ΔE; SD; n = 3) or subjected to Western blot analysis using anti-myc antibody. RANKL was used as a positive control. (C) Raw 264.7 cells were transduced or not with MgR1-CD44ICD and stimulated or not for 30 minutes with LPS (100 μg/mL) or RANKL (100 ng/mL). Nuclear proteins were extracted and incubated for 20 minutes with radiolabeled NF-κB consensus oligonucleotides. Some cells were preincubated with NEMO-binding peptide (NBD; wt; 50 μM) or NBD mutant peptide (mut; 50 μM) for 1 hour at room temperature. The samples were resolved by electrophoresis. (D) Raw 264.7 cells were transduced with MgR1-CD44ICD and stimulated or not with RANKL (100 ng/mL) and then nuclear extracts were analyzed as in panel B, except that some nuclear extracts were preincubated with an antibody directed against p65, myc, or with both p65 and myc. (E) Raw 264.7 cells were transduced with MgR1 empty or encoding CD44ICD and stimulated with RANKL (100 ng/mL) for the indicated times. Total cell extracts were analyzed by Western blotting using antibodies directed against I-κB, phosphorylated I-κB (P-I-κB), and GAPDH.
CD44ICD activates NF-κB. (A) Raw 264.7 cells were cotransfected with pcDNA3-CD44ICD and NF-κB, AP-1 or NFAT luciferase reporter constructs, grown for 2 days, and then either assayed for luciferase activity (*P < .05 and **P < .001 versus 0 μg CD44ICD; SD; n = 3) or subjected to Western blot analysis using anti-myc antibody. RANKL was used as a positive control. (B) Raw 264.7 cells were cotransfected with pcDNA3-CD44FL or pcDNA3-CD44ΔE and NF-κB luciferase reporter construct, grown for 2 days, and either assayed for luciferase activity (**P < .001 versus 0 μg CD44FL or CD44ΔE; SD; n = 3) or subjected to Western blot analysis using anti-myc antibody. RANKL was used as a positive control. (C) Raw 264.7 cells were transduced or not with MgR1-CD44ICD and stimulated or not for 30 minutes with LPS (100 μg/mL) or RANKL (100 ng/mL). Nuclear proteins were extracted and incubated for 20 minutes with radiolabeled NF-κB consensus oligonucleotides. Some cells were preincubated with NEMO-binding peptide (NBD; wt; 50 μM) or NBD mutant peptide (mut; 50 μM) for 1 hour at room temperature. The samples were resolved by electrophoresis. (D) Raw 264.7 cells were transduced with MgR1-CD44ICD and stimulated or not with RANKL (100 ng/mL) and then nuclear extracts were analyzed as in panel B, except that some nuclear extracts were preincubated with an antibody directed against p65, myc, or with both p65 and myc. (E) Raw 264.7 cells were transduced with MgR1 empty or encoding CD44ICD and stimulated with RANKL (100 ng/mL) for the indicated times. Total cell extracts were analyzed by Western blotting using antibodies directed against I-κB, phosphorylated I-κB (P-I-κB), and GAPDH.
Although CD44ICD did not induce a supershift but rather an enhanced shift, suggesting that it does not associate with NF-κB protein complex, we set out to eliminate that possibility. We preincubated nuclear extracts from cells transduced with CD44ICD and stimulated with RANKL, with antibodies directed against the p65 subunit of NF-κB, against myc, and against both p65 and myc. As shown in Figure 6D, the presence of anti-p65 antibody induced a supershift of the protein-DNA complex that was neither modified nor induced by the anti-myc antibody. These data confirmed that CD44ICD does not associate with NF-κB.
To confirm that CD44ICD promotes NF-κB activation, we took advantage of the fact that NF-κB itself stimulates the expression of I-κB. We transduced Raw 264.7 cells with MigR1 alone or encoding CD44ICD. We then treated the transduced cells with RANKL for increasing times and subjected them to Western blotting analysis using antibodies directed against I-κB or its phosphorylated, hence, activated form (P-I-κB). Under these conditions, CD44ICD was associated with an increase in I-κB expression, which became phosphorylated (Figure 6E). These data confirmed that CD44ICD promotes NF-κB activation.
Together, our results strongly suggest that the cleavage of CD44 by PS is a regulated event that promotes NF-κB-dependent macrophage fusion leading to multinucleation.
Because NF-κB is essential for the differentiation of multinucleated osteoclasts,17,18 we asked whether NF-κB is required for the fusion of tissue macrophages. We treated alveolar macrophages cultured in fusogenic conditions with the NF-κB inhibitors Helenalin, CAPE, Bay-11-7082, and the histone deacetylase inhibitor HD. We also used NBD peptide, which blocks osteoclastogenesis.22 Even at concentrations lower than those reported in the literature as effective (0.125 μM for Helenalin; 1 μg/mL for CAPE; 0.2 μM for HD; 3.125 μM for Bay-11-7082), these inhibitors were detrimental to the macrophages, which underwent apoptosis after 1 day of treatment (data not shown). The solvents DMSO and ethanol, in which they were diluted, had no effect. The NBD peptide did not have an effect, even when used at 50 μM and added daily, perhaps as a result of its short half-life in vitro (data not shown). We therefore used the less specific NF-κB inhibitors aspirin and curcumin.37-39 Indeed, Bharti et al recently reported that curcumin, an inhibitor of NF-κB,40 suppresses osteoclastogenesis. Both compounds prevented the fusion of alveolar macrophages in a dose-dependent manner (Figure 7).
Inhibition of NF-κB prevents fusion of macrophages. Rat alveolar macrophages (5 × 106 cells/well in 24-well dishes) were cultured for 3 days in the absence or presence of increasing concentrations of aspirin or curcumin. Note that 10 μM curcumin induced macrophages to detach from the plate (Bar represents 2 mm; *P < .01 and **P < .001 versus control; SD; n = 5).
Inhibition of NF-κB prevents fusion of macrophages. Rat alveolar macrophages (5 × 106 cells/well in 24-well dishes) were cultured for 3 days in the absence or presence of increasing concentrations of aspirin or curcumin. Note that 10 μM curcumin induced macrophages to detach from the plate (Bar represents 2 mm; *P < .01 and **P < .001 versus control; SD; n = 5).
Discussion
We previously reported that rat alveolar macrophages express the standard hemopoietic form of CD44, the surface expression of which is highly but transiently induced at the onset of fusion. We then proposed that CD44 plays a role in the fusion of tissue macrophages.11 We now report that the intracellular domain of CD44 is cleaved by PS and translocates to the nucleus to promote activation of NF-κB and fusion of macrophages.
The observation that inhibitors of PS prevent the fusion of macrophages might explain in part the skeletal phenotype of mice that lack PS1,6 which suggests that PS1, rather than PS2, is required for osteoclastogenesis. Yet, the expression of PS2 is, like that of CD44, induced in alveolar macrophages 24 hours after plating, which suggests that PS2 and CD44 expression is under the control of similar transcriptional mechanisms during fusion.
The dramatic effect of the PS inhibitor DAPT on the shape of macrophages might contribute to the inhibitory effect of DAPT on fusion. It is known that the cytoplasmic tail of CD44 recruits phosphorylated ezrin-radoxin-moesin (ERM) proteins that are linked to filamentous (F) actin and therefore to the actin cytoskeleton (for a review, see Ponta et al41 ). Hence, DAPT and DupE might prevent the fusion of macrophages via 2 complementary mechanisms: one is by preventing the release of CD44ICD, hence CD44ICD-mediated activation of NF-κB, and the other is by favoring the retention of ERM proteins at the plasma membrane; both mechanisms involve CD44ICD.
Bone marrow macrophages and Raw 264.7 cells require stimulation with RANKL to differentiate into osteoclasts. In contrast, alveolar and peritoneal macrophages, which are tissue macrophages, fuse spontaneously in vitro in the sole presence of serum.1,2 Hence, CD44ICD might be sufficient to promote fusion and multinucleation of tissue macrophages. Yet, CD44ICD is not sufficient to induce the fusion of nonfusing cells such as fibroblasts, which remained mononucleated. This implies that activation of NF-κB is essential to promote the fusion of macrophages but insufficient to induce fusion of nonfusogenic cells.
Although our results reveal an accrued amount of nuclear protein binding to NF-κB DNA consensus sequence when CD44ICD expressed exogenously in cells is stimulated with LPS or RANKL, the molecular mechanism by which CD44ICD promotes NF-κB activation remains speculative at this time. One possibility is that CD44ICD potentiates the transcriptional activation of p300/CBP,16 which is known to acetylate RELA. Acetylated RELA has higher DNA-binding affinity for the κB enhancer and does not associate with I-κB (for a review, see Chen and Greene42 ). If this is the case, then CD44ICD promotes NF-κB activation via p300/CBP. However, no direct interaction between CD44ICD and p300/CBP has been identified.16 Yet, the fact that both DAPT and DupE decrease the expression of CD44 and that CD44ICD promotes that of I-κB concur to support a role for CD44ICD in NF-κB-mediated macrophage fusion.
The question as to what mechanism CD44ICD uses to translocate to the nucleus remains open. One possibility is that CD44ICD associates with an acetylase in the cytoplasm, which translocates to the nucleus to then promote NF-κB activation.43-46 Another, though unlikely, possibility is that CD44ICD associates with NF-κB to move to the nucleus, then dissociates from it to associate with an acetylase. But this is all very speculative at this point.
The question as to whether CD44 is required for the fusion of preosteoclasts in vivo is in part answered by 2 recent studies47,48 in which bones from mice that lack CD44 were analyzed by micro-computed tomography. Cao et al47 showed that 7-week-old male mice that lack CD44 have a thicker cortical bone with a smaller medullary area than wild-type male mice. However, the relative osteoclast surface was not affected by the absence of CD44 in these mice. Conversely, Hayer et al48 found no difference between wild-type and CD44-deficient 10-week-old mice, although they did not analyze males and females separately. de Vries et al49 found the same results when they analyzed 7-day-old mice and found no difference in osteoclast number between wild-type and CD44-deficient mice. We, on the other hand, used peripheral quantitative computed tomography (pQCT) and histomorphometry to analyze tibiae from 8-month-old CD44-deficient and wild-type male and female mice and found that male mice have a decreased bone density, whereas density is increased in female when compared to wild-type mice (data not shown). These somewhat contradictory results, which used CD44-deficient mice generated by 3 different laboratories, tend to suggest that the deletion of CD44 is associated with a mild bone phenotype, which could be influenced by the environment. Hence, CD44ICD appears to play a relative minor role in normal bone remodeling. However, the role of CD44 in macrophage fusion in response to bone injury, either mechanical or hormonal such as in estrogen deficiency, or to foreign body, remains to be investigated. Phenotypes often are not revealed in animals when studied under normal physiologic conditions.
Our finding that CD44ICD activates NF-κB might help us understand, in part, the link between inflammation and cancer, which had been suspected for a long time, but was only recently demonstrated by Greten and colleagues50 in a mouse model of colitis-associated cancer. Although soluble CD44 is a well-established indicator of cancer prognosis,51,52 the functional link between CD44 and cancer had not been established. By promoting the activation of NF-κB, CD44 might function as a cornerstone between inflammation and cancer. In addition, NF-κB might promote metastasis indirectly by facilitating the fusion of macrophages with tumor cells.2,53,54 This speculative and old concept remains to be validated. Conversely, NF-κB might promote the repair of tissues by facilitating the fusion of macrophages with somatic cells. This is a new and highly debated issue that has recently emerged in the field of stem cells.2,55
CD44ICD emerges as a novel promoter of NF-κB activation and PS as the regulator of a new intracellular signaling pathway.56 By translocating directly from the plasma membrane to the nucleus, CD44ICD bypasses intracellular signaling pathways that alter gene transcription. This is an exciting new avenue of investigation whereby one plasma membrane protein becomes 4, that is, the intact full-length protein itself and its extracellular, intracellular, and transmembrane domains. In this respect, it will be important to define the role, if any, of the transmembrane domain of CD44.
Cell-to-cell fusion and, in particular, macrophage fusion could become a therapeutic tool for the delivery of genes or drugs in a cell-specific targeted manner. The therapeutic applications of such a strategy encompass cancer, infectious diseases, and genetic disorders. Exploiting CD44ICD in the fusogenicity and the natural motility of macrophages might help deliver drugs and genes in a cell-targeted manner.
Prepublished online as Blood First Edition Paper, September 29, 2005; DOI 10.1182/blood-2005-05-1902.
Supported by the National Institutes of Health (DE12110; A.V.). J.Z.K. was the recipient of a Boehringer Ingelheim Pharmaceutical Fellowship.
W.C. is lead author and generated 90% of the data presented in this manuscript. J.Z.K. subjected fusing alveolar macrophages to oligonucleotide microarray and identified presenilin as a transcript transiently expressed during macrophage fusion; she bred CD44-deficient and wild-type mice and isolated their bones for pQCT analysis. Her contribution to this work is essential. Q.Z. processed the bones from CD44-deficient and wild-type mice and subjected the sections to histomorphometry; although her data are mentioned as “data not shown,” her contribution to this work is essential. H.-Z.K. subjected the bones from CD44-deficient and wild-type mice to pQCT analysis; although the data he generated are mentioned as “data not shown” in “Discussion,” his contribution to this work is essential. C.C. is an expert in confocal microscopy and analyzed the subcellular localization of CD44ΔE-GFP. A.V. is the principal investigator of the laboratory, directed the team working on this project, and wrote the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Drs Jun Li and Randall Barton for their help with microarray analyses and Drs Frédéric Checler, Gerald Crabtree, Bryant Darnay, Shankar Ghosh, Todd Golde, Betty Lamothe, Ira Mellamn, Warren Pear, Joan Steitz, and Jianjiang Ye for generous gifts of reagents. We are grateful to Dr Ira Mellman for providing us with access to his confocal imaging facility. We are grateful to Drs Hongmei Li and Hongji Li, and to Halldor Einarsson for their technical help. We are indebted to Drs Michael Caplan and Anton Bennett for providing helpful suggestions with the manuscript.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal