Previous evidence has shown that platelet-derived growth factor-BB (PDGF-BB) and fibroblast growth factor-2 (FGF-2) directly interact with high affinity, leading to potent reciprocal inhibitory effects on bovine endothelial cells and rat vascular smooth muscle cells. In this study, we report that PDGF-BB inhibits a series of FGF-2–induced events, such as proliferation of human umbilical vein endothelial cells (HUVECs), FGF-2 cellular internalization, phosphorylation of intracellular signaling factors including p38, rac1/cdc42, MKK4, and MKK3/6, and phosphorylation of FGF-receptor 1 (FGF-R1). PDGF-receptor-α (PDGF-Rα) was found to mediate PDGF-BB inhibitory effects because its neutralization fully restored FGF-2 mitogenic activity and internalization. Additional biochemical analyses, coimmunoprecipitation experiments, and FRET analysis showed that FGF-R1 and PDGF-Rα directly interact in vitro and in vivo and that this interaction is somehow increased in the presence of the corresponding ligands FGF-2 and PDGF-BB. These results suggest that FGF-R1/PDGF-Rα heterodimerization may represent a novel endogenous mechanism to modulate the action of these receptors and their ligands and to control endothelial cell function.
Introduction
Fibroblast growth factor-2 (FGF-2) belongs to a 23-member family of growth factors controlling the proliferation and differentiation of many cell types1 as well as angiogenesis and wound healing. It interacts with 4 high-affinity receptors—FGF-R1 (flg), FGF-R2 (bek), FGF-R3, and FGF-R42 —and with low-affinity heparan sulfate proteoglycan receptor.3-5 Spliced variants of these receptors contain 2 or 3 immunoglobulinlike loops in the extracellular-region.6,7
Platelet-derived growth factor (PDGF) controls the growth and survival of different cell types and the development of different tissues. It consists of 5 isoforms of disulfide-bonded homodimers or heterodimers (PDGF-AA, PDGF-BB, PDGF-CC, PDGF-DD, PDGF-AB) with different specificity for the corresponding dimeric receptors. In particular, the homodimer PDGF-Rα is activated by all PDGF isoforms except PDGF-DD, the homodimer PDGF-Rβ is activated by PDGF-BB and PDGF-DD, and the heterodimer PDGF-Rα-β is activated by all isoforms except PDGF-AA.8 Although the PDGF family is known to exert mitogenic and chemotactic action, PDGF-AA and PDGF-Rα are less potent or even inhibitory molecules on endothelial and vascular cells, as we and others have shown.9-13 We have previously observed a direct in vitro high-affinity FGF-2/PDGF-BB interaction14 and strong FGF-2 inhibition both in vitro and in vivo in the presence of either PDGF-BB or PDGF-AA, through a PDGF-Rα–mediated mechanism.13 Consistent with these data, other groups have shown that PDGF-Rα may activate positive and negative signals15 mediated by JNK-1 and p21WAF/CIP1 promoter induction.16 In the present study, we investigated how HUVECs respond in vitro to a FGF-2/PDGF-BB mixture compared with FGF-2 alone, and we identified a novel controlling mechanism involving FGF-R1/PDGF-Rα heterodimerization.
Materials and methods
Cell culture
HUVECs (Clonetics, San Diego, CA) were maintained in endothelial cell basal medium-2 (EBM-2 medium) supplemented with growth factors (EGM-2 kit [Clonetics] containing 2% fetal calf serum, 0.04% hydrocortisone, 0.4% FGF-2, 0.1% vascular endothelial growth factor (VEGF), 0.1% R3-IGF-1, 0.1% ascorbic acid, 0.1% human epidermal growth factor (hEGF), 0.1% GA-1000, and 0.1% heparin. In all experiments, cells were used up to the sixth passage at 80% confluence. Cells were seeded and allowed to adhere for 24 hours, then were starved overnight and used. Porcine aortic endothelial (PAE) cells were grown in Ham nutrient F12 medium (Gibco, Carlsbad, CA) with 10% fetal calf serum, 1% penicillin-streptomycin, and 2% glutamine. Unless otherwise specified, growth factors were always diluted in 0.1% bovine serum albumin (BSA) serum-free medium.
Proliferation assay
HUVEC in vitro proliferation was evaluated as previously reported.17 Briefly, HUVECs plated in 6-well plates (8 × 104 cells/well) were grown for 24 hours in EBM-2 medium supplemented with EGM-2 kit at 37°C in 5% CO2. Then medium was replaced and cells were kept in EBM-2 alone overnight. Cells were then treated with the reported concentrations of human recombinant FGF-2 (R&D Systems, Minneapolis, MN), human recombinant PDGF-BB (R&D Systems), or FGF-2/PDGF-BB mixture (10 ng/mL each), all dissolved in EBM-2 medium + 0.1% BSA for 24 hours at 37°C. Cells were harvested by trypsin-ethylenediaminetetraacetic acid (EDTA) and counted with a hemacytometer.
Anisomycin (Sigma-Aldrich, Poole, United Kingdom) (200 nM, 30-minute pretreatment) was used to specifically activate p38, as reported.18,19 Under such conditions, this molecule did not show toxic effects; rather, it showed some mitogenic effect. Antibodies neutralizing PDGF-Rα and PDGF-Rβ (MAB 322 and AF 385; R&D System) were also tested at 5 μg/mL and 1.25 μg/mL, respectively. All experiments were carried out at least 3 times in duplicate.
Western blot and phosphorylation analyses
HUVECs were treated with FGF-2 (10 ng/mL) with or without PDGF-BB (10 ng/mL) for 30 minutes, rinsed with ice-cold phosphate-buffered saline (PBS; Gibco), and lysed for 15 minutes with 1% triton, 10% glycerol, 100 mM NaCl, 5 mM EDTA, 20 mM HEPES (pH 7.4), 10 mM NaF, 2 mM phenylmethylsulfonyl fluoride (PMSF), 10 μM NaVO3, and 1% protease inhibitor cocktail (Sigma-Aldrich). Lysates were then centrifuged at 17 968g for 15 minutes. After determination of protein concentration with protein assay (Bio-Rad, Hercules, CA), 30 μg total proteins were subjected to 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were electrotransferred to a nitrocellulose membrane (Amersham, Arlington Heights, IL) and blocked with PBS containing 0.1% Tween 20 and 5% nonfat dry milk (Bio-Rad). Membrane staining with Ponceau S solution (Sigma-Aldrich) was also performed to verify the equal loading. Western blot analysis was then carried out by probing the nitrocellulose membrane with antibodies against the phosphorylated forms of p38, rac1/cdc42, MKK4, and MKK3/6 (antibodies 9211, 2461, 9151, 9231; Cell Signaling Technology, Beverly, MA) and against the corresponding unphosphorylated forms to verify the equal loading (antibodies 05-454 [Upstate Biotechology] for p38, 9152 [Cell Signaling Technology] for MKK4, and 9232 [Cell Signaling Technology] for MKK3/6). Bands were detected by incubation with horseradish peroxidase–conjugated secondary antibody (Pierce, Rockford, IL), revealed by means of the enhanced chemiluminescence (ECL) detection system (Amersham) and quantified with a calibrated imaging densitometer (GS 710; Bio-Rad). All experiments were performed at least 3 times.
FGF-R1 phosphorylation assay
HUVECs were stimulated with FGF-2, in the presence or in the absence of PDGF-BB (10 ng/mL each), for 10 minutes. Cells were then rinsed with ice-cold PBS and lysed as reported in “Western blot and phosphorylation analyses” and centrifuged. Lysates containing equal amounts (1 mg) of protein were incubated with 10 μg anti–FGF-R1 antibody, raised against peptide mapping at the carboxy-terminus of FGF-R1, (sc-121; Santa Cruz Biotechnology, Santa Cruz, CA), and kept on orbital shaker overnight at 4°C. Then 60 μL protein G–Sepharose (Santa Cruz Biotechnology) was added, and the orbital shaking was continued for another 2 hours at 4°C. The immune complexes were washed 4 times with lysis buffer and resuspended in SDS-PAGE sample buffer. After 5 minutes of heating at 100°C, samples were separated by 7% SDS-PAGE. Proteins were electrotransferred to a nitrocellulose membrane, incubated in blocking solution (5% nonfat dry milk, PBS, 0.1% Tween 20), washed, and incubated with primary antiphosphotyrosine antibodies (05-321; Upstate Biotech.) and anti–FGF-R1 antibodies raised against peptide mapping within the extracellular domain of FGF-R1 (sc-7945; Santa Cruz Biotechnology). Membranes were then washed with washing buffer (PBS, 0.1% Tween 20), and incubated with the appropriate secondary antibodies. Proteins were detected by ECL (Amersham).
FGF-2 internalization
FGF-2 internalization was investigated as previously reported.20,21 Briefly, FGF-2 (0.5 μM) in PBS was incubated with 4.5 mM EZ-Linksulfo-NHS-biotin (Pierce) for 30 minutes at room temperature (RT). The excess reagent was quenched by 154 mM Tris-HCl (pH 7.4) for 30 minutes at RT. HUVECs were stimulated with biotinylated FGF-2 alone or FGF-2/PDGF-BB mixture (10 ng/mL each) for 1 hour at 37°C in 5% CO2. Cell surface–bound material was extracted with 2 M NaCl in 20 mM HEPES buffer (pH 7.4) and 2 M NaCl in 20 mM acetic acid (pH 4.0) and discarded. The internalized biotin-labeled FGF-2 was then extracted, immobilized onto nitrocellulose membrane, labeled by peroxidase reaction, revealed by ECL (Amersham), and quantified with a calibrated imaging densitometer (GS 710; Bio-Rad).
Solid-phase assay
PDGF-Rα binding to immobilized FGF-R1 was evaluated by solid-phase assay (SPA) as described22 with some modifications. Briefly, microtiter plates (Costar, Cambridge, MA) were coated with 100 μL/well human recombinant FGF-R1β (IIIc)/Fc chimera (R&D Systems, Minneapolis, MN) 10 and 28 μg/mL for the experiments depicted in Figure 4A and 4B, respectively, and diluted in AC7.5 buffer (50 mM Tris-HCl, pH 7.5, 100 mM KCl, 3 mM MgCl2, 1 mM CaCl2) for 4 hours at 4°C. Wells were then blocked in 3% BSA (300 μL/well) in AC7.5 buffer overnight at 4°C. All subsequent operations were carried out at RT in 100 μL/well volume. After washing 3 times with AC7.5/BSA buffer (AC7.5 buffer containing 0.1% Tween-20 and 1 mg/mL BSA), immobilized FGF-R1 was incubated for 4 hours with biotinylated-PDGF-Rα (up to 100 nM) in the presence of 10 M excess unlabeled PDGF-Rα, to assay the dose dependence and to compute total, nonspecific, and specific binding (Figure 4A). A fixed concentration of biotinylated PDGF-Rα (800 nM) was used to assay the interaction in the presence or in the absence of FGF-2/PDGF-BB mixture (800 nM each) (Figure 4B). Wells were then washed 4 times and incubated with 100 μL/well Vectastain ABC Reagent (Vector Laboratories, Burlingame, CA) for 1 hour at RT and finally stained with the ELISA Amplification System (Invitrogen, Carlsbad, CA) (Figure 4A) or with 1 mg/mL P-nitrophenol phosphate in diethanolamine buffer (Figure 4B). Binding of biotinylated-PDGF-Rα to immobilized FGF-R1 was quantified by computing the concentration of bound material according to the reference curve. Specific binding was computed by subtracting nonspecific from total binding at each concentration. Bmax and Kd calculations were performed by a nonlinear regression fit with the GraphPad Prism-4 software. PDGF-Rα biotinylation was achieved as reported for FGF-2 (see “FGF-2 internalization”), and unreacted EZ-Linksulpho-NHS-biotin was discarded by centrifugation in Microcon YM-30 centrifugal filter devices (Millipore, Bedford, MA).
PDGF-Rα transfection
PAE cells were transfected with Lipofectamine Plus Reagent (Invitrogen) diluted in serum-free medium for 3 hours at 37°C in a 5% CO2 environment containing 8 μg DNA encoding wild-type PDGF-Rα (generous gifts of Dr C. H. Heldin, Ludwig Institute for Cancer Research, Uppsala, Sweden)23 or pcDNA3 (Invitrogen) empty vector as control. Medium was then replaced with Ham nutrient F12-10% fetal calf serum (FCS) for 24 hours and with serum-free medium for another 24 hours and used for experiments at 80% confluence. For immunoprecipitation experiments, cells were cotransfected with pEGFP-N1 (Clontech, Palo Alto, CA) reporter vector at a PDGF-Rα vector versus a reporter vector molar ratio of 4:1. Green fluorescent protein (GFP) transfection efficiency was greater than 70%.
FGF-R1/PDGF-Rα coimmunoprecipitation
PAE cells (1.6 × 106) overexpressing PDGF-Rα and, in a different set of experiments, untransfected HUVECs (6 × 106), grown for 24 hours and serum starved for 12 or 6 hours, respectively, were incubated for 10 minutes with FGF-2 with or without PDGF-BB (10 ng/mL each). Cells were then lysed as indicated for Western blot and phosphorylation analyses. PAE and HUVEC lysates containing 1 and 3 mg of total protein respectively, were incubated overnight at 4 °C with 10 μg anti–PDGF-Rα antibody or anti–FGF-R1 antibody (sc-431 or sc-121; Santa Cruz Biotechnology) and subsequently incubated with protein G-Sepharose as indicated in “FGF-R1 phosphorylation assay.” Resin-bound material was separated by 7% SDS-PAGE and probed with anti–FGF-R1 antibody (MAB 125; Chemicon International, Temecula, CA) and/or anti–PDGF-Rα antibody (AF-307-NA; R&D Systems) or antiphosphotyrosine antibody (sc-7020; Santa Cruz Biotechnology).
FRET analysis
PAE cells overexpressing PDGF-Rα were grown on glass coverslips and treated with FGF-2/PDGF-BB (20 ng/mL each) for 15 minutes at 37°C. Cells were then washed 2 times with PBS, fixed with fresh 3% paraformaldehyde for 10 minutes at RT, washed 3 times with PBS, incubated in PBS-2% BSA at RT for 30 minutes, and stained with a monoclonal mouse anti–FGF-R1 antibody (MAB 125; Chemicon International) and/or a polyclonal goat anti–PDGF-Rα antibody (AF-307-NA; R&D Systems) for 1 hour at RT. Cells were then incubated for 1 hour with a secondary antibody Alexa-Fluor 488 chicken anti–mouse IgG (H+L) conjugate (the donor fluorophore) and/or the secondary antibody Alexa-Fluor 546 donkey anti–goat IgG (H+L) conjugate (the acceptor fluorophore) (A-21200 and A-11056; Molecular Probes). Fluorescence resonance energy transfer (FRET) analyses were then carried out with a LSM510 Meta confocal microscope (Zeiss, Oberkochen, Germany) according to the manufacturer's instructions and to a published procedure.24 The emission spectrum of each fluorophore between 488 and 627 nm was determined on a single plane by λ mode function and emission fingerprint technique. Samples were illuminated with 488 nm and 546 nm laser line, respectively, and the emission light was recovered through the whole spectrum (λ mode). For each sample, a single pixel was selected with a pure emission spectrum with a peak at 519 nm and 572 nm for Alexa-Fluor 488 and Alexa-Fluor 546, respectively. All parameters determined in λ mode were then used to evaluate FRET occurrence by exciting samples containing donor and acceptor fluorophores with the 488-nm laser line, in online fingerprint mode, and recovering fluorescence at 572 nm only from pixels emitting spectra exactly matching those stored using λ mode.
Statistical analysis
All experiments were performed at least 3 times in duplicate. Data are reported as the mean plus or minus standard deviation (SD). Student t test was carried out to evaluate statistical significance, and P below .05 was considered significant. Scatchard analysis was carried out with GraphPad-Prism 4 software.
Results
PDGF-BB inhibits FGF-2–induced HUVEC proliferation and intracellular FGF-2 signaling
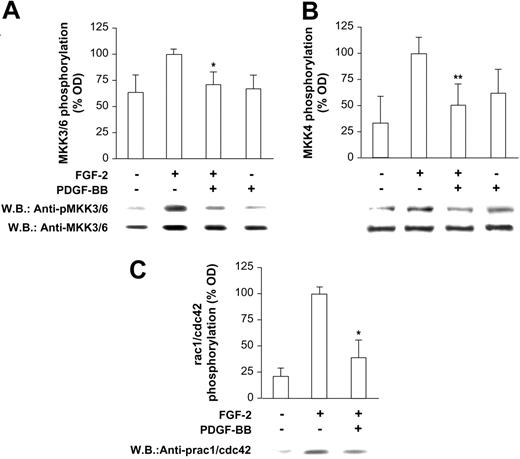
Preliminary dose dependence and time course experiments indicated a strong inhibitory effect of PDGF-BB on FGF-2 mitogenic activity in HUVECs (Figure S1; see the Supplemental Figures link at the top of the online article, at the Blood website). These data agree with previous observations collected on bovine aorta endothelial cells.13 The following experiments were then carried out to identify at which level PDGF-BB exerts its modulating effects on FGF-2. Signaling molecules located at different levels onto the FGF-2 signaling pathway were analyzed. We first investigated p38 because it is known to play a key role at a downstream level of the FGF-2 signaling pathway.25 As reported in Figure 1A, p38 phosphorylation induced by FGF-2 was totally lacking in the presence of PDGF-BB. When p38 was selectively activated by Anisomycin (Sigma-Aldrich) pretreatment (200 nM), the PDGF-BB inhibitory effect was abolished, and FGF-2 mitogenic activity (Figure 1B) and p38 phosphorylation (Figure 1C) were fully restored. Interestingly, Anisomycin (Sigma-Aldrich), at the dose used, did not show any toxic effect; rather it was mitogenic alone and in the presence of PDGF-BB (Figure 1B). Together these data indicated that PDGF-BB inhibition of FGF-2 acts at a level upstream of p38 because its inhibitory effect was overridden by p38 activation and was allowed to rule out nonspecific or toxic effects. Then we investigated the phosphorylation of other intracellular molecules placed upstream of p38, namely MKK3/6, MKK4, and rac1/cdc42. In all cases, a marked reduction of their phosphorylation was found (Figure 2A-C, respectively), strongly supporting the hypothesis that PDGF-BB may inhibit FGF-2 signaling by acting at an early level.
FGF-2–induced p38 phosphorylation in the presence of PDGF-BB. (A) FGF-2–induced p38 phosphorylation was significantly reduced by PDGF-BB (10 ng/mL each) (*P < .001 vs FGF-2). (B) PDGF-BB lost the inhibitory effect of FGF-2 mitogenic activity on selective p38 activation with Anisomycin (*P < .05). (C) Anisomycin pretreatment abolished PDGF-BB reduction of FGF-2–mediated p38 phosphorylation (*P < .001 vs FGF-2). Bars indicate quantification of 3 experiments; representative Western blots are shown below the bar graphs in panels A and C.
FGF-2–induced p38 phosphorylation in the presence of PDGF-BB. (A) FGF-2–induced p38 phosphorylation was significantly reduced by PDGF-BB (10 ng/mL each) (*P < .001 vs FGF-2). (B) PDGF-BB lost the inhibitory effect of FGF-2 mitogenic activity on selective p38 activation with Anisomycin (*P < .05). (C) Anisomycin pretreatment abolished PDGF-BB reduction of FGF-2–mediated p38 phosphorylation (*P < .001 vs FGF-2). Bars indicate quantification of 3 experiments; representative Western blots are shown below the bar graphs in panels A and C.
PDGF-Rα mediates PDGF-BB inhibitory effect on FGF-2
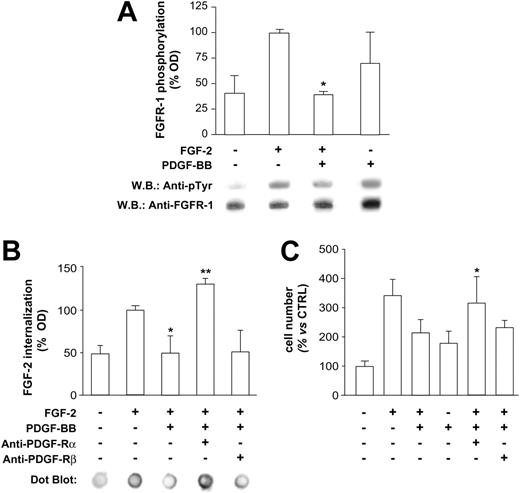
Therefore, additional experiments were carried out investigating events occurring at the membrane level, such as FGF receptor 1 (FGF-R1) phosphorylation and FGF internalization. Western blot analysis of HUVEC immunoprecipitated lysates indicated that FGF-R1 phosphorylation induced by FGF-2 was markedly inhibited in the presence of PDGF-BB (Figure 3A). We then investigated FGF-2 internalization, an event that follows binding to and activation of FGF-R1 and leads FGF-2 to the nucleus, triggering the expression of genes regulating cell cycle and cellular growth.26 As for FGF-R1 phosphorylation, FGF-2 internalization was strongly reduced in the presence of PDGF-BB (Figure 3B). Thus we hypothesized that PDGF-BB may act through a mechanism likely mediated by its receptors. This hypothesis was tested in additional internalization and proliferation experiments, showing that selective neutralization of PDGF-Rα restored FGF-2 internalization and FGF-2-mitogenic effect, whereas selective neutralization of PDGF-Rβ was ineffective (Figure 3B-C, respectively). These data led us to conclude that PDGF-BB exerts its inhibitory effect on FGF-2 by acting, at least in part, at the membrane level, with a mechanism mediated by FGF-R1 and PDGF-Rα and not mediated by PDGF-Rβ.
FGF-R1/PDGF-Rα direct interaction
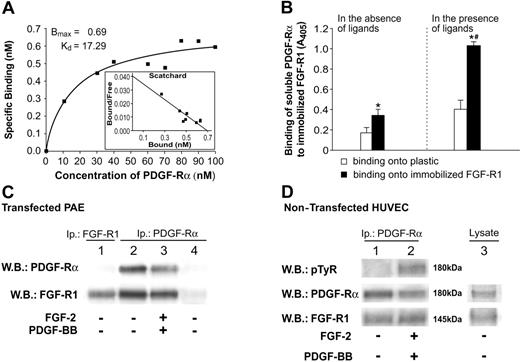
According to these findings and to previously published data,27,28 we then hypothesized that a mechanism able to explain at least in part the PDGF-BB–mediated impairment of FGF-2 activity may involve the formation of a heterocomplex containing FGF-R1 and PDGF-Rα. Direct interaction of these molecules was measured in vitro by SPA experiments. FGF-R1 (10 μg) was immobilized onto plastic and biotin-labeled PDGF-Rα in the presence of 10 M excess unlabeled PDGF-Rα and was dispensed in solution at increasing doses. Under these conditions PDGF-Rα specifically bound FGF-R1 in a concentration-dependent, saturable manner, with a Kd of 17.29 nM. Specific binding was measured by subtraction of nonspecific from total binding and is reported in Figure 4A. Scatchard analysis was also carried out and is depicted in the inset. In other experiments performed at a fixed dose, receptor interaction was assayed in the absence and presence of the corresponding ligands. Figure 4B shows that PDGF-Rα interaction with FGF-R1 is increased by 3.6-fold in the presence of ligands (black bar vs black bar), indicating that FGF-2 and PDGF-BB may modulate the ability of the corresponding receptors to form heterocomplexes. Furthermore, Figure 4B shows that in the absence of ligands, PDGF-Rα binding to immobilized FGF-R1 is significantly higher than binding to immobilized BSA, and in the presence of ligands such difference is markedly increased (black bars vs white bars).
Phosphorylation state of intermediate molecules upstream of p38. FGF-2–induced phosphorylation of (A) MKK3/6, (B) MKK4, and (C) rac 1/cdc 42 was markedly inhibited in the presence of PDGF-BB (*P < .001 vs FGF-2; **P < .05 vs FGF-2). PDGF-BB does not show any significant phosphorylating effect on these molecules, as expected according to data reported in Figure 1B. Bars indicate quantification of 3 experiments; one representative Western blot is shown below the bar graph in each panel.
Phosphorylation state of intermediate molecules upstream of p38. FGF-2–induced phosphorylation of (A) MKK3/6, (B) MKK4, and (C) rac 1/cdc 42 was markedly inhibited in the presence of PDGF-BB (*P < .001 vs FGF-2; **P < .05 vs FGF-2). PDGF-BB does not show any significant phosphorylating effect on these molecules, as expected according to data reported in Figure 1B. Bars indicate quantification of 3 experiments; one representative Western blot is shown below the bar graph in each panel.
Direct binding of these receptors was further investigated in live cells by coimmunoprecipitation experiments, either in PAE cells or in HUVECs. PAE cells express detectable levels of FGF receptors and very low or undetectable levels of PDGF receptors.29 Hence, in these cells, human PDGF-Rα was overexpressed and coimmunoprecipitated with FGF-R1 (Figure 4C, lanes 2-3) as confirmed by FGF-R1 Western blotting analysis (Figure 4C, lane 1). A similar experiment was carried out on untransfected HUVECs, confirming coimmunoprecipitation of the 2 receptors in the presence and in the absence of the corresponding ligands (Figure 4D). Lane 2 of Figure 4D also shows that coimmunoprecipitation of FGF-R 1 and PDGF-Rα, occurring in the presence of ligands, does not impair PDGF-Rα tyrosine phosphorylation.
These experiments indicate that FGF-R1 and PDGF-Rα interact in vitro and in live cells. The presence of the corresponding ligands increases the heterodimerization in the SPA assay.
FRET confocal analysis
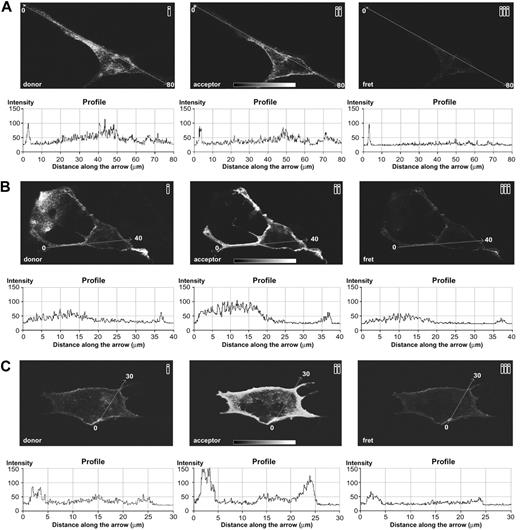
FRET microscopy is an ideal technique to highlight short-range (less than 10 nm) in vivo protein–protein interactions; therefore, additional experiments were carried out with this technique to confirm that the 2 receptors interact in live cells. Single-plane confocal images of PAE cells transfected with human PDGF-Rα are reported in Figure 5. Figure 5B-C shows 2 representative examples of cells treated with the FGF-2/PDGF-BB mixture, whereas Figure 5A reports cells treated with culture medium only, as control. FRET between PDGF-Rα and FGF-R1 occurred in PAE cells treated with FGF-2/PDGF-BB mixture (Figure 5B-C, section 3), whereas it was undetectable in the absence of ligands (Figure 5A, section 3). FRET and SPA experiments show similar results at the qualitative and quantitative levels. In fact, FRET signal quantification in the presence of ligands indicated a roughly 2-fold increase compared with the background (profiles beneath sections 3 of Figure 5B-C), in agreement with the SPA measurements (Figure 4B, right, black bar vs white bar). Further, in the absence of the ligands, the FRET signal was strongly reduced (or absent) as observed in SPA (Figure 4B, black bar vs black bar).
Additional experiments carried out on untransfected melanoma cells (SK-Mel 110) confirmed the FRET occurrence between PDGF-Rα and FGF-R1 in the presence of the corresponding ligands (Figure S2), further supporting the finding that FGF-R1 and PDGF-Rα directly interact.
Discussion
FGF-2 receptors and PDGF-BB receptors are transmembrane proteins transducing signals from the extracellular environment to a number of different intracellular molecules.30 Several studies show that α and β receptors of PDGF have distinct functions, thus accounting for different effects exerted by PDGF family members; increasing evidence refers to such receptors as cancer drug targets.31 PDGF-Rβ is well known to activate angiogenic signals,31,32 and PDGF-BB is known to potentiate FGF-2 angiogenic activity,33 likely through PDGF-Rβ. The role of PDGF-Rα is somehow more complex because it triggers inducing signals and inhibitory signals.14,16 We previously showed, both in vitro and in vivo, that FGF-2 and PDGF-BB interact with high affinity,14 leading to reciprocal inhibitory effects mediated, at least in part, by PDFG-Rα and FGF-R1.13,34 These data represent the first evidence indicating that PDGF-BB may acquire in vivo antiangiogenic effects mediated by PDGF-Rα and highlight that PDGF activity may be controlled or even counteracted by the expression levels and activation state of the corresponding α and β receptors.
FGF-R1 phosphorylation, FGF-2 internalization, and PDGF-Rα role. (A) On HUVEC lysates, FGF-R1 phosphorylation was evaluated by immunoprecipitation and was found to be markedly inhibited by FGF-2/PDGF-BB treatment (10 minutes) compared with FGF-2 alone (*P < .001 vs FGF-2). Apparently, PDGF-BB alone induces FGF-R1 phosphorylation, according to data previously reported,36 although a statistical significance was not reached in our experiments (B) FGF-2 internalization was abolished by PDGF-BB (*P < .05 vs FGF-2) and PDGF-Rα–neutralizing antibody restored FGF-2 internalization, whereas neutralizing PDGF-Rβ had no effect (**P < .01 vs FGF-2/PDGF-BB). (C) PDGF-BB inhibitory effect on FGF-2 mitogenic activity (*P < .05 vs FGF-2) was abolished by neutralizing PDGF-Rα (**P < .05 vs FGF-2/PDGF-BB), whereas PDGF-Rβ neutralization was ineffective. Bars indicate quantification of 3 experiments; 1 representative experiment is reported for each panel.
FGF-R1 phosphorylation, FGF-2 internalization, and PDGF-Rα role. (A) On HUVEC lysates, FGF-R1 phosphorylation was evaluated by immunoprecipitation and was found to be markedly inhibited by FGF-2/PDGF-BB treatment (10 minutes) compared with FGF-2 alone (*P < .001 vs FGF-2). Apparently, PDGF-BB alone induces FGF-R1 phosphorylation, according to data previously reported,36 although a statistical significance was not reached in our experiments (B) FGF-2 internalization was abolished by PDGF-BB (*P < .05 vs FGF-2) and PDGF-Rα–neutralizing antibody restored FGF-2 internalization, whereas neutralizing PDGF-Rβ had no effect (**P < .01 vs FGF-2/PDGF-BB). (C) PDGF-BB inhibitory effect on FGF-2 mitogenic activity (*P < .05 vs FGF-2) was abolished by neutralizing PDGF-Rα (**P < .05 vs FGF-2/PDGF-BB), whereas PDGF-Rβ neutralization was ineffective. Bars indicate quantification of 3 experiments; 1 representative experiment is reported for each panel.
FGF-R1/PDGF-Rα interaction examined in SPA and coimmunoprecipitation. (A) Solid-phase assay (SPA) was performed to analyze PDGF-Rα binding to immobilized FGF-R1. Up to 100 nM biotinylated-PDGF-Rα recombinant protein, in the presence of 10 M excess of unlabeled PDGF-Rα, was incubated onto immobilized FGF-R1 (10 μg/mL), and a colorimetric assay was carried out. Specific binding was calculated by subtracting nonspecific from total binding. Bmax and Kd values were computed by GraphPad Prism 4 software, applying a nonlinear regression fit. Scatchard analysis is also reported (inset). (B) Interaction of soluble PDGF-Rα (800 nM each) to plastic (□) or to FGF-R1–immobilized factor (28 μg/mL) (▪) is reported as OD. Left: PDGF-Rα interaction in the absence of ligands. Right: PDGF-Rα interaction in the presence of ligands. Results are reported as mean ± SD of 3 experiments. *P < .05 (▪ vs □). #P < .05 (▪ vs ▪). (C) Lysates of PAE cells transfected with human PDGF-Rα were immunoprecipitated with anti–PDGF-Rα antibody and analyzed by immunoblotting, probed with anti–FGF-R1 antibody (bottom bands), and reprobed with anti–PDGF-Rα antibody (top bands). FGF-R1/PDGF-Rα coimmunoprecipitation was found in the absence (lane 2) and in the presence (lane 3) of FGF-2/PDGF-BB. Lane 4 reports PAE cells transfected with pcDNA3 empty vector, as control. They expressed very low levels of PDGF-Rα; thus, no signal was evident in Western blot for PDGF-Rα or for FGF-R1. Lane 1 represents the positive control for FGF-R1; it shows Western blot of cell lysates immunoprecipitated and immunoblotted with anti–FGF-R1 antibody and allows identification of FGF-R1 as the band observed in lanes 2 and 3. PDGF R-α was not detected in lane 1 as opposed to lanes 2 and 3, likely for a different steric impediment of the antibody couple used in lane 1 compared with the antibody couple used in lanes 2 and 3. (D) Lysates of untransfected HUVECs were immunoprecipitated with anti–PDGF-Rα antibodies and were analyzed by immunoblotting, probed with anti–FGF-R1 antibody (bottom bands), and reprobed with anti–PDGF-Rα antibodies (middle bands). FGF-R1/PDGF-Rα complex was found in untreated (lane 1) and FGF-2/PDGF-BB–treated (lane 2) cells. Immunoblotting with antiphosphotyrosine antibody (top bands) shows the activation of PDGF-Rα in lane 2. Lane 3 shows expression levels of FGF-R1 and PDGF-Rα by Western blot analysis of a cellular HUVEC lysate.
FGF-R1/PDGF-Rα interaction examined in SPA and coimmunoprecipitation. (A) Solid-phase assay (SPA) was performed to analyze PDGF-Rα binding to immobilized FGF-R1. Up to 100 nM biotinylated-PDGF-Rα recombinant protein, in the presence of 10 M excess of unlabeled PDGF-Rα, was incubated onto immobilized FGF-R1 (10 μg/mL), and a colorimetric assay was carried out. Specific binding was calculated by subtracting nonspecific from total binding. Bmax and Kd values were computed by GraphPad Prism 4 software, applying a nonlinear regression fit. Scatchard analysis is also reported (inset). (B) Interaction of soluble PDGF-Rα (800 nM each) to plastic (□) or to FGF-R1–immobilized factor (28 μg/mL) (▪) is reported as OD. Left: PDGF-Rα interaction in the absence of ligands. Right: PDGF-Rα interaction in the presence of ligands. Results are reported as mean ± SD of 3 experiments. *P < .05 (▪ vs □). #P < .05 (▪ vs ▪). (C) Lysates of PAE cells transfected with human PDGF-Rα were immunoprecipitated with anti–PDGF-Rα antibody and analyzed by immunoblotting, probed with anti–FGF-R1 antibody (bottom bands), and reprobed with anti–PDGF-Rα antibody (top bands). FGF-R1/PDGF-Rα coimmunoprecipitation was found in the absence (lane 2) and in the presence (lane 3) of FGF-2/PDGF-BB. Lane 4 reports PAE cells transfected with pcDNA3 empty vector, as control. They expressed very low levels of PDGF-Rα; thus, no signal was evident in Western blot for PDGF-Rα or for FGF-R1. Lane 1 represents the positive control for FGF-R1; it shows Western blot of cell lysates immunoprecipitated and immunoblotted with anti–FGF-R1 antibody and allows identification of FGF-R1 as the band observed in lanes 2 and 3. PDGF R-α was not detected in lane 1 as opposed to lanes 2 and 3, likely for a different steric impediment of the antibody couple used in lane 1 compared with the antibody couple used in lanes 2 and 3. (D) Lysates of untransfected HUVECs were immunoprecipitated with anti–PDGF-Rα antibodies and were analyzed by immunoblotting, probed with anti–FGF-R1 antibody (bottom bands), and reprobed with anti–PDGF-Rα antibodies (middle bands). FGF-R1/PDGF-Rα complex was found in untreated (lane 1) and FGF-2/PDGF-BB–treated (lane 2) cells. Immunoblotting with antiphosphotyrosine antibody (top bands) shows the activation of PDGF-Rα in lane 2. Lane 3 shows expression levels of FGF-R1 and PDGF-Rα by Western blot analysis of a cellular HUVEC lysate.
In the present study, we report such a PDGF-BB inhibitory effect on human endothelial cells and show PDGF-BB–dependent marked impairment of the FGF-2 signal transduction, including p38 phosphorylation. Activating p38 by Anisomycin (Sigma-Aldrich) treatment reverted the PDGF-BB inhibitory effect, suggesting that PDGF-BB inhibits FGF-2 by acting at a level upstream of p38. Phosphorylation of other FGF-2 pathway molecules located upstream of p38, including FGF-R1, was also inhibited in the presence of PDGF-BB. Although the role of Rac-1 phosphorylation in FGF-2 signaling must be further clarified, data reported in Figure 2 and Figure 3 indicate that PDGF-BB inhibits FGF-2–dependent phosphorylation of different intracellular molecules. This strongly suggests cross-talk of the 2 factors at an early stage, likely at the membrane level. Interestingly, PDGF-BB itself induced FGF-R1 phosphorylation (Figure 3A), as other authors have recently hypothesized.35 Although no statistical significance was achieved compared with controls, these experiments suggested PDGF-BB and FGF-2 may both phosphorylate FGF-R1, and they supported the direct interference of PDGF-BB at an early stage of the FGF-2 signaling pathway. Previous evidence indicates that PDGF-BB and FGF-2 form nonfunctional heterodimers,14 and other data collected in vitro and in vivo indicate that PDGF-Rα is involved in the PDGF-BB-mediated inhibition of FGF-2.13,14,34 In accordance with these previous observations, in the present study PDGF-Rα neutralization was found to restore FGF-2 internalization and mitogenic effect, which were inhibited in the presence of PDGF-BB. In these experiments, PDGF-Rβ neutralization was ineffective. All these findings highlight the inhibitory role of PDGF-Rα as opposed to PDGF-Rβ, further indicating their distinct functions despite the overlapping signaling pathways.36
Previous evidence reports the heterodimerization of the PDGF receptor and epidermal growth factor (EGF) receptor,27,28 showing that cross-talk of different factors may be mediated, at least in some cases, by heterodimerization of their receptors. FGF-R1 and PDGF-Rα are both tyrosine kinase receptors and type 1 membrane proteins; the former belongs to the FGF receptor family and has 3 immunoglobulinlike C2-type domains, whereas the latter belongs to the CSF-1/PDGF receptor subfamily and has 5 immunoglobulinlike C2-type domains. We thus hypothesized that PDGF-Rα may directly interact with FGF-R1, accounting, at least in part, for the inhibitory effects observed previously and reported in the present study. Confirming such hypothesis, the 2 purified receptors showed marked specific, dose-dependent, and saturable interactions in the SPA assays (Figure 4A). In live cells, coimmunoprecipitation assays (Figure 4C-D) and FRET analyses confirmed the occurrence of direct binding (Figure 5). FGF-R1/PDGF-Rα interaction was always observed in the presence of the corresponding ligands (PDGF-BB and FGF-2), in SPA, coimmunoprecipitation, and FRET analyses. Interestingly, in the absence of ligands, heterodimerization was significantly reduced in SPA and undetectable in FRET. It is possible that FGF-R1 and PDGF-Rα were not close enough (less than or equal to 10 nm) to give a FRET signal, unless they were engaged by their ligands. This observation prompted us to hypothesize a receptor heterodimerization phenomenon controlled by the presence of the corresponding ligands. Given the high-affinity interaction reported for the FGF-2/PDGF-BB heterodimer,14 we propose here the intriguing concept that heterodimerization of FGF-2 and PDGF-BB ligands under appropriate conditions may control the heterodimerization of the corresponding receptors. The rate of receptor and ligand heterodimerization may well depend on their expression level. At lower concentrations, FGF-2 and PDGF-BB may dimerize less and therefore impair heterodimerization of the corresponding receptors. According to this hypothesis, ligand/receptor-complex formation may be modulated in vivo under physiologic or pathologic conditions, or both, depending on their expression levels and may explain, at least in part, the positive synergistic effect of FGF-2 and PDGF37 and the reciprocal inhibitory action.13,14,34 On the other hand, the angiogenic potential of such factors is known to be markedly inhibited in vivo. In fact, despite their continuous expression, no angiogenesis is observed under physiologic conditions in vivo. Therefore, it is reasonable to hypothesize a mechanism controlling these factors or their receptors, based on their expression levels, controlling their ability to form homocomplexes (activating signals) or heterocomplexes (inhibitory signals). This mechanism may parallel other known endogenous inhibitory mechanisms discussed in detail in several studies38 (see also references in Nyberg et al38 ), which include matrix-derived inhibitors (such as endostatin and fibronectin fragments) and nonmatrix-derived inhibitors, such as platelet factor 4, interferons, TIMPs, and others. According to our hypothesis, altering the physiologic equilibrium of the ligands and receptor expression levels might alter the formation of the corresponding heterocomplexes, accounting for the altered angiogenesis observed in pathologic conditions such as atherosclerosis and tumor growth. We thus conclude that such ligand/receptor complex formation may represent a novel endogenous mechanism to control cell response and to modulate the activity of FGF-2 and PDGF growth factors.
FGF-R1/PDGF-Rα interaction examined by FRET in single-plane confocal images of PAE cells transfected with human PDGF-Rα. (A) Cells grown in culture medium without growth factors. Donor-fluorophore signal, corresponding to the FGF-R1 (section 1) and acceptor-fluorophore signal corresponding to PDGF-Rα (section 2) are shown; no FRET between these receptors was detected (section 3). (B-C) Cells treated with FGF-2/PDGF-BB mixture (20 ng/mL each). Donor-fluorophore signal, corresponding to the FGF-R1 (section 1) and the acceptor-fluorophore signal corresponding to PDGF-Rα (section 2), are shown; ligand-induced FRET is evident in section 3. Graphs reported below each panel show the signal quantification measured along the arrow. Original magnification, 100 ×; objective lens used was 40×/1.0 numeric aperture. Slides were marked with an aqueous medium with antifading agents (Biomedia, Foster City, CA). Images were processed using Adobe Photoshop version 5 (Adobe Systems, San Jose, CA).
FGF-R1/PDGF-Rα interaction examined by FRET in single-plane confocal images of PAE cells transfected with human PDGF-Rα. (A) Cells grown in culture medium without growth factors. Donor-fluorophore signal, corresponding to the FGF-R1 (section 1) and acceptor-fluorophore signal corresponding to PDGF-Rα (section 2) are shown; no FRET between these receptors was detected (section 3). (B-C) Cells treated with FGF-2/PDGF-BB mixture (20 ng/mL each). Donor-fluorophore signal, corresponding to the FGF-R1 (section 1) and the acceptor-fluorophore signal corresponding to PDGF-Rα (section 2), are shown; ligand-induced FRET is evident in section 3. Graphs reported below each panel show the signal quantification measured along the arrow. Original magnification, 100 ×; objective lens used was 40×/1.0 numeric aperture. Slides were marked with an aqueous medium with antifading agents (Biomedia, Foster City, CA). Images were processed using Adobe Photoshop version 5 (Adobe Systems, San Jose, CA).
Prepublished online as Blood First Edition Paper, December 1, 2005; DOI 10.1182/blood-2005-04-1524.
Supported in part by Istituto Superiore di Sanità (ISS) grant CS-135-2003 and Ministero della Istruzione dell'Università e della Ricerca (MIUR) grant RF2003.
D.F. designed and performed the research and wrote the paper; M.S.A., K.R., and G.R. performed the research; M.C.C. designed the research and conducted general supervision; A.F. designed the research, analyzed the data, wrote the paper, and supervised the research.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal