Abstract
Interleukin-12 (IL-12) is a heterodimeric cytokine produced by dendritic cells (DCs) in response to Toll-like receptor (TLR) ligation. While the mechanisms regulating IL-12p40 chain gene expression are well characterized, molecular events involved in IL-12p35 chain gene activation remain to be clarified. Since IL-12p35 mRNA was induced in human DCs activated through TLR3 or TLR4 but not TLR2, we investigated the potential role of interferon regulatory factor 3 (IRF-3) in IL-12p35 gene transactivation. First, a binding site for IRF-3 named interferon-stimulated response element-1 (ISRE-1) was identified in the human IL-12p35 promoter region between nucleotides -251 and -240. The ISRE-1 site was required for IL-12p35 gene activation in RAW 264.7 cells stimulated by lipopolysaccharide (LPS) or PolyI:C. Ectopic expression of IRF-3 was found to up-regulate IL-12p35 gene activation in the same system. Furthermore, chromatin immunoprecipitation (ChIP) studies demonstrated that IRF-3 is recruited to ISRE-1 site in TLR4- or TLR3-stimulated human DCs. Finally, experiments on DCs from IRF-3-deficient mice established that TLR4-induced IL-12p35 mRNA and IL-12p70 synthesis are impaired in absence of IRF-3. We conclude that IRF-3 binds to a critical cis-acting element in the IL-12p35 gene promoter and thereby represents a key factor for the induction of IL-12p70 synthesis in DCs.
Introduction
Interleukin-12 (IL-12) is a heterodimeric cytokine playing an important role in the induction of T-helper 1 (Th1) and cytotoxic T-cell responses.1 In cancer, exogenously administered as well as endogenously produced IL-12 was shown to exert antitumor and antimetastatic effects.2 For example, IL-12 was shown to provide resistance against experimentally induced T-cell lymphoma.3 Dendritic cells (DCs) represent a major source of IL-12 upon ligation of Toll-like receptors (TLRs) by microbial products.1,4 TLR ligands such as monophosphoryl lipid A (MPL; a TLR4 ligand) are indeed used as adjuvants in recently developed anticancer vaccines such as those preventing uterine cervix epithelioma.5 Initial studies on IL-12 synthesis regulation focused on IL-12p40 chain gene expression but there is accumulating evidence that IL-12p35 gene transcription is also tightly regulated. Recently, IL-12p35 gene transcription upon TLR engagement was found to depend on nuclear translocation of p65 and c-Rel nuclear factor κB (NF-κB) members as well as on recruitment of Sp1, the latter depending on remodeling of a strategically positioned nucleosome.6-8 Furthermore, IL-12p35 gene expression was shown to be enhanced by IFN-γ via the recruitment of interferon regulatory factor 1 (IRF-1) and ICSBP.7,9
TLRs elicit distinct signaling cascades depending on the recruitment of various TIR (Toll/IL-1 receptor)-containing adaptors.10 The myeloid differentiation factor 88 (MyD88) coupled to most TLRs drives activation of NF-κB, mitogen-activated protein kinases, and IRF-5.11,12 On the other hand, recruitment of TIR domain-containing adaptor inducing IFN-β (TRIF) associated with TLR3 and TLR4 results in activation and nuclear translocation of IRF-3, a critical transcription factor for IFN-β gene expression.13,14 Herein, we provide evidence that beside its role in the induction of IFN-β synthesis, IRF-3 also binds to a critical cis-element in the IL-12p35 promoter and thereby represents a key transcription factor for the synthesis of bioactive IL-12 upon TLR4 or TLR3 ligation.
Materials and methods
Mice
IRF-3-deficient mice14 were obtained from the Riken BioResource Center (Ibaraki, Japan) with the approval of T. Taniguchi. Mice were bred and maintained in specific pathogen-free conditions according to institutional guidelines.
Cells and reagents
Human monocyte-derived DCs were generated from peripheral blood mononuclear cells (PBMCs) in the presence of recombinant human GM-CSF (hGM-CSF) and hIL-4, as previously described.15 DCs were generated from mouse bone marrow in the presence of recombinant murine GM-CSF (mGM-CSF) according to a protocol described by Lutz et al16 with slight modifications.17 The DC population obtained with this protocol routinely contained greater than 95% of CD11c+ DCs as assessed by flow cytometry analysis. The RAW 264.7 murine macrophage cell line (LGC Promochem, Teddington, United Kingdom) was maintained in DMEM medium supplemented with 5% FCS, nonessential amino acids, 2 mM glutamine, and penicillin/streptomycin. Ultra-Pure lipopolysaccharide (LPS) from Escherichia coli (0111:B4) and peptidoglycan (PGN) were obtained from Cayla (Toulouse, France). Polyinosine-polycytidylic acid (PolyI:C) and Pam3CSK4 (S-[2.3-bis(palmitoyloxy)-(2-RS)-propyl]-N-palmitoyl-(R)-Cys-(S)-Ser-Lys4-OH) trihydrochloride were purchased from GE Healthcare (Gent, Belgium) and EMC Microcollections (Tübingen, Germany), respectively. Monophosphoryl lipid A (MPL) was kindly provided by GlaxoSmithKline Biologicals (Rixensart, Belgium). Cycloheximide (CHX) and recombinant mIFN-γ (rmIFN-γ) were obtained from Sigma-Aldrich (Bornem, Belgium) and R&D Systems (Abingdon, United Kingdom), respectively.
Plasmid constructs
The luciferase reporter p35-lucWT plasmid was previously described.8 The plasmids p35lucMut A and Mut B are derivatives of p35-lucWT in which the interferon-stimulated response element 1 (ISRE-1) site was altered by the QuickChange Site-directed Mutagenesis Method (Stratagene, La Jolla, CA). IRF-1wt (IRF-1wild type), IRF-3wt, and IRF-3(5D) expression vectors were obtained from R. Lin (McGill University, Montreal, QC, Canada).
Quantification of cytokine production
Murine IL-12p70 levels were determined in cell-free supernatants by specific enzyme-linked immunosorbent assay (ELISA; Duoset; R&D Systems) with detection limits of 15 pg/mL.
RNA purification and real-time reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted using a MagnaPure LC RNA-High Performance Isolation Kit (Roche Diagnostics, Brussels, Belgium). Reverse transcription and real-time PCR reactions were then carried out using LightCycler-RNA Master Hybridization Probes (one-step procedure) on a Lightcycler apparatus (Roche Diagnostics). Primer sequences are described in Table 1.
Oligonucleotide sequences used for PCR
. | Oligonucleotides, 5′-3′ . |
|---|---|
| RT-PCR | |
| Human IL-12p35 | |
| Forward | GCCACAGGTCTGCATCCA |
| Reverse | GACCTGGCGGGCTGAGTA |
| Probe | 6FAM-TGGACACATGCTGAGCCGGC-TAMRA |
| Human IL-12p40 | |
| Forward | AGCCTCCTCCTTGTGGCTA |
| Reverse | TGTGCTGGTTTTATCTTTTGTG |
| Probe | 6FAM-CCCTGGCACCCAGCACAATG-TAMRA |
| Human IF144 | |
| Forward | TCTGTTTTCCAAGGGCATGT |
| Reverse | GCCATCTTTCCCGTCTCTAA |
| Probe | 6FAM-AGTGCCCACCAAAGCCTGATGC-BHQ1 |
| Human β-actin | |
| Forward | AGCCTCCTCCTTGTGGCTA |
| Reverse | TGTGCTGGTTTTATCTTTTGTG |
| Probe | 6FAM-CCCTGGCACCCAGCACAATG-TAMRA |
| Murine IL-12p35 | |
| Forward | CTTAGCCAGTCCCGAAACCT |
| Reverse | TTGGTCCCGTGTGATGTCT |
| Probe | 6FAM-TCTGGCCGTCTTCACCATGTCA-BHQ1 |
| Murine IL12p40 | |
| Forward | GTTCAACATCAAGAGCAGTAGCA |
| Reverse | CTGCAGACAGAGACGCCATT |
| Probe | 6FAM-CCCTGACTCTCGGGCAGTGACA-TAMRA |
| Murine β-actin | |
| Forward | TCCTGAGCGCAAGTACTCTGT |
| Reverse | CTGATCCACATCTGCTGGAAG |
| Probe | BHQ2-ATCGGTGGCTCCATCCTGGC-Pulsar650 |
| ChIP | |
| Human IL-12p35 promoter | |
| Forward | GCGAACATTTCGCTTTCATT |
| Reverse | ACTTTCCCGGGACTCTGGT |
| Human IL-12p40 promoter | |
| Forward | GCATACAGTTGTTCCATCC |
| Reverse | CTCTACTCCTTTCTGATGGA |
| Human GAPDH promoter | |
| Forward | TACTAGCGGTTTTACGGGCG |
| Reverse | TCGAACAGGAGGAGCAGAGAGCGA |
. | Oligonucleotides, 5′-3′ . |
|---|---|
| RT-PCR | |
| Human IL-12p35 | |
| Forward | GCCACAGGTCTGCATCCA |
| Reverse | GACCTGGCGGGCTGAGTA |
| Probe | 6FAM-TGGACACATGCTGAGCCGGC-TAMRA |
| Human IL-12p40 | |
| Forward | AGCCTCCTCCTTGTGGCTA |
| Reverse | TGTGCTGGTTTTATCTTTTGTG |
| Probe | 6FAM-CCCTGGCACCCAGCACAATG-TAMRA |
| Human IF144 | |
| Forward | TCTGTTTTCCAAGGGCATGT |
| Reverse | GCCATCTTTCCCGTCTCTAA |
| Probe | 6FAM-AGTGCCCACCAAAGCCTGATGC-BHQ1 |
| Human β-actin | |
| Forward | AGCCTCCTCCTTGTGGCTA |
| Reverse | TGTGCTGGTTTTATCTTTTGTG |
| Probe | 6FAM-CCCTGGCACCCAGCACAATG-TAMRA |
| Murine IL-12p35 | |
| Forward | CTTAGCCAGTCCCGAAACCT |
| Reverse | TTGGTCCCGTGTGATGTCT |
| Probe | 6FAM-TCTGGCCGTCTTCACCATGTCA-BHQ1 |
| Murine IL12p40 | |
| Forward | GTTCAACATCAAGAGCAGTAGCA |
| Reverse | CTGCAGACAGAGACGCCATT |
| Probe | 6FAM-CCCTGACTCTCGGGCAGTGACA-TAMRA |
| Murine β-actin | |
| Forward | TCCTGAGCGCAAGTACTCTGT |
| Reverse | CTGATCCACATCTGCTGGAAG |
| Probe | BHQ2-ATCGGTGGCTCCATCCTGGC-Pulsar650 |
| ChIP | |
| Human IL-12p35 promoter | |
| Forward | GCGAACATTTCGCTTTCATT |
| Reverse | ACTTTCCCGGGACTCTGGT |
| Human IL-12p40 promoter | |
| Forward | GCATACAGTTGTTCCATCC |
| Reverse | CTCTACTCCTTTCTGATGGA |
| Human GAPDH promoter | |
| Forward | TACTAGCGGTTTTACGGGCG |
| Reverse | TCGAACAGGAGGAGCAGAGAGCGA |
EMSAs
ISREwt probe was generated by end labeling the double-stranded oligonucleotide 5′-ACTGCGAACATTTCGCTTTCATTTTGGGCCG-3′ (corresponding to nucleotide [nt] -259 to -229) and electrophoretic mobility shift assays (EMSAs) were performed as previously described.8 For competition analysis, we used increasing concentrations (12.5- to 50-fold molar excess) of unlabeled ISREwt, ISREMut A, ISREMut B, and ISRE from ISG15 (5′-GATCCTCGGGAAAGGGAAACCGAAACTGAAGCC-3′); mutated ISRE from ISG15 (ISG15Mut; 5′-GATCCTCGGGAAAGGGAGGCCGAGGCTGAAGCC-3′); and NF-κB consensus (5′-AGTTGAGGGACTTTCCCAGGC-3′). IRF-1 Cons (5′-GGAAGCGAAAATGAAATTGACT-3′) and IRF-1MUT (5′-GGAAGCGAGGATGAGGTTGACT-3′) were obtained from Tebu-bio Boechout, Belgium).
Transient transfection and luciferase assays
RAW 264.7 cells were transfected using FuGENE-6 (Roche Diagnostics) and stimulated with LPS and IFN-γ as previously described.8 When indicated, cells were stimulated for 20 hours with PolyI:C (20 μg/mL). Forty-eight hours after transfection, cells were harvested and promoter activities were analyzed using the Dual Luciferase Reporter Assay system (Promega, Leiden, The Netherlands). Promoter activities were then normalized to Renilla luciferase activities.
Immunofluorescence microscopy
DCs were seeded onto glass coverslips (2 × 105 cells/100 μL), allowed to adhere, then cultured in the absence or presence of LPS (1 μg/mL) or PolyI:C (10 μg/mL). Cells were then fixed with 2% paraformaldehyde and stored in methanol (-20°C). For the staining, cells were washed in PBS, permeabilized, and blocked with 2% bovine serum albumin (BSA) containing 0.1% Triton X-100. Cells were next incubated with primary mouse monoclonal anti-IRF-3 antibody (clone SL-12, 5 μg/mL, 60 minutes; BD Biosciences, Erembodegem, Belgium) and then with fluorescently labeled secondary antibody (anti-mouse Alexa 488, 2 μg/mL, 60 minutes; Invitrogen, Merelbeke, Belgium). Nuclei were counterstained with Toto-3 (Invitrogen). The coverslips were then washed and mounted with Vectashield antifade (Labconsult, Brussels, Belgium) before being analyzed by confocal microscopy. Images were captured using a Leica TCS SP2 AOBS confocal laser scanning microscope based on the Leica DM IRE2 inverted microscope and equipped with a 100×/1.40 APO oil-immersion objective lens (Leica, Heidelberg, Germany). Images were processed using PowerPoint software (Microsoft, Redmond, WA).
Detection of IRF-3 DNA-binding activity
IRF-3 binding activity in nuclear extracts was measured with Trans-am IRF transcription factor assay kit (Active Motif Europe, Rixensart, Belgium) according to the manufacturer's protocol. Nuclear extracts (5 μg) were incubated with plate-coated ISRE consensus oligonucleotide. Plates were washed and anti-IRF-3 antibody was added to the wells. Antibody binding was detected with a secondary HRP-conjugated antibody and developed with TMB substrate. The intensity of the reaction was measured at 450 nm.
ChIP assay
The chromatin immunoprecipitation (ChIP) experiments were performed using the ChIP-IT kit from Active Motif according to manufacturer's instructions. For each experimental condition, 107 human DCs were cross-linked by 1% formaldehyde for 5 minutes. Isolated nuclei were then subjected to sonication (15 × 10 s, 10% maximum power on a Branson sonicator [VWR International, Leuven, Belgium]). DNA fragments obtained ranged between 200 and 500 bp. Chromatin fractions were precleared with protein G beads followed by immunoprecipitation with 4 μg rabbit polyclonal IRF-3 antibody (Active Motif) or control. Cross-linking was reversed and purified DNA was subjected to PCR amplification (32 cycles) using Taq polymerase from Roche Diagnostics. The samples were analyzed by electrophoresis on a 3% agarose gel stained with SybrGold (Invitrogen). Primer sequences are detailed in Table 1.
Results
Differential induction of IL-12p35 by TLR ligands in human monocyte-derived DCs
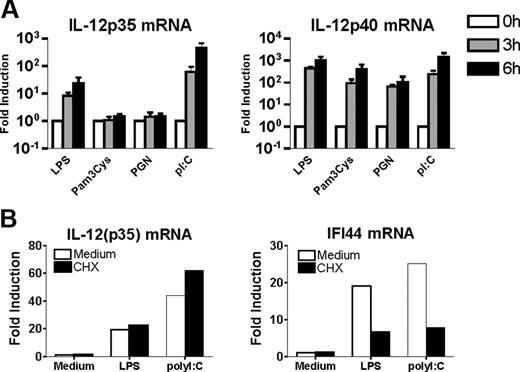
In a first series of experiments, we compared the ability of distinct TLR agonists to induce IL-12p40 and IL-12p35 gene expression in human monocyte-derived DCs. In agreement with previous studies,18,19 we found that TLR2 engagement by Pam3CSK4 or PGN readily triggered IL12p40 but not IL12p35 gene transcription (Figure 1A). In contrast, ligation of TLR3 by PolyI:C or TLR4 by LPS induced both IL-12p40 and IL-12p35 mRNA accumulation as expected (Figure 1A). Since the MyD88-independent pathway coupled to TLR4 and TLR3 leads to synthesis of IFN-β, which was recently shown to enhance IL-12p35 chain synthesis in an autocrine manner,19 we determined the impact of de novo protein synthesis inhibition on the induction of IL-12p35 mRNA accumulation by LPS or Poly I:C. As shown in Figure 1B, transcriptional activation of the IL-12p35 gene was not altered by the addition of CHX at a dose that totally inhibited IFN-β protein synthesis. As control, we verified that CHX suppressed the induction of IFI44 gene (interferon-induced protein 44), which belongs to the set of genes dependent on type 1 IFN synthesis (Figure 1B). Taken together, these data are compatible with the hypothesis that IL-12p35 gene is a primary response gene downstream of the MyD88-independent pathway shared by TLR3 and TLR4. Because of the critical role of IRF-3 in this signaling cascade,20 further experiments were conducted to investigate the possible involvement of IRF-3 in the transcriptional activation of the human IL-12p35 gene.
IL-12p35 is a TLR3/TLR4 primary response gene. Monocyte-derived DCs were either incubated in medium alone or stimulated with LPS (1 μg/mL), PGN (10 μg/mL), Pam3CSK4 (10 μg/mL), or PolyI:C (10 μg/mL). (A) Total RNA was extracted after 3 and 6 hours and analyzed by real-time RT-PCR. Levels were normalized using β-actin mRNA as reference and compared with unstimulated conditions. Data are shown as mean ± SEM of 6 independent experiments on different donors. (B) DCs were pretreated with CHX (10 μg/mL) for 1 hour before LPS or PolyI:C addition for 3 hours. Total RNA was then extracted and analyzed by real-time RT-PCR. One representative donor of 5 is shown.
IL-12p35 is a TLR3/TLR4 primary response gene. Monocyte-derived DCs were either incubated in medium alone or stimulated with LPS (1 μg/mL), PGN (10 μg/mL), Pam3CSK4 (10 μg/mL), or PolyI:C (10 μg/mL). (A) Total RNA was extracted after 3 and 6 hours and analyzed by real-time RT-PCR. Levels were normalized using β-actin mRNA as reference and compared with unstimulated conditions. Data are shown as mean ± SEM of 6 independent experiments on different donors. (B) DCs were pretreated with CHX (10 μg/mL) for 1 hour before LPS or PolyI:C addition for 3 hours. Total RNA was then extracted and analyzed by real-time RT-PCR. One representative donor of 5 is shown.
Specific binding of IRF-3 to ISRE-1 site, a critical cis-acting element of the human IL-12p35 promoter
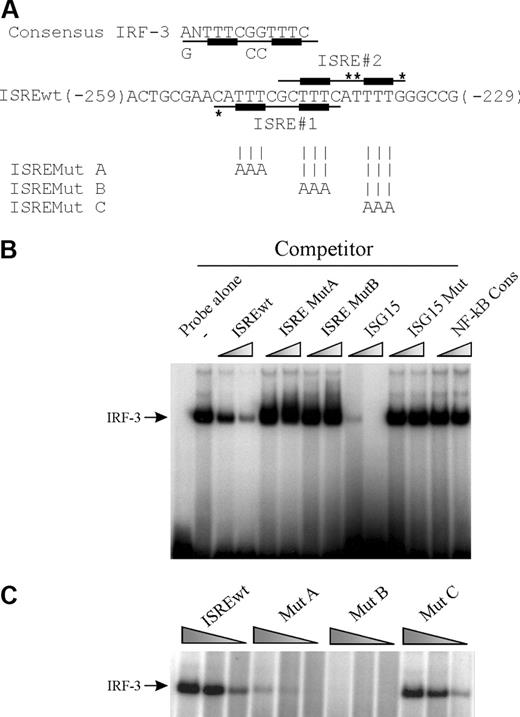
We analyzed the nucleotide sequence of the IL-12p35 promoter region for the presence of putative IRF-3 binding sites.21 Two overlapping potential ISRE sites (named ISRE-1 and -2) were found between nucleotides -251 and -234. ISRE-2 (-248/-235; Figure 2A) corresponds to the IRF-E site, known to bind IRF-1 in response to IFN-γ.7 We designed a double-stranded oligonucleotide (designated ISREwt) encompassing ISRE-1 (-251/-240) and ISRE-2 (-248/-235; Figure 2A). As shown in Figure 2B, N-terminal IRF-3 strongly bound the ISREwt probe. Competition experiments with an excess of different double-stranded oligonucleotides indicated that this binding was specific: it was inhibited in the presence of homologous unlabeled ISREwt or the ISRE element from the ISG15 promoter, whereas it was not affected by a heterologous NF-κB consensus competitor or by mutated versions of ISG15 or IL-12p35 ISRE oligonucleotides (Figure 2B). Moreover, as shown in Figure 2C, recombinant IRF-3 failed to bind or bound only weakly to oligonucleotide constructs in which mutations were introduced in the ISRE-1 site (ISRE Mut B and ISRE Mut A, respectively), whereas mutation in the ISRE-2 site (ISRE Mut C) did not affect IRF-3 binding.
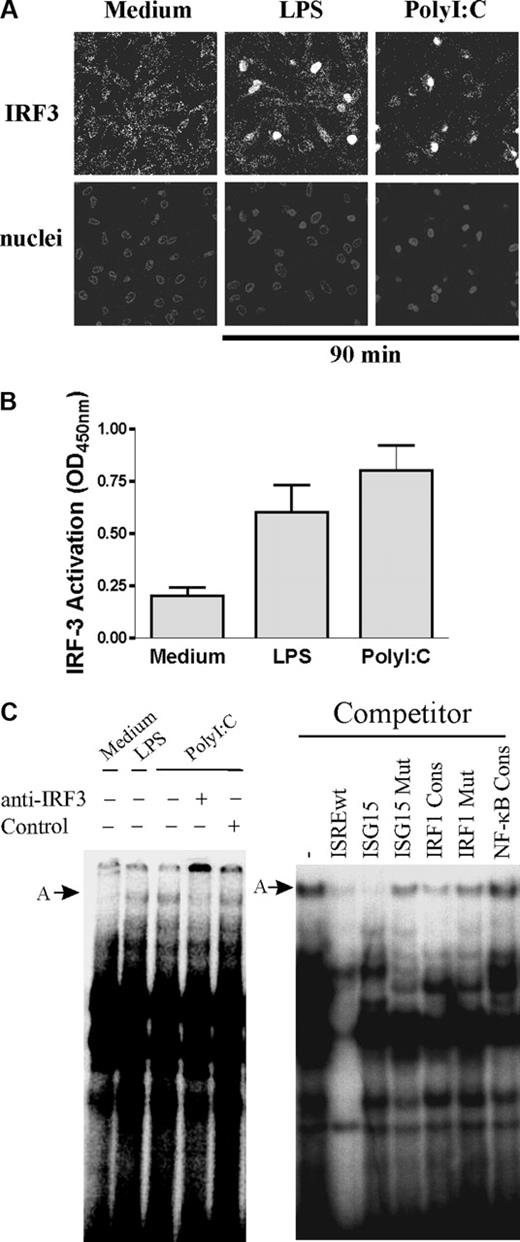
We then examined activation of IRF-3 in human DCs in response to LPS or PolyI:C. As shown in Figure 3A, we observed translocation of endogenous IRF-3 to the nuclei (assessed by immunofluorescence) in response to both TLR3 and TLR4 ligands. We quantified DNA-binding activity of IRF-3 using an ELISA-based assay (Figure 3B). IRF-3 activation upon LPS or PolyI:C was found to be comparable. ISREwt was then tested in EMSA experiments for DNA-protein interactions with nuclear extracts from human DCs (Figure 3C). Upon activation, IRF-3 associates with other transcription factors and coactivators such as CBP, thereby forming multicomponent complexes.22 A high-molecular-weight complex was observed in response to LPS and PolyI:C (Figure 3C left panel, complex A). Addition of anti-IRF-3 antibody interfered with the formation of this complex. Furthermore, complex A was found to be specific (Figure 3C right panel): its formation was inhibited in the presence of homologous unlabeled ISREwt or ISG15 ISRE, whereas it was not affected by a heterologous NF-κB consensus competitor or by a mutated version of ISG15 oligonucleotide. In the presence of an IRF-1 binding sequence (IRF-1 Cons), we observed only slight inhibition of complex A formation. Taken together, these experiments indicate that both purified and endogenous IRF-3 physically interact in vitro with the ISRE-1 site of the human IL-12p35 promoter.
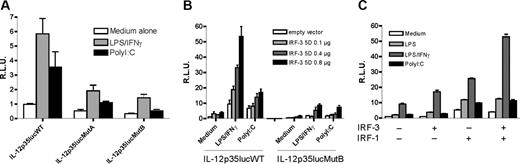
To examine the functional role of the ISRE-1 site in the transcriptional activity of the IL-12p35 promoter, mutations A and B were introduced by site-directed mutagenesis in the context of the -1121/+66 human IL-12p35 reporter plasmid. In RAW cells transiently transfected with the wild-type IL-12p35lucWT construct, we found that IL-12p35 promoter activity was induced in response to either LPS+IFN-γ or PolyI:C. Mutation A and to a greater extent mutation B significantly reduced IL-12p35 promoter activation (Figure 4A). We next cotransfected a plasmid expressing a constitutively active form of IRF-323 (IRF-3 5D) and assessed thereby the impact of IRF-3 overexpression on IL-12p35 promoter activity. As shown in Figure 4B, IRF-3 5D did not significantly up-regulate IL-12p35 promoter activity in absence of stimulation, indicating that IRF-3 alone is not sufficient to induce IL-12p35 transcriptional activation. However, upon LPS/IFN-γ and, to a lesser extent, PolyI:C stimulation, we observed a dose-dependent up-regulation of IL-12p35 promoter activity by IRF-3 5D, demonstrating a positive regulatory role of IRF-3. Mutation B negatively affected the response to transcriptional activation by IRF-3 5D (Figure 4B). As ISRE-1 site overlaps ISRE-2 site, which was shown to bind IRF-1,7 we looked for possible synergy between IRF-3 and IRF-1 on IL-12p35 promoter activity. We therefore cotransfected IL-12p35 reporter construct with IRF-3wt and IRF-1wt expression vectors. We observed an up-regulation of IL-12p35 promoter activity by either IRF-3wt or IRF-1wt (Figure 4C). A further enhancement of IL-12p35 promoter activity was observed when the 2 expression vectors were combined in LPS/IFN-γ-stimulated cells but not in resting, LPS-, or PolyI:C-stimulated cells. We concluded from this set of experiments that the ISRE-1 element is critically involved in IL-12p35 transcriptional activation induced by LPS/IFN-γ, PolyI:C, or IRF-3.
Physical characterization of the ISRE-1 site within the IL-12p35 proximal promoter region. (A) IRF-3 consensus DNA-binding sequence is shown.21 The oligonucleotide used as probe in EMSA (named ISREwt) encompasses 2 overlapping putative ISRE sites (ISRE-1 and -2, indicated by the underlined nucleotides). An asterisk indicates nucleotides that differ from the consensus. For ISREMut A, B, and C probes, only the bases that are changed compared with ISREwt are indicated. (B) Competition assays. Recombinant N-terminal IRF-3 (2 ng) was incubated with radiolabeled ISREwt probe in the absence or presence of 12.5- and 50-fold excess of the indicated unlabeled competitor. (C) EMSA was performed with decreasing amounts of IRF-3 protein (10, 2, and 0.4 ng) and radiolabeled ISREwt, ISRE Mut A, Mut B, and Mut C probes.
Physical characterization of the ISRE-1 site within the IL-12p35 proximal promoter region. (A) IRF-3 consensus DNA-binding sequence is shown.21 The oligonucleotide used as probe in EMSA (named ISREwt) encompasses 2 overlapping putative ISRE sites (ISRE-1 and -2, indicated by the underlined nucleotides). An asterisk indicates nucleotides that differ from the consensus. For ISREMut A, B, and C probes, only the bases that are changed compared with ISREwt are indicated. (B) Competition assays. Recombinant N-terminal IRF-3 (2 ng) was incubated with radiolabeled ISREwt probe in the absence or presence of 12.5- and 50-fold excess of the indicated unlabeled competitor. (C) EMSA was performed with decreasing amounts of IRF-3 protein (10, 2, and 0.4 ng) and radiolabeled ISREwt, ISRE Mut A, Mut B, and Mut C probes.
IRF-3 activation and binding to IL-12p35 ISRE site in human DCs. (A) DCs were incubated with medium alone, LPS, or PolyI:C for 90 minutes. Subcellular localization of IRF-3 was analyzed by immunofluorescence using anti-IRF-3 (top panels). Nuclei were counterstained with Toto-3 (bottom panels). One representative donor of 3 is shown. (B) Assessment of IRF-3 activation in DCs. DCs were left untreated or stimulated for 90 minutes with LPS or PolyI:C. IRF-3 binding activity in nuclear extracts (5 μg protein) was assessed using an ELISA-based technique (IRF-3 TransAM; Active Motif). Results are presented as mean ± SEM from 3 different donors. OD indicates optical density. (C) Endogenous IRF-3 binds to IL-12p35 ISRE site. DCs were left untreated or stimulated with LPS or PolyI:C for 90 minutes. Nuclear extracts (10 μg protein) were incubated with radiolabeled ISREwt probe from the p35 promoter. When indicated, PolyI:C-treated extracts were incubated with polyclonal IRF-3 antibody or control antibody. To ensure specificity of the binding, nuclear extracts from PolyI:C-stimulated DCs were incubated with radiolabeled ISREwt probe in the presence of a 50-fold molar excess of the indicated unlabeled competitor.
IRF-3 activation and binding to IL-12p35 ISRE site in human DCs. (A) DCs were incubated with medium alone, LPS, or PolyI:C for 90 minutes. Subcellular localization of IRF-3 was analyzed by immunofluorescence using anti-IRF-3 (top panels). Nuclei were counterstained with Toto-3 (bottom panels). One representative donor of 3 is shown. (B) Assessment of IRF-3 activation in DCs. DCs were left untreated or stimulated for 90 minutes with LPS or PolyI:C. IRF-3 binding activity in nuclear extracts (5 μg protein) was assessed using an ELISA-based technique (IRF-3 TransAM; Active Motif). Results are presented as mean ± SEM from 3 different donors. OD indicates optical density. (C) Endogenous IRF-3 binds to IL-12p35 ISRE site. DCs were left untreated or stimulated with LPS or PolyI:C for 90 minutes. Nuclear extracts (10 μg protein) were incubated with radiolabeled ISREwt probe from the p35 promoter. When indicated, PolyI:C-treated extracts were incubated with polyclonal IRF-3 antibody or control antibody. To ensure specificity of the binding, nuclear extracts from PolyI:C-stimulated DCs were incubated with radiolabeled ISREwt probe in the presence of a 50-fold molar excess of the indicated unlabeled competitor.
Effect of ISRE-1 site mutation on IL-12p35 gene expression. (A) RAW 264.7 cells were transiently transfected with 1 μg IL-12p35lucWT, IL-12p35lucMutA or IL-12p35lucMutB, and 80 ng pRL-TK as an internal control plasmid. Cells were stimulated with either IFN-γ (100 ng/mL) and LPS (1 μg/mL) or PolyI:C (20 μg/mL). Values represent the mean ± SEM of 6 independent experiments performed in triplicates. (B) Ectopic expression of IRF-3 5D up-regulates IL-12p35 promoter activity. RAW cells were cotransfected with 400 ng of reporter plasmid and increasing amounts of IRF-3 5D. The total amount of DNA transfected was maintained constant by complementing with the empty vector. (C) Transient transfection of IL-12p35lucWT (400 ng) in RAW cells and expression plasmids encoding IRF-1wt (400 ng) and IRF-3wt (400 ng) as indicated. Results represent the mean ± SEM of triplicates. A representative experiment of 3 is shown. RLU indicates relative light units.
Effect of ISRE-1 site mutation on IL-12p35 gene expression. (A) RAW 264.7 cells were transiently transfected with 1 μg IL-12p35lucWT, IL-12p35lucMutA or IL-12p35lucMutB, and 80 ng pRL-TK as an internal control plasmid. Cells were stimulated with either IFN-γ (100 ng/mL) and LPS (1 μg/mL) or PolyI:C (20 μg/mL). Values represent the mean ± SEM of 6 independent experiments performed in triplicates. (B) Ectopic expression of IRF-3 5D up-regulates IL-12p35 promoter activity. RAW cells were cotransfected with 400 ng of reporter plasmid and increasing amounts of IRF-3 5D. The total amount of DNA transfected was maintained constant by complementing with the empty vector. (C) Transient transfection of IL-12p35lucWT (400 ng) in RAW cells and expression plasmids encoding IRF-1wt (400 ng) and IRF-3wt (400 ng) as indicated. Results represent the mean ± SEM of triplicates. A representative experiment of 3 is shown. RLU indicates relative light units.
IRF-3 is recruited to the IL-12p35 promoter in LPS- and PolyI:C-stimulated DCs
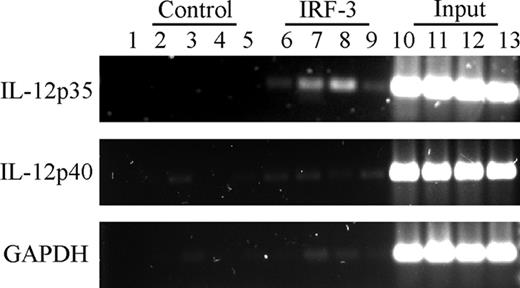
To determine if IRF-3 was recruited to the endogenous IL-12p35 promoter region in response to TLR signaling, we performed ChIP experiments on human DCs stimulated with LPS, PolyI:C, or Pam3CSK4. We used primers encompassing the ISRE-1 site. As illustrated in Figure 5, IRF-3 binding to the IL-12p35 promoter region was readily detectable in response to either TLR4 or TLR3 engagement (lanes 7 and 8). On the other hand, TLR2 ligation by Pam3CSK4 (lane 9), which does not lead to IRF-3 translocation or IL-12p35 activation, did not induce IRF-3 binding to the IL-12p35 promoter. No recruitment of IRF-3 to the IL-12p40 promoter was detected. As additional control, we performed PCR using primers specific for the GAPDH promoter and again no signal was observed when chromatin samples were immunoprecipitated with anti-IRF-3 antibody under this condition. These results therefore establish that TLR3- and TLR4-induced signaling results in the recruitment of IRF-3 to the promoter region of the endogenous IL-12p35 gene.
Inducible recruitment of IRF-3 to the endogenous IL-12p35 locus in human DCs (ChIP assay). Monocyte-derived DCs were either incubated with medium alone (lanes 2, 6, and 10) or stimulated with LPS (1 μg/mL; lanes 3, 7, and 11), PolyI:C (10 μg/mL; lanes 4, 8, and 12), or Pam3CSK4 (10 μg/mL; lanes 5, 9, and 13). After 3 hours, cells were treated with 1% formaldehyde to cross-link proteins bound to DNA. After sonication, chromatin samples were immunoprecipitated with anti-IRF-3 rabbit polyclonal antibodies (lanes 6-9) or control antibodies (lanes 2-5). Purified DNA was amplified using primers specific for IL-12p35, IL-12p40, or GAPDH promoter regions. For each sample, 2% of the cross-link-released chromatin was saved and used as input control (lanes 10-13). Lane 1 indicates PCR reaction without sample.
Inducible recruitment of IRF-3 to the endogenous IL-12p35 locus in human DCs (ChIP assay). Monocyte-derived DCs were either incubated with medium alone (lanes 2, 6, and 10) or stimulated with LPS (1 μg/mL; lanes 3, 7, and 11), PolyI:C (10 μg/mL; lanes 4, 8, and 12), or Pam3CSK4 (10 μg/mL; lanes 5, 9, and 13). After 3 hours, cells were treated with 1% formaldehyde to cross-link proteins bound to DNA. After sonication, chromatin samples were immunoprecipitated with anti-IRF-3 rabbit polyclonal antibodies (lanes 6-9) or control antibodies (lanes 2-5). Purified DNA was amplified using primers specific for IL-12p35, IL-12p40, or GAPDH promoter regions. For each sample, 2% of the cross-link-released chromatin was saved and used as input control (lanes 10-13). Lane 1 indicates PCR reaction without sample.
Impaired IL-12p35 mRNA and IL-12p70 production in bone marrow-derived DCs from IRF-3-deficient mice
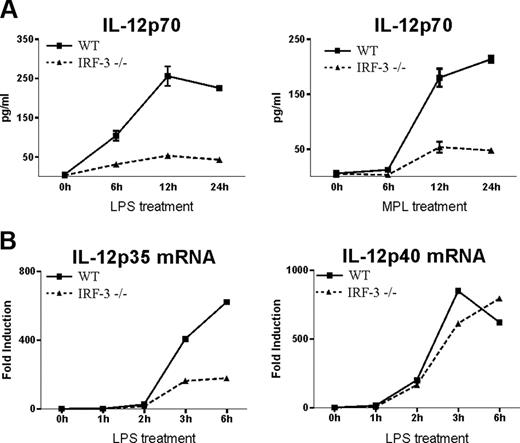
To further investigate the role of IRF-3 in the induction of IL-12p70 synthesis, we compared the levels of IL-12p70 produced upon LPS stimulation of bone marrow-derived DCs from IRF-3-deficient and wild-type mice. As shown in Figure 6A, IL-12p70 synthesis by IRF-3-/- DCs was profoundly depressed in comparison with DCs from wild-type mice (Figure 6A). Importantly, LPS-induced IL-12p35 mRNA levels were greatly decreased in IRF-3-/- DCs compared with wild-type DCs, whereas IL-12p40 mRNA levels were similar in both types of DCs (Figure 6B). These experiments establish that IRF-3 is required for efficient IL-12p35 gene expression and thereby plays an essential role in the induction of bioactive IL-12p70 synthesis. Finally, we compared IL-12p70 levels in wild-type and IRF-3-/- DCs upon stimulation with MPL, a nontoxic derivative of the lipid A region of LPS, currently used as vaccine adjuvant. As shown in Figure 6A, induction of IL-12p70 production by MPL was also shown to be IRF-3 dependent.
IRF-3 is required for efficient IL-12p35 synthesis in mouse DCs. Bone marrow-derived DCs from C57BL/6 wild-type (wt) or IRF-3-/- mice were stimulated at day 7 by LPS (100 ng/mL) or MPL (1 μg/mL) for the indicated time period. (A) Supernatants were collected and analyzed for IL-12(p70) levels by ELISA. Results represent the mean ± SEM of triplicates. A representative experiment of 3 is shown. (B) Total RNA was extracted and analyzed by real-time RT-PCR. One representative experiment of 3 is shown.
IRF-3 is required for efficient IL-12p35 synthesis in mouse DCs. Bone marrow-derived DCs from C57BL/6 wild-type (wt) or IRF-3-/- mice were stimulated at day 7 by LPS (100 ng/mL) or MPL (1 μg/mL) for the indicated time period. (A) Supernatants were collected and analyzed for IL-12(p70) levels by ELISA. Results represent the mean ± SEM of triplicates. A representative experiment of 3 is shown. (B) Total RNA was extracted and analyzed by real-time RT-PCR. One representative experiment of 3 is shown.
Discussion
There is accumulating evidence that IRF family members are critically involved in TLR signaling. Indeed, recent reports established that IRF-5 activated by MyD88 controls the synthesis of proinflammatory cytokines,12 whereas IRF-7 was shown to be critical for type I interferon synthesis in response to ligands of TLR9 subfamily members.24 IRF-3, which is activated downstream of TRIF,13 is known to be required for IFN-β activation in response to TLR3 and TLR4 agonists.20 Herein, we demonstrate another key function for IRF-3, namely to promote IL-12 synthesis by inducing IL-12p35 gene expression in response to TLR3 or TLR4 engagement. Beside its role in innate antiviral responses, IRF-3 is therefore expected to be involved in the induction of protective or pathogenic Th1 responses elicited by microbial products.
Together with recent studies, our findings establish that both MyD88-dependent and TRIF-dependent signals are required for IL-12p70 synthesis upon TLR4-mediated DC activation. Whereas the MyD88-dependent pathway is required for IL-12p40 gene expression,25 TRIF-dependent signaling appears critical for IRF-3-dependent IL-12p35 gene expression. First, as shown in this report, IRF-3 directly binds and transactivates the IL-12p35 gene promoter. Second, IRF-3 induces the expression of IFN-β, which was recently shown to act in an autocrine manner to promote IL-12p35 synthesis.19 The direct action of IRF-3 on the IL-12p35 gene promoter requires cooperation with other transcription factors, such as Sp1, NF-κB, and other IRF members including IRF-1, as suggested by the additive effect of IRF-3 and IRF-1 in RAW cell line (Figure 4C). It also probably involves the recruitment of transcriptional coactivators (such as CBP/p30026 ) and chromatin remodeling complexes, as shown for the IFN-β gene promoter.27 Indeed, we previously demonstrated that disruption of a strategically positioned nucleosome (nuc-2), located in the vicinity of the ISRE-1 site, is required for the interaction of Sp1 with a critical binding site in the IL-12p35 gene promoter.8 Interestingly, we observed that deficient IL-12p35 gene induction in LPS-stimulated DCs from human newborns is associated with both impaired nuc-2 remodeling and decreased IRF-3 activation (Goriely et al28 ; and V. A., Ezra Aksoy, and S. G., manuscript in preparation).
Of note, TLR4-triggered IL-12p40 expression was shown to be decreased in macrophages and DCs from TRIF-deficient mice.13,29 In contrast, it was not affected in DCs from IRF-3-deficient mice (Figure 6B), suggesting that IRF-3 is not implicated in TRIF-dependent IL-12p40 activation.
Our findings might be relevant to recently described viral immune evasion strategies, as several viruses appear to disrupt TRIF-induced IRF-3 activation upon TLR3 engagement by viral dsRNA.30-35 We suggest that by inhibiting IRF-3 activation, viruses escape not only type 1 IFN-dependent innate immune responses but also IL-12-dependent adaptive T-cell responses. Furthermore, the involvement of IRF-3 in IL-12 synthesis could contribute to the regulatory role of this transcription factor in cancer. Indeed, IRF-3 was shown to display direct tumor suppressor activity in vitro.36-39 In vivo, this effect is also mediated through modulation of the immune response.40 IL-12 plays an important role in the control of neoplastic transformation2 and it was recently shown that IRF-1-mediated host defense against lymphoid malignancies was dependent on IL-12 production.3 Interestingly, human herpesvirus-8 (HHV8) was shown to repress IRF-3 activation through expression of vIRF-1.41 We therefore suggest that deficient IRF-3 activation might contribute to the impaired IL-12 production, which was previously proposed to be involved in the pathogenesis of HHV8-associated lymphoproliferative disorders.42
Prepublished online as Blood First Edition Paper, October 11, 2005; DOI 10.1182/blood-2005-06-2416.
Supported by the Fonds National de la Recherche Scientifique (FNRS, Belgium) and an Interuniversity Attraction Pole of the Belgian Federal Science Policy. The Institute for Medical Immunology is sponsored by the government of the Walloon Region and GlaxoSmithKline Biologicals. S.G. is a postdoctoral Researcher of the FNRS. V.F. is a Research Associate of the FNRS.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank T. Taniguchi for providing the IRF-3-/- mice.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal