Innate immunity is involved in the biology of graft versus host disease and common airway diseases. We screened 15 genes in this pathway using a linkage disequilibrium-based approach to identify potential candidate genes that may be involved in the development of airflow obstruction after hematopoietic cell transplantation. Sixty-nine single-nucleotide polymorphisms were selected for assessment in a discovery cohort (n = 363). Significant associations were validated in a validation cohort (n = 209). Expression of the candidate gene was demonstrated by detecting gene transcript and protein in malignant and normal small airway epithelial cells. In the discovery cohort, 133 patients developed significant airflow decline. Four patient and donor bactericidal/permeability-increasing (BPI) haplotypes were associated with a 2-fold to 3-fold increased risk of developing significant airflow decline (P values, .004-.038). This association was confirmed in the validation cohort, which had 66 patients with significant airflow decline, with 9 significant haplotypes (P values, .013-.043). BPI gene transcript and protein were detected in airway epithelial cells. These results suggest mutations in the BPI gene significantly influence the risk of developing rapid airflow decline after hematopoietic cell transplantation and may represent a novel therapeutic target for this form of airway disease.
Introduction
New-onset airflow obstruction, known as bronchiolitis obliterans syndrome in its severest form, affects up to 26% of patients who receive an allogeneic hematopoietic cell transplantation (HCT).1 Despite all the advances in the management of HCT over the last 2 decades, the prevalence of this syndrome remains high, often resulting in long-lasting and sometimes devastating effects. Although the allograft reaction has been repeatedly associated with this syndrome in clinical studies, this risk is modest,2-7 suggesting that other unknown variables are important in the pathogenesis of this syndrome.
While it is well established that attack of the recipient's tissues by alloreactive T cells of donor origin is a key component in the pathogenesis of graft versus host disease (GVHD),8-10 other mononuclear cells also play a significant role.11 When primed by Th1 cytokines like IFN-γ and IL-2, or activated by lipopolysaccharide through the innate immune system, monocytes and dendritic cells can produce proinflammatory and anti-inflammatory cytokines that can further amplify or maintain the graft versus host reaction.12-14 Innate immunity is also involved in the development of airway diseases. Lipopolysaccharide is ubiquitous to the environment and has been found to cause chronic airway inflammation in various airway diseases such as asthma and emphysema.15-23 Based upon these data, we postulated that inflammation induced by lipopolysaccharide via the innate immunity pathway may contribute to the development of rapid airflow decline after transplantation. Using a candidate pathway haplotype-based approach, we screened genes in the innate immunity pathway to identify genes that are associated with an increased risk of developing significant airflow decline after HCT.
Patients, materials, and methods
Patient selection and clinical characteristics
All stem cell recipients and their donors gave written informed consent according to protocols approved by the institutional review board at the Seattle Cancer Care Alliance/Fred Hutchinson Cancer Research Center. All patients who received their first allogeneic HCT between January 1, 1990, and December 31, 2002, excluding patients who did not have a pretransplantation pulmonary function test (PFT) or at least 1 posttransplantation PFT, were eligible for study. “Discovery” and “validation” cohorts were randomly selected for study based upon donor and recipient DNA specimen availability.
Clinical variables that we previously identified as significant clinical risk factors (age at transplantation, extent of GVHD, and pretransplantation 1-second forced expiratory volume [FEV1]) were retrieved from a prospectively collected clinical database.1 The patient's underlying disease state was categorized as low, intermediate, or high risk.24 Low-risk diseases included chronic myeloid leukemia in chronic phase, refractory anemia, aplastic anemia, and Blackfan-Diamond syndrome. Intermediate-risk diseases included chronic myeloid leukemia in accelerated phase or in chronic phase after blast phase, acute leukemia or lymphoma in remission, refractory anemia with excess blasts, chronic lymphocytic leukemia, and paroxysmal nocturnal hemoglobinuria. High-risk diseases included chronic myeloid leukemia in blast phase, juvenile chronic myeloid leukemia, acute leukemia or lymphoma in relapse, refractory anemia with excess blasts in transformation, and myeloma. Acute GVHD was graded based upon stages of organ involvement using standard criteria and categorized as no (grades 0-II) or yes (grades III-IV) as previously reported.24-26 The diagnosis and staging of chronic GVHD were established by using clinical, histologic, and laboratory criteria published previously27 and were characterized according to the presence or absence of clinical extensive chronic GVHD. Acute and chronic GVHD were then integrated and categorized as no acute or chronic GVHD, acute GVHD alone, de novo chronic GVHD (not preceded by acute GVHD), quiescent-onset chronic GVHD (preceded by acute GVHD that was followed by a period of quiescence), or progressive-onset chronic GHVD (preceded by acute GVHD without a period of quiescence). Other clinical variables were also retrieved for comparison purposes. Patient race in the discovery cohort was classified as white or nonwhite, which included Hispanic, Asian, African-American, and American Indian patients. Donor race in the discovery cohort was not classified due to lack of race data for 15% of the donors. All patients received a myeloablative conditioning regimen.
Airflow decline phenotype
Pulmonary function testing has been previously described in detail.1 All PFTs were performed according to the American Thoracic Society guidelines28 and expressed as a percent of the predicted values of FEV1 and forced vital capacity (FVC) calculated using published equations.29,30 If a bronchodilator was administered, the postbronchodilator value was used. All PFTs, including the pretransplantation and all posttransplantation PFTs for each patient, were used to generate a least squares regression line to determine the annualized rate of FEV1 decline. We previously used this approach in a 10-year epidemiologic evaluation of posttransplantation lung function decline to develop a phenotype that characterized the change in airflow after HCT.1 This study demonstrated that an annualized rate of FEV1 decline of more than 5% per year significantly influenced a patient's mortality risk after HCT, indicating that a cutoff at this level had significant clinical implications. Therefore, patients who experienced an annualized rate of FEV1 decline of more than 5% per year with the lowest documented posttransplantation FEV1/FVC ratio of less than 0.8 were considered to have developed the significant airflow decline phenotype.
DNA, single-nucleotide polymorphism selection, and genotyping
Donor and recipient DNA were extracted (QIAamp DNA Blood Mini Kit; Qiagen, Valencia, CA) from B-lymphoblastoid cell lines immortalized by Epstein-Barr virus transformation.31 Well-characterized candidate genes from the innate immunity pathway were selected based upon the availability of sequence data from healthy subjects of European decent generated by SeattleSNPs and the Innate Immunity Program for Genomic Application: toll-like receptor 2 (TLR2), toll-like receptor 3 (TLR3), toll-like receptor 4 (TLR4), toll-like receptor 6 (TLR6), CD14, bactericidal/permeability-increasing protein (BPI), lipopolysaccharide binding protein (LBP), toll-interleukin-1 receptor (TIR) domain-containing adapter protein (TIRAP), interleukin-1 receptor-associated kinase 4 (IRAK4), tumor necrosis factor receptor-associated factor 6 (TRAF6), tumor necrosis factor-alpha (TNF-α), interleukin-1 beta (IL-1β), interleukin-1 receptor antagonist (IL-1rn), interleukin-6 (IL-6), interleukin-10 (IL-10). All single-nucleotide polymorphism (SNPs) with a frequency of 10% or more were identified, and a set of maximally informative tagSNPs were selected using LDSelect,32 which is based upon an algorithim that identifies linkage disequilibrium-selected tagSNPs using the r2 linkage disequilibrium statistic (threshold more than 0.64).33 SNPs with a minor allele frequency (MAF) of 0.1 or more but existing as singletons were not selected.
Two high-throughput genotyping platforms were used for genotyping. The discovery cohort was genotyped using primer extension of multiplex polymerase chain reaction (PCR) products, with detection of the allele-specific extension products by matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI-TOF; Sequenom, San Diego, CA).34 Genotyping of the validation cohort was performed using the Illumina Beadarray platform.35 This is based upon the assembly of up to 50 000 beads, each bead about 3 μm in diameter and representing a different oligonucleotide probe sequence. The beads are placed in microwells located at the end of fiber-optic bundles and serve as the substrate for a high-density microarray.35 Only genes that were considered to exhibit evidence of significant association with the airflow decline phenotype in the discovery cohort were analyzed in the validation cohort. Data quality was assessed by random duplicate samples and/or sex discrimination.
Airway epithelial cell analyses
Cells and culture. A549 cells (American Type Culture Collection [ATCC]; Manassas, VA), a human epithelial cell line isolated from a lung carcinoma, and small airway epithelial cells (SAECs; Cambrex, East Rutherford, NJ), isolated from distal small airways (less than 1 mm diameter) immediately after death from humans without known lung disease, were cultured according to the manufacturer's protocol and subcultured in 6-well plates until 80% confluence. Whole blood drawn from healthy volunteers was anticoagulated with sodium citrate (3 mg/mL) and incubated in a CO2 incubator with lipopolysaccharide (10 ng/mL) for 1 hour at 37°C. The white cell layer was separated by centrifugation and used as positive controls.
RNA isolation and RT-PCR. Total RNA was isolated using the RNA easy Mini Kit (Qiagen) and treated with the RNAse free DNAse kit (Quiagen) according to the manufacturer's protocol. RNA was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA), and PCR was carried out using Platinum Taq Polymerase (Invitrogen). Reverse transcription-PCR (RT-PCR) was carried out using a human BPI sense (5′-GCACCTGTTCCTGATGGG-3′) and antisense (5′-AGCACAAATGGAAATTTCTTG-3′) primer set yielding a 255 bp product.36 An 18S primer set yielding a 314 bp product was used as a control.37 The amplified products were separated on TBE-1% agarose gels and detected with ethidium bromide. As a positive control, RNA was isolated from the buffy coat of whole blood stimulated with lipopolysaccharide and analyzed for BPI mRNA and 18S expression.
Flow cytometry. A549 cells and SAECs were trypsinized and fixed in Cytofix/Cytoperm Buffer (PharMingen, San Diego, CA) according to manufacturer's protocol. White blood cells were treated with red blood cell (RBC) lysis buffer (Sigma, St Louis, MO), fixed in Cytofix/Cytoperm Buffer, and frozen at -70°C in 10% DMSO/90% fetal bovine serum. After 5 minutes of thawing, white blood cells were refixed for 5 minutes. Cell samples were incubated with monoclonal antibody to BPI (4E3, a gift from Dr Wim A. Buurman38 ), mouse IgG control (R&D Systems, Minneapolis, MN), and/or mouse anti-human anticytokeratin monoclonal antibody (Dako, Carpinteria, CA), followed by FITC-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, Bar Harbor, ME), resuspended in FACS buffer, and analyzed by flow cytometry.
Statistical methods
The Pearson χ2 test and univariate linear regression were used to assess categorical and continuous clinical variables, respectively. All patient and donor tagSNP haplotypes were analyzed using the Hplus software.39 This approach evaluates phenotypic association with gene-based haplotypes while incorporating uncertainties due to unphased genotype data and adjustment for covariates that have been previously determined to be associated with development of significant airflow decline. Patient and donor haplotypes were assessed in independent multivariate models. Genes with 2 or more haplotypes that were significantly associated with the airflow decline phenotype in the discovery cohort, identified by a P value of less than .05, were reassessed in the validation cohort.
Results
Population characteristics
There were 363 patient donor pairs in the discovery cohort and 209 patient donor pairs in the validation cohort, with 133 and 66 patients who developed significant airflow decline, respectively. Overall, the clinical characteristics of these cohorts were similar, except the patients in the discovery cohort were older, more likely to receive a related matched donor, and were followed for a shorter time (Table 1).
Comparison of the clinical characteristics of the discovery and validation cohorts
Characteristics . | Discovery cohort . | Validation cohort . | P . |
|---|---|---|---|
| No. patients | 363 | 209 | |
| Median age ± standard deviation, y | 37.3 ± 10.5 | 34.6 ± 13.3 | .009 |
| Patient sex, no. (%) | .589 | ||
| Male | 208 (57) | 125 (60) | |
| Female | 155 (43) | 84 (40) | |
| Donor sex, no. (%) | .967 | ||
| Male | 221 (61) | 128 (61) | |
| Female | 142 (39) | 81 (39) | |
| Patient race, no. (%) | .524 | ||
| White | 320 (88) | 188 (90) | |
| Nonwhite* | 43 (12) | 21 (10) | |
| Reason for transplantation, no. (%) | .781 | ||
| Low-risk disease | 173 (48) | 97 (46) | |
| Intermediate-risk disease | 100 (27) | 63 (30) | |
| High-risk disease | 90 (25) | 49 (24) | |
| Total body irradiation, no. (%) | .535 | ||
| No | 73 (20) | 35 (17) | |
| Yes | 290 (80) | 174 (83) | |
| Pretransplantation FEV1, no. (%) | .1 | ||
| More than 80% | 330 (91) | 178 (85) | |
| 70% to 80% | 19 (5) | 22 (10) | |
| 60% to 70% | 11 (3) | 8 (4) | |
| Less than 60% | 3 (1) | 1 (< 1) | |
| GVHD no. (%) | .51 | ||
| No acute or chronic | 141 (39) | 74 (35) | |
| Acute only | 0 (0) | 0 (0) | |
| De novo chronic | 23 (6) | 12 (6) | |
| Quiescent-onset chronic | 187 (43) | 91 (43) | |
| Progressive-onset chronic | 42 (12) | 33 (16) | |
| Median follow-up time ± standard deviation, y | 3.1 ± 2.8 | 6.0 ± 4.5 | < .001 |
| Patients with significant airflow decline, no. (%) | 133 (37) | 66 (32) | .236 |
Characteristics . | Discovery cohort . | Validation cohort . | P . |
|---|---|---|---|
| No. patients | 363 | 209 | |
| Median age ± standard deviation, y | 37.3 ± 10.5 | 34.6 ± 13.3 | .009 |
| Patient sex, no. (%) | .589 | ||
| Male | 208 (57) | 125 (60) | |
| Female | 155 (43) | 84 (40) | |
| Donor sex, no. (%) | .967 | ||
| Male | 221 (61) | 128 (61) | |
| Female | 142 (39) | 81 (39) | |
| Patient race, no. (%) | .524 | ||
| White | 320 (88) | 188 (90) | |
| Nonwhite* | 43 (12) | 21 (10) | |
| Reason for transplantation, no. (%) | .781 | ||
| Low-risk disease | 173 (48) | 97 (46) | |
| Intermediate-risk disease | 100 (27) | 63 (30) | |
| High-risk disease | 90 (25) | 49 (24) | |
| Total body irradiation, no. (%) | .535 | ||
| No | 73 (20) | 35 (17) | |
| Yes | 290 (80) | 174 (83) | |
| Pretransplantation FEV1, no. (%) | .1 | ||
| More than 80% | 330 (91) | 178 (85) | |
| 70% to 80% | 19 (5) | 22 (10) | |
| 60% to 70% | 11 (3) | 8 (4) | |
| Less than 60% | 3 (1) | 1 (< 1) | |
| GVHD no. (%) | .51 | ||
| No acute or chronic | 141 (39) | 74 (35) | |
| Acute only | 0 (0) | 0 (0) | |
| De novo chronic | 23 (6) | 12 (6) | |
| Quiescent-onset chronic | 187 (43) | 91 (43) | |
| Progressive-onset chronic | 42 (12) | 33 (16) | |
| Median follow-up time ± standard deviation, y | 3.1 ± 2.8 | 6.0 ± 4.5 | < .001 |
| Patients with significant airflow decline, no. (%) | 133 (37) | 66 (32) | .236 |
FEV1 expressed as a percent of predicted normal values.
Nonwhite includes African American, Asian, Hispanic, and Native American
TagSNP allelic analyses
From the 15 innate immunity genes, 69 tagSNPs were selected for analysis (summarized in Table S1, available on the Blood website; see the Supplemental Tables link at the top of the online articles). Allelic analyses in multivariate models demonstrated there were 4 patient tagSNP genotypes that had a P value of less than .05 (Table S2). These were located on the IL-1β gene: tagSNP 5277 (P = .005) and tagSNP 8234 (P = .005); and on the BPI gene: tagSNP 33065 (P = .038) and tagSNP 36045 (P = .025). There were also 2 donor tagSNP genotypes that had a P value of less than .05. One was located on the BPI gene: tagSNP 36045 (P = .008); the other was located on the LBP gene: tagSNP 541 (P = .017).
TagSNP haplotype analyses
In 15 genes, 93 tagSNP haplotypes were inferred among the patients and donors using these 69 tagSNPs (Table S3). There were 3 genes with one tagSNP haplotype each that were associated with airflow decline at a P value of less than .05 (donor TIRAP haplotype, P = .011; patient IL-1β haplotype, P = .036; and patient LBP haplotype, P = .014), but only the BPI gene met the criteria for assessment in the validation cohort.
Patient BPI TagSNP haplotypes
There were 14 patient BPI tagSNP haplotypes in the discovery cohort. Haplotypes 6 and 13 were significantly associated with a 3-fold increase in the risk of developing the airflow decline phenotype (Table 2; P = .004 and .038, respectively), with 3 additional haplotypes that trended toward significant association with an increase in the risk of developing the phenotype (haplotype 10, P = .095; haplotype 11, P = .077; haplotype 14, P = .063). Similar associations were evident in the subanalysis restricted to white patients (Table 2; haplotype 6, P = .011; haplotype 13, P = .051). In the validation cohort, the association between the patient BPI haplotypes and the airflow decline phenotype was confirmed with 4 BPI haplotypes (Table 2; haplotype 2, P = .013; haplotype 7, P = .027; haplotype 9, P = .025; haplotype 11, P = .03). Although some of the patient haplotypes in the discovery and validation cohorts were the same, the haplotypes with a P value of less than .05 were similar but not the same across these cohorts.
Patient BPI haplotypes in the discovery and validation cohorts and their associated risk of significant airflow decline after transplantation
. | tagSNPs . | . | . | . | . | . | . | . | Haplotype frequencies . | . | . | . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype . | 2738 . | 7258 . | 9083 . | 10214 . | 17016 . | 23356 . | 33065 . | 36045 . | Controls . | Cases . | Odds ratio (95% CI) . | P values . | ||||||||||||
| Discovery cohort: all patients | ||||||||||||||||||||||||
| 1 | C | G | G | A | A | T | G | G | 0.251 | 0.183 | Referent | — | ||||||||||||
| 2 | G | A | G | A | G | C | A | A | 0.137 | 0.123 | 1.22 (0.68-2.21) | .506 | ||||||||||||
| 3 | C | A | G | A | G | C | A | A | 0.095 | 0.105 | 1.55 (0.84-2.84) | .157 | ||||||||||||
| 4 | C | A | G | A | G | C | G | G | 0.081 | 0.083 | 1.27 (0.64-2.5) | .491 | ||||||||||||
| 5 | C | G | C | A | A | C | G | G | 0.06 | 0.06 | 1.48 (0.67-3.28) | .333 | ||||||||||||
| 6 | C | G | G | A | A | T | A | A | 0.054 | 0.102 | 3.1 (1.44-6.68) | .004 | ||||||||||||
| 7 | C | G | C | A | A | T | G | G | 0.041 | 0.025 | 1.03 (0.35-3.02) | .956 | ||||||||||||
| 8 | C | G | G | G | G | C | A | G | 0.041 | 0.042 | 1.22 (0.54-2.77) | .636 | ||||||||||||
| 9 | C | G | G | G | G | C | A | A | 0.035 | 0.041 | 1.66 (0.64-4.19) | .281 | ||||||||||||
| 10 | C | G | G | G | G | C | G | G | 0.026 | 0.031 | 2.31 (0.86-6.18) | .095 | ||||||||||||
| 11 | C | A | G | A | G | C | A | G | 0.024 | 0.035 | 2.68 (0.9-8.0) | .077 | ||||||||||||
| 12 | C | G | G | A | A | C | A | G | 0.019 | 0.006 | 0.27 (0.02-3.99) | .343 | ||||||||||||
| 13 | C | G | G | A | A | C | A | A | 0.018 | 0.041 | 3.28 (1.07-10.07) | .038 | ||||||||||||
| 14 | G | A | G | A | G | C | G | G | 0.016 | 0.034 | 3.0 (0.94-9.54) | .063 | ||||||||||||
| Discovery cohort: white patients only | ||||||||||||||||||||||||
| 1 | C | G | G | A | A | T | G | G | 0.247 | 0.183 | Referent | — | ||||||||||||
| 2 | G | A | G | A | G | C | A | A | 0.143 | 0.128 | 1.2 (0.65-2.2) | .568 | ||||||||||||
| 3 | C | A | G | A | G | C | A | A | 0.098 | 0.108 | 1.67 (0.9-3.1) | .102 | ||||||||||||
| 4 | C | A | G | A | G | C | G | G | 0.079 | 0.083 | 1.25 (0.61-2.55) | .536 | ||||||||||||
| 5 | C | G | C | A | A | C | G | G | 0.066 | 0.071 | 1.61 (0.75-3.47) | .225 | ||||||||||||
| 6 | C | G | G | A | A | T | A | A | 0.058 | 0.099 | 2.77 (1.27-6.07) | .011 | ||||||||||||
| 7 | C | G | G | G | G | C | A | G | 0.047 | 0.044 | 1.15 (0.5-2.68) | .741 | ||||||||||||
| 8 | C | G | C | A | A | T | G | G | 0.041 | 0.023 | 0.9 (0.29-2.82) | .861 | ||||||||||||
| 9 | C | A | G | A | G | C | A | G | 0.029 | 0.036 | 1.72 (0.58-5.15) | .329 | ||||||||||||
| 10 | C | G | G | G | G | C | A | A | 0.029 | 0.034 | 1.51 (0.5-4.54) | .462 | ||||||||||||
| 11 | C | G | G | G | G | C | G | G | 0.026 | 0.028 | 2.14 (0.77-5.93) | .144 | ||||||||||||
| 12 | C | G | G | A | A | C | A | G | 0.022 | 0.006 | 0.31 (0.02-4.24) | .382 | ||||||||||||
| 13 | G | A | G | A | G | C | G | G | 0.021 | 0.047 | 3.07 (0.99-9.5) | .051 | ||||||||||||
| 14 | C | G | G | A | A | C | A | A | 0.017 | 0.04 | 2.95 (0.89-9.85) | .078 | ||||||||||||
| Validation cohort: all patients | ||||||||||||||||||||||||
| 1 | C | G | G | A | A | T | G | G | 0.238 | 0.133 | Referent | — | ||||||||||||
| 2 | G | A | G | A | G | C | A | A | 0.12 | 0.191 | 2.83 (1.25-6.38) | .013 | ||||||||||||
| 3 | C | A | G | A | G | C | G | G | 0.098 | 0.103 | 1.71 (0.57-5.14) | .343 | ||||||||||||
| 4 | C | A | G | A | G | C | A | A | 0.088 | 0.037 | 1.0 (0.36-2.78) | .994 | ||||||||||||
| 5 | C | G | G | A | A | T | A | A | 0.074 | 0.043 | 1.24 (0.34-4.5) | .742 | ||||||||||||
| 6 | C | G | G | A | A | C | A | G | 0.046 | 0.041 | 1.41 (0.35-5.72) | .629 | ||||||||||||
| 7 | C | G | G | G | G | C | A | A | 0.043 | 0.08 | 3.28 (1.14-9.44) | .027 | ||||||||||||
| 8 | C | G | C | A | A | C | G | G | 0.036 | 0.064 | 1.82 (0.52-6.35) | .345 | ||||||||||||
| 9 | C | G | C | A | A | T | G | G | 0.031 | 0.04 | 4.57 (1.16-17.95) | .03 | ||||||||||||
| 10 | G | A | G | A | G | C | G | G | 0.025 | 0.036 | 2.29 (0.51-10.33) | .282 | ||||||||||||
| 11 | C | G | G | G | G | C | A | G | 0.018 | 0.043 | 5.5 (1.24-24.38) | .025 | ||||||||||||
. | tagSNPs . | . | . | . | . | . | . | . | Haplotype frequencies . | . | . | . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype . | 2738 . | 7258 . | 9083 . | 10214 . | 17016 . | 23356 . | 33065 . | 36045 . | Controls . | Cases . | Odds ratio (95% CI) . | P values . | ||||||||||||
| Discovery cohort: all patients | ||||||||||||||||||||||||
| 1 | C | G | G | A | A | T | G | G | 0.251 | 0.183 | Referent | — | ||||||||||||
| 2 | G | A | G | A | G | C | A | A | 0.137 | 0.123 | 1.22 (0.68-2.21) | .506 | ||||||||||||
| 3 | C | A | G | A | G | C | A | A | 0.095 | 0.105 | 1.55 (0.84-2.84) | .157 | ||||||||||||
| 4 | C | A | G | A | G | C | G | G | 0.081 | 0.083 | 1.27 (0.64-2.5) | .491 | ||||||||||||
| 5 | C | G | C | A | A | C | G | G | 0.06 | 0.06 | 1.48 (0.67-3.28) | .333 | ||||||||||||
| 6 | C | G | G | A | A | T | A | A | 0.054 | 0.102 | 3.1 (1.44-6.68) | .004 | ||||||||||||
| 7 | C | G | C | A | A | T | G | G | 0.041 | 0.025 | 1.03 (0.35-3.02) | .956 | ||||||||||||
| 8 | C | G | G | G | G | C | A | G | 0.041 | 0.042 | 1.22 (0.54-2.77) | .636 | ||||||||||||
| 9 | C | G | G | G | G | C | A | A | 0.035 | 0.041 | 1.66 (0.64-4.19) | .281 | ||||||||||||
| 10 | C | G | G | G | G | C | G | G | 0.026 | 0.031 | 2.31 (0.86-6.18) | .095 | ||||||||||||
| 11 | C | A | G | A | G | C | A | G | 0.024 | 0.035 | 2.68 (0.9-8.0) | .077 | ||||||||||||
| 12 | C | G | G | A | A | C | A | G | 0.019 | 0.006 | 0.27 (0.02-3.99) | .343 | ||||||||||||
| 13 | C | G | G | A | A | C | A | A | 0.018 | 0.041 | 3.28 (1.07-10.07) | .038 | ||||||||||||
| 14 | G | A | G | A | G | C | G | G | 0.016 | 0.034 | 3.0 (0.94-9.54) | .063 | ||||||||||||
| Discovery cohort: white patients only | ||||||||||||||||||||||||
| 1 | C | G | G | A | A | T | G | G | 0.247 | 0.183 | Referent | — | ||||||||||||
| 2 | G | A | G | A | G | C | A | A | 0.143 | 0.128 | 1.2 (0.65-2.2) | .568 | ||||||||||||
| 3 | C | A | G | A | G | C | A | A | 0.098 | 0.108 | 1.67 (0.9-3.1) | .102 | ||||||||||||
| 4 | C | A | G | A | G | C | G | G | 0.079 | 0.083 | 1.25 (0.61-2.55) | .536 | ||||||||||||
| 5 | C | G | C | A | A | C | G | G | 0.066 | 0.071 | 1.61 (0.75-3.47) | .225 | ||||||||||||
| 6 | C | G | G | A | A | T | A | A | 0.058 | 0.099 | 2.77 (1.27-6.07) | .011 | ||||||||||||
| 7 | C | G | G | G | G | C | A | G | 0.047 | 0.044 | 1.15 (0.5-2.68) | .741 | ||||||||||||
| 8 | C | G | C | A | A | T | G | G | 0.041 | 0.023 | 0.9 (0.29-2.82) | .861 | ||||||||||||
| 9 | C | A | G | A | G | C | A | G | 0.029 | 0.036 | 1.72 (0.58-5.15) | .329 | ||||||||||||
| 10 | C | G | G | G | G | C | A | A | 0.029 | 0.034 | 1.51 (0.5-4.54) | .462 | ||||||||||||
| 11 | C | G | G | G | G | C | G | G | 0.026 | 0.028 | 2.14 (0.77-5.93) | .144 | ||||||||||||
| 12 | C | G | G | A | A | C | A | G | 0.022 | 0.006 | 0.31 (0.02-4.24) | .382 | ||||||||||||
| 13 | G | A | G | A | G | C | G | G | 0.021 | 0.047 | 3.07 (0.99-9.5) | .051 | ||||||||||||
| 14 | C | G | G | A | A | C | A | A | 0.017 | 0.04 | 2.95 (0.89-9.85) | .078 | ||||||||||||
| Validation cohort: all patients | ||||||||||||||||||||||||
| 1 | C | G | G | A | A | T | G | G | 0.238 | 0.133 | Referent | — | ||||||||||||
| 2 | G | A | G | A | G | C | A | A | 0.12 | 0.191 | 2.83 (1.25-6.38) | .013 | ||||||||||||
| 3 | C | A | G | A | G | C | G | G | 0.098 | 0.103 | 1.71 (0.57-5.14) | .343 | ||||||||||||
| 4 | C | A | G | A | G | C | A | A | 0.088 | 0.037 | 1.0 (0.36-2.78) | .994 | ||||||||||||
| 5 | C | G | G | A | A | T | A | A | 0.074 | 0.043 | 1.24 (0.34-4.5) | .742 | ||||||||||||
| 6 | C | G | G | A | A | C | A | G | 0.046 | 0.041 | 1.41 (0.35-5.72) | .629 | ||||||||||||
| 7 | C | G | G | G | G | C | A | A | 0.043 | 0.08 | 3.28 (1.14-9.44) | .027 | ||||||||||||
| 8 | C | G | C | A | A | C | G | G | 0.036 | 0.064 | 1.82 (0.52-6.35) | .345 | ||||||||||||
| 9 | C | G | C | A | A | T | G | G | 0.031 | 0.04 | 4.57 (1.16-17.95) | .03 | ||||||||||||
| 10 | G | A | G | A | G | C | G | G | 0.025 | 0.036 | 2.29 (0.51-10.33) | .282 | ||||||||||||
| 11 | C | G | G | G | G | C | A | G | 0.018 | 0.043 | 5.5 (1.24-24.38) | .025 | ||||||||||||
For discovery cohort, n = 363; for white patients, n = 320; for validation cohort, n = 209. Haplotypes with a P ≤ .05 are shown in italics. All haplotypes were analyzed in multivariate models that included age at transplantation, pretransplantation FEV1, extent of GVHD and duration of follow-up as covariates.
CI indicates confidence interval; —, no P value for the group.
Donor BPI tagSNP haplotypes
There were 15 donor BPI tagSNP haplotypes in the discovery cohort. Haplotypes 2 and 13 were significantly associated with a 2- to 3-fold increase in the risk of developing the airflow decline phenotype (Table 3; P = .012 and .009, respectively). Haplotypes 4 and 5 also trended toward significant association with development of the phenotype (P = .081 and .07, respectively). In the validation cohort, the association between the BPI gene and the airflow decline phenotype was confirmed with 5 significant haplotypes (Table 3; haplotype 2, P = .041; haplotype 4, P = .028; haplotype 5, P = .043; haplotype 9, P = .029; haplotype 11, P = .033). Haplotype 6 also trended toward significant association with the phenotype (P = .077). As with the patient haplotypes, some of the donor haplotypes in the discovery and validation cohorts were the same. However, the haplotypes with a P value of .05 or less were similar but not the same across these cohorts.
Donor BPI haplotypes in the discovery and validation cohorts and their associated risk of significant airflow decline after transplantation
. | tagSNPs . | . | . | . | . | . | . | . | Haplotype frequencies . | . | . | . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype . | 2738 . | 7258 . | 9083 . | 10214 . | 17016 . | 23356 . | 33065 . | 36045 . | Controls . | Cases . | Odds ratio (95% CI) . | P values . | ||||||||||||
| Discovery cohort: all donors (N = 363) | ||||||||||||||||||||||||
| 1 | C | G | G | A | A | T | G | G | 0.224 | 0.175 | Referent | — | ||||||||||||
| 2 | G | A | G | A | G | C | A | A | 0.119 | 0.169 | 2.16 (1.18-3.93) | .012 | ||||||||||||
| 3 | C | A | G | A | G | C | A | A | 0.114 | 0.117 | 1.32 (0.71-2.45) | .379 | ||||||||||||
| 4 | C | A | G | A | G | C | G | G | 0.075 | 0.1 | 1.93 (0.92-4.04) | .081 | ||||||||||||
| 5 | C | G | G | A | A | T | A | A | 0.059 | 0.072 | 2.13 (0.94-4.81) | .07 | ||||||||||||
| 6 | C | G | C | A | A | C | G | G | 0.04 | 0.04 | 1.87 (0.73-4.78) | .189 | ||||||||||||
| 7 | C | G | G | G | G | C | A | G | 0.04 | 0.035 | 1.03 (0.37-2.89) | .959 | ||||||||||||
| 8 | G | A | G | A | G | C | G | G | 0.036 | 0.035 | 1.24 (0.37-4.16) | .726 | ||||||||||||
| 9 | C | A | G | A | G | C | A | G | 0.036 | 0.023 | 0.87 (0.1-3.87) | .854 | ||||||||||||
| 10 | C | G | G | A | A | C | A | G | 0.037 | 0.014 | 0.5 (0.13-1.93) | .312 | ||||||||||||
| 11 | C | G | G | G | G | C | G | G | 0.03 | 0.017 | 0.95 (0.14-3.7) | .938 | ||||||||||||
| 12 | C | G | C | A | A | T | G | G | 0.024 | 0.038 | 2.4 (0.76-7.55) | .134 | ||||||||||||
| 13 | C | G | G | G | G | C | A | A | 0.021 | 0.04 | 2.83 (1.39-10.51) | .009 | ||||||||||||
| 14 | C | G | G | A | A | C | A | A | 0.015 | 0.023 | 2.42 (0.71-8.29) | .159 | ||||||||||||
| 15 | G | A | G | A | G | C | A | G | 0.013 | 0.016 | 1.44 (0.22-9.48) | .705 | ||||||||||||
| Validation cohort: all donors | ||||||||||||||||||||||||
| 1 | G | G | G | A | A | T | A | A | 0.075 | 0.019 | Referent | — | ||||||||||||
| 2 | G | G | G | A | A | T | G | G | 0.217 | 0.284 | 5.99 (1.08-33.23) | .041 | ||||||||||||
| 3 | G | A | G | A | G | C | A | A | 0.129 | 0.085 | 2.68 (0.4-17.88) | .31 | ||||||||||||
| 4 | G | A | G | A | G | C | G | G | 0.096 | 0.133 | 6.96 (1.24-39.06) | .028 | ||||||||||||
| 5 | C | A | G | A | G | C | A | A | 0.091 | 0.107 | 6.36 (1.06-38.32) | .043 | ||||||||||||
| 6 | G | G | G | G | G | C | A | A | 0.05 | 0.055 | 5.84 (0.82-41.38) | .077 | ||||||||||||
| 7 | G | A | G | A | G | C | A | G | 0.05 | 0.033 | 3.28 (0.49-22.05) | .222 | ||||||||||||
| 8 | C | A | G | A | G | C | G | G | 0.039 | 0.042 | 4.68 (0.55-39.98) | .158 | ||||||||||||
| 9 | G | G | C | A | A | T | G | G | 0.038 | 0.055 | 8.57 (1.25-58.72) | .029 | ||||||||||||
| 10 | G | G | G | G | G | C | A | G | 0.038 | 0.027 | 3.21 (0.37-27.78) | .289 | ||||||||||||
| 11 | G | G | C | A | A | C | G | G | 0.035 | 0.06 | 8.0 (1.19-53.89) | .033 | ||||||||||||
. | tagSNPs . | . | . | . | . | . | . | . | Haplotype frequencies . | . | . | . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype . | 2738 . | 7258 . | 9083 . | 10214 . | 17016 . | 23356 . | 33065 . | 36045 . | Controls . | Cases . | Odds ratio (95% CI) . | P values . | ||||||||||||
| Discovery cohort: all donors (N = 363) | ||||||||||||||||||||||||
| 1 | C | G | G | A | A | T | G | G | 0.224 | 0.175 | Referent | — | ||||||||||||
| 2 | G | A | G | A | G | C | A | A | 0.119 | 0.169 | 2.16 (1.18-3.93) | .012 | ||||||||||||
| 3 | C | A | G | A | G | C | A | A | 0.114 | 0.117 | 1.32 (0.71-2.45) | .379 | ||||||||||||
| 4 | C | A | G | A | G | C | G | G | 0.075 | 0.1 | 1.93 (0.92-4.04) | .081 | ||||||||||||
| 5 | C | G | G | A | A | T | A | A | 0.059 | 0.072 | 2.13 (0.94-4.81) | .07 | ||||||||||||
| 6 | C | G | C | A | A | C | G | G | 0.04 | 0.04 | 1.87 (0.73-4.78) | .189 | ||||||||||||
| 7 | C | G | G | G | G | C | A | G | 0.04 | 0.035 | 1.03 (0.37-2.89) | .959 | ||||||||||||
| 8 | G | A | G | A | G | C | G | G | 0.036 | 0.035 | 1.24 (0.37-4.16) | .726 | ||||||||||||
| 9 | C | A | G | A | G | C | A | G | 0.036 | 0.023 | 0.87 (0.1-3.87) | .854 | ||||||||||||
| 10 | C | G | G | A | A | C | A | G | 0.037 | 0.014 | 0.5 (0.13-1.93) | .312 | ||||||||||||
| 11 | C | G | G | G | G | C | G | G | 0.03 | 0.017 | 0.95 (0.14-3.7) | .938 | ||||||||||||
| 12 | C | G | C | A | A | T | G | G | 0.024 | 0.038 | 2.4 (0.76-7.55) | .134 | ||||||||||||
| 13 | C | G | G | G | G | C | A | A | 0.021 | 0.04 | 2.83 (1.39-10.51) | .009 | ||||||||||||
| 14 | C | G | G | A | A | C | A | A | 0.015 | 0.023 | 2.42 (0.71-8.29) | .159 | ||||||||||||
| 15 | G | A | G | A | G | C | A | G | 0.013 | 0.016 | 1.44 (0.22-9.48) | .705 | ||||||||||||
| Validation cohort: all donors | ||||||||||||||||||||||||
| 1 | G | G | G | A | A | T | A | A | 0.075 | 0.019 | Referent | — | ||||||||||||
| 2 | G | G | G | A | A | T | G | G | 0.217 | 0.284 | 5.99 (1.08-33.23) | .041 | ||||||||||||
| 3 | G | A | G | A | G | C | A | A | 0.129 | 0.085 | 2.68 (0.4-17.88) | .31 | ||||||||||||
| 4 | G | A | G | A | G | C | G | G | 0.096 | 0.133 | 6.96 (1.24-39.06) | .028 | ||||||||||||
| 5 | C | A | G | A | G | C | A | A | 0.091 | 0.107 | 6.36 (1.06-38.32) | .043 | ||||||||||||
| 6 | G | G | G | G | G | C | A | A | 0.05 | 0.055 | 5.84 (0.82-41.38) | .077 | ||||||||||||
| 7 | G | A | G | A | G | C | A | G | 0.05 | 0.033 | 3.28 (0.49-22.05) | .222 | ||||||||||||
| 8 | C | A | G | A | G | C | G | G | 0.039 | 0.042 | 4.68 (0.55-39.98) | .158 | ||||||||||||
| 9 | G | G | C | A | A | T | G | G | 0.038 | 0.055 | 8.57 (1.25-58.72) | .029 | ||||||||||||
| 10 | G | G | G | G | G | C | A | G | 0.038 | 0.027 | 3.21 (0.37-27.78) | .289 | ||||||||||||
| 11 | G | G | C | A | A | C | G | G | 0.035 | 0.06 | 8.0 (1.19-53.89) | .033 | ||||||||||||
For discovery cohort, n = 363; for validation cohort, n = 209. Haplotypes with a P ≤ 05 are shown in italics. All haplotypes were analyzed in multivariate models that included age at transplantation, pretransplantation; FEV1, extent of GVHD, and duration of follow-up as covariates.
— indicates no P value for the group.
Lipopolysaccharide binding protein TagSNP haplotypes
Among the patient tagSNP haplotypes in the discovery cohort, the LBP gene, which is immediately adjacent to the BPI gene on chromosome 20, also had 1 haplotype that was significantly associated with a 2.8-fold increase in the risk of developing significant airflow decline after transplantation (Table 4; haplotype 8, P = .014). There were 2 additional patient LBP haplotypes that trended toward significant association with the phenotype (haplotype 2, P = .091; haplotype 10, P = .069). Subanalysis restricted to white patients revealed the same significant haplotype (Table 4; haplotype 6, P = .022). Among donors, one LBP haplotype trended toward significant association with the phenotype (Table 4; haplotype 6, P = .055), but no haplotypes had a P value of less than .05. Given the proximity of the LBP gene to the BPI gene and the number of haplotypes that trended toward significant association, LBP was also assessed in the validation cohort. However, no association was observed in the validation cohort.
Patient LBP haplotypes in the discovery cohort and their associated risk of significant airflow decline after transplantation
. | tagSNPs . | . | . | . | . | . | . | . | . | . | Haplotype frequencies . | . | . | . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype . | 541 . | 1598 . | 1756 . | 5584 . | 7400 . | 13506 . | 17002 . | 25276 . | 28559 . | 33158 . | Controls . | Cases . | Odds ratio (95% CI) . | P values . | ||||||||||||||
| All patients | ||||||||||||||||||||||||||||
| 1 | C | A | A | G | T | G | G | C | T | T | 0.325 | 0.265 | Referent | — | ||||||||||||||
| 2 | T | A | A | G | C | G | A | C | T | C | 0.224 | 0.259 | 1.44 (0.94-2.19) | .091 | ||||||||||||||
| 3 | C | A | A | G | C | G | A | C | T | C | 0.061 | 0.06 | 1.42 (0.72-2.79) | .312 | ||||||||||||||
| 4 | C | A | A | G | T | G | G | C | T | C | 0.051 | 0.05 | 1.27 (0.54-2.95) | .586 | ||||||||||||||
| 5 | C | T | G | G | C | G | A | C | T | C | 0.051 | 0.05 | 1.14 (0.55-2.37) | .724 | ||||||||||||||
| 6 | C | A | A | G | T | G | G | C | A | T | 0.037 | 0.04 | 1.38 (0.61-3.13) | .445 | ||||||||||||||
| 7 | C | A | A | G | T | G | A | C | T | C | 0.037 | 0.01 | 0.55 (0.19-1.54) | .254 | ||||||||||||||
| 8 | T | A | A | G | T | T | A | T | T | C | 0.035 | 0.077 | 2.8 (1.24-6.34) | .014 | ||||||||||||||
| 9 | C | T | A | G | T | G | A | C | T | C | 0.019 | 0.015 | 1.19 (0.27-5.3) | .824 | ||||||||||||||
| 10 | C | T | G | G | C | G | A | C | T | T | 0.008 | 0.028 | 4.8 (0.88-26.11) | .069 | ||||||||||||||
| White patients only | ||||||||||||||||||||||||||||
| 1 | C | A | A | G | T | G | G | C | T | T | 0.337 | 0.274 | Referent | — | ||||||||||||||
| 2 | T | A | A | G | C | G | A | C | T | C | 0.225 | 0.263 | 1.38 (0.89-2.13) | .149 | ||||||||||||||
| 3 | C | A | A | G | C | G | A | C | T | C | 0.063 | 0.067 | 1.52 (0.77-3.02) | .23 | ||||||||||||||
| 4 | C | T | G | G | C | G | A | C | T | C | 0.05 | 0.051 | 1.08 (0.52-2.24) | .838 | ||||||||||||||
| 5 | C | A | A | G | T | G | G | C | T | C | 0.05 | 0.047 | 1.14 (0.46-2.88) | .775 | ||||||||||||||
| 6 | T | A | A | G | T | T | A | T | T | C | 0.04 | 0.083 | 2.62 (1.15-5.94) | .022 | ||||||||||||||
| 7 | C | A | A | G | T | G | A | C | T | C | 0.037 | 0.011 | 0.71 (0.26-1.95) | .509 | ||||||||||||||
| 8 | C | A | A | G | T | G | G | C | A | T | 0.036 | 0.038 | 1.24 (0.52-2.95) | .624 | ||||||||||||||
| Donors | ||||||||||||||||||||||||||||
| 1 | C | A | A | G | T | G | G | C | T | T | 0.299 | 0.264 | Referent | — | ||||||||||||||
| 2 | T | A | A | G | C | G | A | C | T | C | 0.231 | 0.273 | 1.32 (0.87-1.99) | .194 | ||||||||||||||
| 3 | C | A | A | G | C | G | A | C | T | C | 0.07 | 0.047 | 0.76 (0.36-1.58) | .458 | ||||||||||||||
| 4 | C | A | A | G | T | G | G | C | T | C | 0.057 | 0.06 | 1.22 (0.61-2.46) | .571 | ||||||||||||||
| 5 | C | A | A | G | T | G | G | C | A | T | 0.046 | 0.042 | 1.05 (0.43-2.6) | .913 | ||||||||||||||
| 6 | C | A | A | G | T | G | A | C | T | C | 0.036 | 0 | 0.28 (0.06-1.33) | .11 | ||||||||||||||
| 7 | T | A | A | G | T | T | A | T | T | C | 0.035 | 0.065 | 2.2 (0.98-4.91) | .055 | ||||||||||||||
| 8 | C | T | G | G | C | G | A | C | T | C | 0.035 | 0.053 | 1.94 (0.82-4.59) | .129 | ||||||||||||||
| 9 | C | T | A | G | T | G | A | C | T | C | 0.024 | 0.018 | 0.94 (0.26-3.38) | .921 | ||||||||||||||
| 10 | T | A | G | A | C | G | G | C | T | C | 0.02 | 0.014 | 0.71 (0.18-2.88) | .635 | ||||||||||||||
. | tagSNPs . | . | . | . | . | . | . | . | . | . | Haplotype frequencies . | . | . | . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype . | 541 . | 1598 . | 1756 . | 5584 . | 7400 . | 13506 . | 17002 . | 25276 . | 28559 . | 33158 . | Controls . | Cases . | Odds ratio (95% CI) . | P values . | ||||||||||||||
| All patients | ||||||||||||||||||||||||||||
| 1 | C | A | A | G | T | G | G | C | T | T | 0.325 | 0.265 | Referent | — | ||||||||||||||
| 2 | T | A | A | G | C | G | A | C | T | C | 0.224 | 0.259 | 1.44 (0.94-2.19) | .091 | ||||||||||||||
| 3 | C | A | A | G | C | G | A | C | T | C | 0.061 | 0.06 | 1.42 (0.72-2.79) | .312 | ||||||||||||||
| 4 | C | A | A | G | T | G | G | C | T | C | 0.051 | 0.05 | 1.27 (0.54-2.95) | .586 | ||||||||||||||
| 5 | C | T | G | G | C | G | A | C | T | C | 0.051 | 0.05 | 1.14 (0.55-2.37) | .724 | ||||||||||||||
| 6 | C | A | A | G | T | G | G | C | A | T | 0.037 | 0.04 | 1.38 (0.61-3.13) | .445 | ||||||||||||||
| 7 | C | A | A | G | T | G | A | C | T | C | 0.037 | 0.01 | 0.55 (0.19-1.54) | .254 | ||||||||||||||
| 8 | T | A | A | G | T | T | A | T | T | C | 0.035 | 0.077 | 2.8 (1.24-6.34) | .014 | ||||||||||||||
| 9 | C | T | A | G | T | G | A | C | T | C | 0.019 | 0.015 | 1.19 (0.27-5.3) | .824 | ||||||||||||||
| 10 | C | T | G | G | C | G | A | C | T | T | 0.008 | 0.028 | 4.8 (0.88-26.11) | .069 | ||||||||||||||
| White patients only | ||||||||||||||||||||||||||||
| 1 | C | A | A | G | T | G | G | C | T | T | 0.337 | 0.274 | Referent | — | ||||||||||||||
| 2 | T | A | A | G | C | G | A | C | T | C | 0.225 | 0.263 | 1.38 (0.89-2.13) | .149 | ||||||||||||||
| 3 | C | A | A | G | C | G | A | C | T | C | 0.063 | 0.067 | 1.52 (0.77-3.02) | .23 | ||||||||||||||
| 4 | C | T | G | G | C | G | A | C | T | C | 0.05 | 0.051 | 1.08 (0.52-2.24) | .838 | ||||||||||||||
| 5 | C | A | A | G | T | G | G | C | T | C | 0.05 | 0.047 | 1.14 (0.46-2.88) | .775 | ||||||||||||||
| 6 | T | A | A | G | T | T | A | T | T | C | 0.04 | 0.083 | 2.62 (1.15-5.94) | .022 | ||||||||||||||
| 7 | C | A | A | G | T | G | A | C | T | C | 0.037 | 0.011 | 0.71 (0.26-1.95) | .509 | ||||||||||||||
| 8 | C | A | A | G | T | G | G | C | A | T | 0.036 | 0.038 | 1.24 (0.52-2.95) | .624 | ||||||||||||||
| Donors | ||||||||||||||||||||||||||||
| 1 | C | A | A | G | T | G | G | C | T | T | 0.299 | 0.264 | Referent | — | ||||||||||||||
| 2 | T | A | A | G | C | G | A | C | T | C | 0.231 | 0.273 | 1.32 (0.87-1.99) | .194 | ||||||||||||||
| 3 | C | A | A | G | C | G | A | C | T | C | 0.07 | 0.047 | 0.76 (0.36-1.58) | .458 | ||||||||||||||
| 4 | C | A | A | G | T | G | G | C | T | C | 0.057 | 0.06 | 1.22 (0.61-2.46) | .571 | ||||||||||||||
| 5 | C | A | A | G | T | G | G | C | A | T | 0.046 | 0.042 | 1.05 (0.43-2.6) | .913 | ||||||||||||||
| 6 | C | A | A | G | T | G | A | C | T | C | 0.036 | 0 | 0.28 (0.06-1.33) | .11 | ||||||||||||||
| 7 | T | A | A | G | T | T | A | T | T | C | 0.035 | 0.065 | 2.2 (0.98-4.91) | .055 | ||||||||||||||
| 8 | C | T | G | G | C | G | A | C | T | C | 0.035 | 0.053 | 1.94 (0.82-4.59) | .129 | ||||||||||||||
| 9 | C | T | A | G | T | G | A | C | T | C | 0.024 | 0.018 | 0.94 (0.26-3.38) | .921 | ||||||||||||||
| 10 | T | A | G | A | C | G | G | C | T | C | 0.02 | 0.014 | 0.71 (0.18-2.88) | .635 | ||||||||||||||
For all patients, n = 363; for white patients, n = 320; for donors, n = 363. Haplotypes with a P ≤ .05 are shown in italics. All haplotypes were analyzed in multivariate models that included age at transplantation, pretransplantation; FEV1, extent of GVHD, and duration of follow-up as covariates.
— indicates no P value for the group.
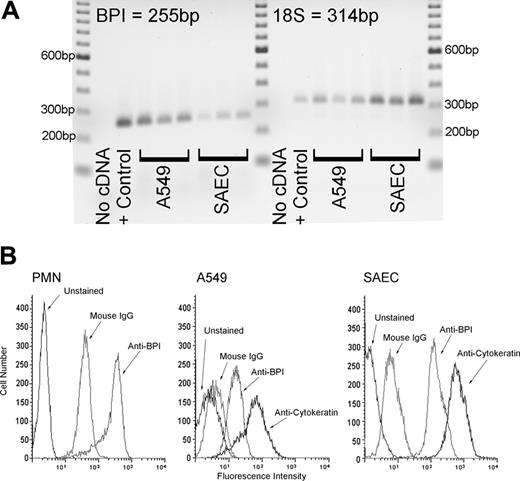
Expression of BPI by airway epithelial cells
We used A549 epithelial cells and SAECs to determine whether airway epithelial cells expressed the BPI gene. The BPI transcript was readily detectable in both cell types as well as in white blood cells that served as positive controls (Figure 1A). Demonstration of BPI protein production by the A549 epithelial cells, SAECs, and human white blood cells was confirmed by flow cytometric analysis (Figure 1B).
Discussion
Severe airflow obstruction (AFO) was first recognized in the early 1980s as a serious and often fatal complication of bone marrow transplantation.40-43 Although earlier studies have estimated that the prevalence ranges from 6% to 20%, a recent study by our group, using the same definition of significant airflow decline as used in this current genetic analysis, determined that the prevalence is closer to 26% to 30%. The same study also demonstrated that this definition has significant clinical implications; the estimated attributable mortality rates at 3, 5, and 10 years were 9%, 12%, and 18%, respectively.1 Although multiple factors have been associated with development of HCT-related AFO (methotrexate use,7 hypogammaglobulinemia,44 and respiratory infections45 ), the risk factor most commonly associated with the development of severe AFO, in both univariate and multivariable analyses, is GVHD,1-7,27 suggesting that AFO may represent an immunologic process that is either a pulmonary manifestation of GVHD or a pulmonary syndrome that is immunologically driven by mechanisms similar to those involved in GVHD.
There are an abundance of data supporting the relationship between lipopolysaccharide and AFO in nontransplantation patients. It is well known that because lipopolysaccharide is ubiquitous, everyone is constantly exposed to at least low levels of environmental lipopolysaccharide.15,16 One of the hypothesized consequences of this constant exposure is the development of asthma, a form of obstructive airway disease. For instance, the concentration of lipopolysaccharide in the domestic setting has been directly related to the clinical severity of asthma17 and an increased risk of repeated wheezing during the first year of life.18 Investigations into the mechanism of lipopolysaccharide-induced AFO have demonstrated that lipopolysaccharide activates macrophages and airway epithelial cells (AECs), results in release of proinflammatory cytokines, and leads to recruitment of polymorphonuclear cells to the airspace.22,23 Studies conducted on emphysema patients have also found that lipopolysaccharide-induced inflammatory signaling is increased and may be involved in elastin destruction by causing increased release of elastolytic enzymes such as matrix metalloproteinase.19,20 There are also a number of human studies confirming that variations in the genes of the lipopolysaccharide signaling pathway influence the AFO phenotype. Studies by Arbour et al46 have demonstrated that the Asp299Gly polymorphism in the TLR4 gene dampens lipopolysaccharide signaling through TLR4 but also has a significant influence on the AFO phenotype; subjects with the recessive allele did not exhibit significant AFO after inhalation of lipopolysaccharide. Transfection studies confirmed these findings, demonstrating that the TLR4 mutation interrupts TLR4-mediated signaling and that the wild-type allele rescues the lipopolysaccharide hyporesponsive phenotype. These studies indicate lipopolysaccharide-induced inflammation plays a central role in the pathogenesis of many types of obstructive airway diseases. Given the role that lipopolysaccharide signaling also plays in GVHD after HCT, it is highly likely that these same mechanisms are also important in the development of HCT-related AFO.
Using 2 different patient and donor cohorts, we discovered and validated statistically significant associations between a cluster of patient and donor tagSNP haplotypes at the BPI locus and HCT-related AFO. BPI and LBP belong to the same family of complex lipid transport proteins that play critical roles in the innate immune response to bacterial infection.47,48 The function of BPI is antagonistic to LBP. While LBP catalyzes transfer of lipopolysaccharide monomers to CD14 to promote TLR4-dependent cell activation and release of inflammatory mediators, BPI antagonizes the action of LBP by binding the lipid A region of lipopolysaccharide to facilitate aggregation of lipopolysaccharide, thus inhibiting transfer of lipopolysaccharide to CD14.49-52 BPI is also capable of inhibiting bacterial growth, activating bacterial phospholipid hydrolysis, and penetrating the inner bacterial membrane to dissipate electrochemical gradients required for bacterial viability.53,54
The neutrophil has been considered the primary source of BPI.55 However, BPI has also been detected as an inducible molecule on the surface of intestinal and oral epithelial cells36 and reported to be expressed by the A549 malignant epithelial cell line.36 Bcause both the patients' and the donors' BPI haplotypes were associated with the airflow decline phenotype, we pursued studies to determine if the BPI gene and protein is transcribed and translated by airway epithelial cells. Our study clearly demonstrated for the first time that primary epithelial cells cultured from normal human small airways constitutively express the BPI gene and produce the BPI protein. Because airway epithelial cells are regarded as the first line of host defense against bacteria or their products,56-58 and influx of neutrophils into airways can result in significant inflammation and obstruction,59,60 it is plausible that the extent of BPI mobilization within the airway and/or its intrinsic functional properties may be a critical determinant in the development of lipopolysaccharide-triggered airway disease.
Human airway epithelial cells express the BPI gene and produce the BPI protein. (A) This panel shows that A549 human lung epithelial cells and human SAECs constitutively express the BPI gene. Buffy coat from whole blood stimulated with lipopolysaccharide served as a positive control. Total RNA was isolated, and BPI mRNA was examined by RT-PCR. 18S ribosomal RNA was used as a loading control. (B) This panel shows that by flow cytometric analysis, A549 cells, SAECs, and neutrophils all stained positively for BPI protein. Permeabilized cells were stained with monoclonal antibody to BPI, murine IgG control, and/or murine anti-human anticytokeratin monoclonal antibody, followed by FITC-conjugated goat antimouse antibody. Fluorescence intensity was well above background staining with control IgG.
Human airway epithelial cells express the BPI gene and produce the BPI protein. (A) This panel shows that A549 human lung epithelial cells and human SAECs constitutively express the BPI gene. Buffy coat from whole blood stimulated with lipopolysaccharide served as a positive control. Total RNA was isolated, and BPI mRNA was examined by RT-PCR. 18S ribosomal RNA was used as a loading control. (B) This panel shows that by flow cytometric analysis, A549 cells, SAECs, and neutrophils all stained positively for BPI protein. Permeabilized cells were stained with monoclonal antibody to BPI, murine IgG control, and/or murine anti-human anticytokeratin monoclonal antibody, followed by FITC-conjugated goat antimouse antibody. Fluorescence intensity was well above background staining with control IgG.
Ample evidence also exists supporting the possibility that there is a genetic influence on the risk of developing AFO. Multiple familial aggregation and twin studies have shown that there is a genetic component to the development of obstructive airway disease in nontransplantation subjects.61-69 Genomewide linkage analyses have also identified several chromosomal regions to be linked with several phenotypic aspects of lung function.70-73 More recently, innate immunity genetic variants have been associated with an increased risk of developing GVHD after HCT74-77 and allograft rejection after lung transplantation,78,79 a process also known as bronchiolitis obliterans and considered similar to the syndrome observed in our patients.
Our search for genes that play a critical role in airway diseases likely benefited from several advantages unique to the HCT population. First, most HCT patients have normal lung function prior to transplantation,80 minimizing the potential effects of previously existing pulmonary disease. Second, all patients are exposed to the potential causal pathway of the airway disease at a well-defined time point (ie, conditioning regimen and transplantation). In addition, the airflow decline phenotype occurs frequently, generally within the first year after transplantation. These features increase the likelihood of identifying subjects who develop this phenotype. Finally, genetic analysis of the “chimeric” subjects, while adding some complexity, provides a unique opportunity to dissect the pathobiology of airway disease by permitting independent analyses of the patient and donor genotypes. Future studies will need to focus on developing methods to assess the potential effects of genome-genome interaction.
Perhaps the most striking feature of our data is that the BPI locus was found to be associated with the AFO phenotype in 2 independent populations of patient and donor pairs, yet the tagSNP haplotypes found significant in the discovery cohort are similar but not identical to the significant tagSNP haplotypes found in the validation cohort. However, this is not unusual for several reasons. First, the haplotype differences suggest that the functional mutation or mutations is not represented by one of the tagSNPs and that these mutations may be in varying degrees of linkage disequilibrium with the significant haplotypes we found. Second, recent studies have demonstrated that in some genes there are many rare mutations that affect gene function and that these rare mutations, when considered as a group, represent a measurable phenotype in the population as a whole.81,82 Perhaps the best example of this is the cystic fibrosis transmembrane conductance regulator (CFTR) gene. More than 1000 mutations, most of which are very rare, have been identified on this gene and determined to cause cystic fibrosis.83 In CFTR haplotype analyses, it is clear that the number of mutations found in association with particular haplotypes is related to its frequency among the population, reflecting the different degrees of linkage disequilibrium between the causal mutation and the many possible haplotypes.84 When considered in light of our BPI data, although we have identified a locus of potential importance in this disease process, future sequencing analysis of the entire BPI gene, along with the flanking regions that may contain regulatory sites, is warranted to fully characterize the genetic variation in our diseased population and identify all of the potential functional mutations.
The results of this study have intriguing therapeutic implications. Based upon the biology of BPI, a loss-of-function BPI mutation is likely the culprit responsible for our findings. If this is true, therapeutic options may be already available. BPI expression by epithelial cells can be enhanced by endogenous aspirin-triggered lipoxins, dampening endotoxin-mediated signaling in epithelia and enhancing killing of Gram-negative bacteria.36 A recombinant form of BPI is also available.85 Reformulation of this product for direct delivery to airways of BPI-deficient individuals may reduce their risk of developing HCT-related airway disease. Ultimately, if these findings are reproducible in nontransplantation populations, this represents an opportunity for the development of novel therapeutic options to treat other more common obstructive airway diseases such as asthma and chronic obstructive pulmonary disease.
Prepublished online as Blood First Edition Paper, November 22, 2005; DOI 10.1182/blood-2005-06-2338.
Supported by National Institutes of Health (NIH) grants HL71914, CA18029, CA15704, CA106320, AI33484, K23HL69860, an American Lung Association of Washington Research Grant, and the Amy Strelzer Manasevit Research Award from the National Marrow Donor Program.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal