Abstract
Host response to viral infection involves distinct effectors of innate and adaptive immunity, whose mobilization needs to be coordinated to ensure protection. Here we show that influenza virus triggers, in human blood dendritic-cell (DC) subsets (ie, plasmacytoid and myeloid DCs), a coordinated chemokine (CK) secretion program with 3 successive waves. The first one, occurring at early time points (2 to 4 hours), includes CKs potentially attracting effector cells such as neutrophils, cytotoxic T cells, and natural killer (NK) cells (CXCL16, CXCL1, CXCL2, and CXCL3). The second one occurs within 8 to 12 hours and includes CKs attracting effector memory T cells (CXCL8, CCL3, CCL4, CCL5, CXCL9, CXCL10, and CXCL11). The third wave, which occurs after 24 to 48 hours, when DCs have reached the lymphoid organs, includes CCL19, CCL22, and CXCL13, which attract naive T and B lymphocytes. Thus, human blood DC subsets carry a common program of CK production, which allows for a coordinated attraction of the different immune effectors in response to viral infection.
Introduction
Upon microbial invasion, the immune system must first sense the microbe's presence and then rapidly deliver an appropriate response. This requires the precise coordination of several simultaneous tasks, including (1) influx of immediate immune effectors, such as neutrophils and natural killer (NK) cells, (2) activation of antigen-specific memory B and T lymphocytes into effector cells; and (3) priming of naive lymphocytes that leads to recruitment of more effector cells and, upon primary infection, establishment of immune memory. This complex program is thought to be coordinated by dendritic cells (DCs).1-3
DCs constitute a complex system of cells with common and unique functions. Human blood contains 2 DC subsets, myeloid (mDCs) and plasmacytoid (pDCs).3,4 mDCs demonstrate remarkable plasticity and, depending on cytokine environment, can differentiate into either macrophages (with macrophage–colony-stimulating factor [M-CSF]), or distinct subsets of tissue-localized DCs, that is, epithelial Langerhans cells (with interleukin 15 [IL-15] or transforming growth factor β [TGF-β]) or interstitial DCs (with IL-4). Upon activation, mDCs secrete IL-12 and mature into antigen-presenting cells able to prime T cells. pDCs are considered poor antigen-presenting cells, but are major producers of type I interferons upon viral activation.5
DCs attract immune effectors through chemokines (CKs)6-9 and regulate their maturation and function through cell-cell contact, and/or soluble factors.1-3 Several studies analyzed CK secretion by DCs either generated ex vivo and exposed to bacterial products,10 or sorted from the blood and exposed to influenza virus upon culture with granulocyte macrophage–colony-stimulating factor (GM-CSF) and IL-4 (mDCs) or GM-CSF and IL-3 (pDCs).11,12 Furthermore, secretion of a limited set of CKs was analyzed at a single time point after viral exposure.13 Thus, there is a paucity of knowledge about the kinetics of CK response of primary blood DC subsets exposed to live influenza virus. As DCs need to coordinate the different steps of an immune response, we surmised that viral infection might trigger a sequential program of CK secretion that may permit such coordination.
Materials and methods
DC purification, staining and culture
pDCs and mDCs were purified from a healthy donor's buffy coats. Ficoll-enriched peripheral blood mononuclear cells (PBMCs) were depleted of lineage-positive cells with CD3, CD14, CD19, CD16, CD56, and glycophorin A microbeads (Miltenyi Biotec, Auburn, CA). After staining with lineage cocktail–fluorescein isothiocyanate (FITC), CD11c–allophycocyanin (APC) and CD123–phycoerythrin (PE; BD Biosciences, San Jose, CA) and HLA-DR-QR (Sigma, St Louis, MO) monoclonal antibodies (mAbs), cells were sorted on a FACSVantage (Becton Dickinson, San Jose, CA) to at least 99% purity.
For viral infection, purified pDCs and mDCs were cultivated in 96-well U-bottom plates (25 000 cells/well) in 200 μL complete medium (RPMI 1640, 2 mM l-glutamine, penicillin/streptomycin, and 10% fetal calf serum), containing 1:125 hemagglutinin (HA) titer per milliliter of live influenza virus A/PR/8/34 (Charles River Laboratories, Wilmington, MA). At different time points cell pellets and supernatant were collected and stored at –80°C.
For phenotype analysis, DCs were stained as described in the first paragraph, but replacing the CD123-PE with CD123 biotin/streptavidin-Alexa Fluor 405 (Molecular Probes, Eugene, OR), sorted on a FACSAria (Becton Dickinson), and stained before or after influenza virus infection using PE-conjugated CD83, CD40 (Beckman Coulter); CD80, and CD86 (BD Biosciences).
Microarray sample preparation and Genechip array hybridization
Cell pellets (250 000 cells) were resuspended in RLT buffer (Qiagen, Valencia, CA) and frozen at –80°C. Total RNA was extracted with RNeasy columns (Qiagen), and analyzed with the 2100 Bioanalyser (Agilent, Palo Alto, CA). Amplification was as follows: total RNA (300 ng) was reverse transcribed for 2 hours at 42°C in the presence of 1 μM T7-(dT)24 oligonucleotide (Operon, Huntsville, AL), 1× first strand buffer, 10 mM dithiothreitol (DTT), 0.75 mM deoxynucleoside triphosphate (dNTP), 20 U/μL Superscript II reverse transcriptase (all from Invitrogen, Carlsbad, CA), and 2 U/μL RNAse inhibitor (Ambion, Austin, TX). Second strand synthesis was achieved by incubation for 2 hours at 16°C, after addition of 1× second strand buffer, 0.2 mM dNTP, 0.07 U/μL Escherichia coli DNA ligase, 0.27 U/μL E coli DNA polymerase I, 0.013 U/μL E coli RNAse H, and then addition of 0.13 U/μL T4 DNA polymerase (all from Invitrogen) for 10 minutes at 16°C. Double-stranded cDNA was precipitated with 0.5 M ammonium acetate and 70% ethanol, and first round of in vitro transcription was performed for 6 hours at 37°C, in the presence of 1× Megascript buffer, 30 mM dNTP, 1× Megascript T7 enzyme mix (all from Ambion). Amplified cRNA was cleaned up using an RNeasy mini kit column (Qiagen), and quantified by migration on a 2100 Bioanalyser. Second reverse transcription was performed as for the first round but using 100 pM of a random hexamer pd(N)6 (Roche, Milan, Italy), and after treatment with 0.2 U/μL of RNAse H at 37°C for 20 minutes and enzyme heat inactivation, second strand synthesis was performed by addition of 0.13 μM T7-(dT)24 oligonucleotide, 1× second strand buffer, 0.2 mM dNTP, 0.27 U/μL E coli DNA polymerase I, and incubation at 16°C for 2 hours. Double-strand cDNA was then purified using the cDNA cleanup module from Affymetrix, and in vitro transcription was performed using the Enzo RNA transcript labeling kit (Affymetrix, Santa Clara, CA). Finally, biotin-labeled cRNA was purified using the cRNA cleanup module (Affymetrix), and stored at –80°C until hybridization. Purified biotin-labeled cRNA was then fragmented and hybridized on the human Genechip expression arrays U133 A and B (Affymetrix), according to protocols provided by the manufacturer.
Microarray data analysis
Raw intensity values from each chip were first prescaled to the 500 target intensity value in Affymetrix Microarray suite (MAS 5.0), before being imported in GeneSpring 6.1 (Silicon Genetics, Santa Clara, CA). Chip quality was evaluated by analyzing variability within the parameters of MAS expression report: scale factor, average background, percentage of present calls, 3′ bias for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and actin, and spike control expression; we verified that the value for each chip was included within the mean plus or minus 2 standard deviations. In GeneSpring, for per chip normalization, each measurement on each chip was normalized to the 50th percentile of the measurements on that chip; for per gene normalization, the measurement of each probe was normalized to the median of the measurements of that probe in the different chips. Before applying any statistical test, the normalized intensities were then log transformed, and gene lists were filtered, first on flags, excluding genes that were called absent in all chips, and then on confidence using a t test P value with no multiple testing correction, selecting genes whose expression was varying significantly across the experiment. Finally, samples were grouped by conditions (pDCs versus mDCs, and noninfected versus virus infected), for statistical group comparison using analysis of variance test (ANOVA) with a Bonferroni multiple testing correction. To analyze gene expression of selected gene list, chips were normalized with a per chip normalization without any per gene normalization or any log transformation. We thus arbitrarily considered as significantly expressed genes whose normalized intensity was 3 times higher than the 50th percentile.
Luminex and ELISA
CXCL8, CCL3, CCL4, CCL5, CXCL9, and CXCL10 were assessed by Luminex (Biosource). Samples were treated according to manufacturer's protocols. CCL19, CCL22, CXCL1, CXCL13, and CXCL16 expression was analyzed by enzyme-linked immunosorbent assay (ELISA; R&D Systems, Minneapolis, MN). All samples were treated in duplicates according to the manufacturer's protocol.
Cell-migration experiments and analysis
Cell migration experiments were performed by placing 29 μL/well of DCs culture supernatant, in 96-well ChemoTX lower plates (Neuroprobe, Gaithersburg, MD); and 150 × 103 PBMCs per well on the top of the ChemoTX membrane (5.7-mm diameter, 3-μm pore size). After 45 minutes at 37°C with 5% CO2, adherent cells on the bottom of the membrane were detached. After incubation at 4°C for 30 minutes, plates were centrifuged and cells were pooled and stained with CD3-PERCP, CD4-APC, CD8-APC, CCR7-PE, and CD45RA-FITC mAbs to analyze T-lymphocyte migration; CD19-APC, CD27-PE, and anti–IgD-FITC (Dako, Glostrup, Denmark) to analyze B-cell migration; CD16-FITC, CD14-PE, CD3-PERCP, and CD56-APC mAbs to analyze monocyte, macrophage, NK, and NKT cell migration; LIN-FITC, HLA-DR-PERCP, CD11c-APC, and CD123-PE mAbs, to analyze DC migration. All mAbs were from BD Biosciences. Statistical comparison of cell migration was done using 2-tailed paired t test.
Results
Influenza virus triggers a common CK production program in human pDCs and mDCs
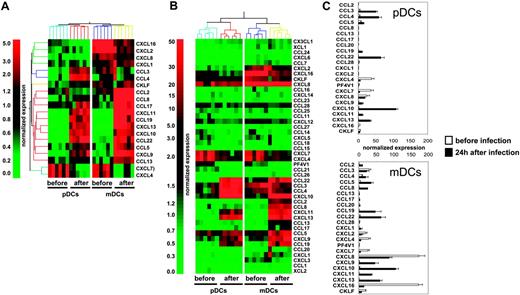
To determine the global CK production program triggered in human DC subsets upon viral exposure, we used gene microarray analysis in mDCs and pDCs purified from 6 healthy donors, before and 24 hours after in vitro exposure to influenza virus (Influenza A/PR/8/34). Statistical comparison and analysis of fold-change expression values showed that influenza virus up-regulated 4712 probe sets in pDCs and 4560 probe sets in mDCs (data not shown). Twenty of these differentially expressed transcripts encoded CKs (Figure 1A), leading us to carry out a systematic analysis of influenza virus–induced CK transcript expression in mDCs and pDCs (44 CK probe sets were present on the U133 chips). Transcripts with expression levels at least 3-fold over the 50th percentile were considered significant (Figure 1B). Thus, 4 of 44 CKs were significantly expressed in both DC subsets before activation (CXCL4, CXCL7, CXCL8, and CXCL16; Figure 1B-C). mDCs expressed more CXCL8 (× 7) and CXCL16 (× 36) than pDCs, whereas levels of CXCL4 and CXCL7 were comparable (Figure 1B-C and Table S1, which is available on the Blood website; see the Supplemental Materials link at the top of the online article). Moreover, mDCs, which displayed a more activated phenotype based on CD86, CD83, and HLA-DR expression (Figure S1), also expressed CCL3 and CXCL2. CXCL4 is a major product of platelets14-16 previously not identified in DCs. Although the lack of reagents does not permit demonstration of the production of CXCL4 by DCs, the purity of DC preparations determined by FACS analysis and the absence of platelet-specific gene expression exclude a possible platelet contamination.
Upon exposure to influenza virus for 24 hours, expression of these 4 CKs was reduced and 12 others were turned on in both pDCs and mDCs (Figure 1B-C; Table S1). These included CKs attracting polymorphonuclear cells (PMNCs): CXCL1, CXCL2, and CXCL317 ; CKs involved in the inflammatory response: CCL3, CCL4, and CCL517,18 ; interferon-inducible CKs CXCL9, CXCL10, and CXCL1117 ; as well as CKs expressed in lymphoid tissues CCL19, CCL22, and CXCL13.17 The spectrum of CKs induced by influenza virus exposure was remarkably similar between the 2 DC subsets. Only 2 of 44 CK transcripts, the monocyte-attracting CKs CCL2 and CCL8,17 were uniquely expressed in mDCs. The comparative analysis of the 2 DC subsets showed that mDCs expressed higher levels of CCL5 (× 2.4), CXCL9 (× 3.8), CXCL11 (× 3.1), CCL19 (× 4.3), and CXCL13 (× 1.8); whereas pDCs expressed higher levels of the inflammatory CKs CCL3 (× 2.3), and CCL4 (× 3).
Transcription of CK genes in blood DC subsets exposed to influenza virus. (A) Total RNA was extracted from pDCs and mDCs of 6 healthy donors after sort (before), and of 6 other donors 24 hours after culture with influenza virus (after). Amplified cRNA was hybridized on Affymetrix HG-U133 chips. Gene expression was analyzed with GeneSpring 6.1 software. Each probe set was normalized with a per chip normalization to the 50th percentile, and a per gene normalization to the median of each gene. Within the most differentially expressed probe sets (P < .05 with Bonferroni multiple testing correction, or > 2 fold-change), we identified a set of 20 chemokine probes. (B) Transcription of 44 different chemokines. After per-chip normalization to the 50th percentile, the normalized intensity values of each probe set were used as a measurement of its expression. (C) Normalized expression of the most significantly transcribed chemokines (3-fold over the 50th percentile). Each bar is representative of mean normalized intensity plus or minus standard error for chemokine's gene expression in pDCs and mDCs from 6 healthy donors before infection or 6 healthy donors after infection.
Transcription of CK genes in blood DC subsets exposed to influenza virus. (A) Total RNA was extracted from pDCs and mDCs of 6 healthy donors after sort (before), and of 6 other donors 24 hours after culture with influenza virus (after). Amplified cRNA was hybridized on Affymetrix HG-U133 chips. Gene expression was analyzed with GeneSpring 6.1 software. Each probe set was normalized with a per chip normalization to the 50th percentile, and a per gene normalization to the median of each gene. Within the most differentially expressed probe sets (P < .05 with Bonferroni multiple testing correction, or > 2 fold-change), we identified a set of 20 chemokine probes. (B) Transcription of 44 different chemokines. After per-chip normalization to the 50th percentile, the normalized intensity values of each probe set were used as a measurement of its expression. (C) Normalized expression of the most significantly transcribed chemokines (3-fold over the 50th percentile). Each bar is representative of mean normalized intensity plus or minus standard error for chemokine's gene expression in pDCs and mDCs from 6 healthy donors before infection or 6 healthy donors after infection.
Influenza virus triggers a coordinated CK production program with 3 successive waves
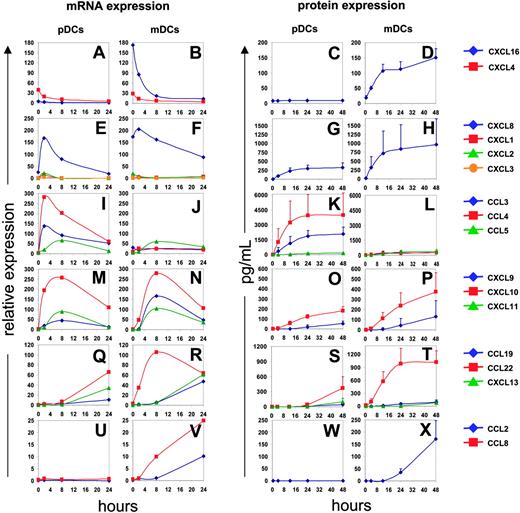
To analyze the kinetics of CK expression, purified DCs (25 000 cells) were exposed to influenza virus (1/125 HA titer per μL); pellets and supernatants were collected at 4, 8, 16, 24, and 48 hours (Figure 2). Before viral exposure, CXCL4 and CXCL16 were actively transcribed in both subsets (Figure 2A-B). The CXCL16 protein was, however, detected in mDC but not pDC supernatants (Figure 2D). CXCL16, which, like CX3CL1, is expressed as a transmembrane protein,19 is cleaved by a metalloprotease and attracts effector cells expressing the CXCR6 receptor.20,21 The expression of CXCL4 has been found earlier in platelets14,15 and monocytes.22 The transcription of both CXCL4 and CXCL16 is down-regulated upon viral exposure.
In the early postviral exposure phase, 2 to 4 hours, DCs transcribe mainly CXCL1, CXCL2, CXCL3, and CXCL8, which are known to attract PMNCs.23 The peak of their transcription occurred approximately 2 hours after exposure and subsequently decreased (Figure 2E-F). CXCL8 protein could be detected in supernatants of both pDCs and mDCs 4 hours after exposure, and reached a maximum at 12 hours (Figure 2G-H).
At intermediate phase (4 to 8 hours) after viral exposure, the transcription of the inflammatory CKs CCL3, CCL4, and CCL5 (Figure 2I-J) and the interferon-regulated CKs CXCL9, CXCL10, and CXCL11 was induced in both mDCs and pDCs (Figure 2M-N). Transcription of these genes reached its peak at 8 hours, while corresponding protein expression was detected in supernatants at 12 hours for CCL3 and CCL4 (Figure 2K-L), and 24 hours for CCL5, CXCL9, and CXCL10 (Figure 2K-L, O-P). All of the CKs produced at the intermediate time points are known to attract activated memory T cells.24 Furthermore, the CKs CCL3, CCL4, and CCL5, are chemotactic for CCR5 expressing monocytes and DCs.18
Finally, at late time points (> 12 hours after exposure), CCL19, CCL22, and CXCL13 transcripts were expressed by both pDCs and mDCs (Figure 2Q-R). The CKs could be detected in supernatants 24 to 48 hours after viral exposure (Figure 2S-T). These CKs attract lymphocytes. CCL19 is chemotactic for CCR7-expressing cells,25 that is, naive and central memory T cells (CD45RA+/–, CCR7+),26,27 as well as mature DCs.28,29 CCL22 attracts Th2 effector memory T cells.30 CXCL13 is the main CK responsible for B-cell attraction.31,32 Finally, 2 monocyte chemoattractants, CCL2 and CCL8, were expressed predominantly by mDCs (Figure 2U-X). Interestingly, we did not observe CCL17 expression by mDCs, which seems in contrast to an earlier study.11 A possible explanation might come from different experimental approaches, with different methods to isolate mDCs and activation using another strain of inactivated influenza virus.
Kinetics of CK expression. Blood DCs isolated from 4 donors were exposed to influenza virus. Total RNA was processed and gene expression was analyzed after per chip normalization as in Figure 1 (left). Protein expression was evaluated by multiplex cytokine analysis (Luminex; CCL2, CCL3, CCL4, CCL5, CXCL8, CXCL9, and CXCL10) or by ELISA (CXCL16, CCL19, CCL22, and CXCL13) (right). CXCL16 and CXCL4 are expressed before infection and their expression constantly decreases upon viral stimulation (A-D). CXCL1, CXCL2, CXCL3, and CXCL8 are transiently expressed between 2 and 8 hours, longer for CXCL8 (E-H). Inflammatory chemokines (CCL3, CCL4, CCL5) and interferon-dependent chemokines (CXCL9, CXCL10, CXCL11) expression is up-regulated between 2 and 8 hours and then decreases progressively (I-P). CCL19, CCL22, and CXCL13 are significantly expressed after 8 to 24 hours (Q-T). Finally, CCL2 and CCL8 are expressed only by infected mDCs (U-X).
Kinetics of CK expression. Blood DCs isolated from 4 donors were exposed to influenza virus. Total RNA was processed and gene expression was analyzed after per chip normalization as in Figure 1 (left). Protein expression was evaluated by multiplex cytokine analysis (Luminex; CCL2, CCL3, CCL4, CCL5, CXCL8, CXCL9, and CXCL10) or by ELISA (CXCL16, CCL19, CCL22, and CXCL13) (right). CXCL16 and CXCL4 are expressed before infection and their expression constantly decreases upon viral stimulation (A-D). CXCL1, CXCL2, CXCL3, and CXCL8 are transiently expressed between 2 and 8 hours, longer for CXCL8 (E-H). Inflammatory chemokines (CCL3, CCL4, CCL5) and interferon-dependent chemokines (CXCL9, CXCL10, CXCL11) expression is up-regulated between 2 and 8 hours and then decreases progressively (I-P). CCL19, CCL22, and CXCL13 are significantly expressed after 8 to 24 hours (Q-T). Finally, CCL2 and CCL8 are expressed only by infected mDCs (U-X).
Influenza virus–triggered DC subsets attract monocytes, NK cells, and effector memory CD4+ T cells
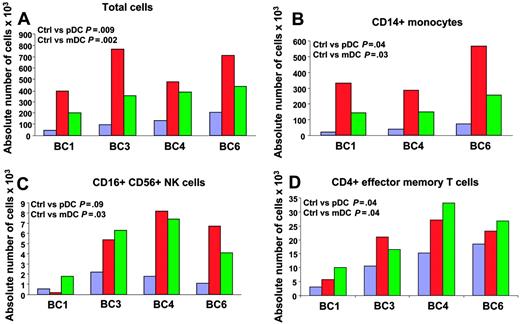
We next analyzed the attraction of different immune effectors using an in vitro migration assay. Supernatants of DC subsets, exposed to influenza virus for 24 hours, were added to the lower part of a migration chamber. PBMCs, isolated from 4 healthy volunteers, were added on top of a 3-um pore diameter filter. Migration of different immune cells was analyzed by viable cell count and flow cytometry after a 45-minute incubation to determine which immune effectors are attracted in the early phase after viral exposure. As shown in Figure 3A, supernatants of DCs exposed to influenza virus attracted more PBMCs (mean ± SD = 587 × 103 ± 181 × 103 for pDCs and 344 × 103 ± 99 × 103 for mDCs; P = .009 and .002, respectively; n = 4) than culture medium containing influenza virus (mean ± SD = 121 × 103 ± 67 × 103). Supernatants of pDCs attracted more PBMCs than supernatants of mDCs (P = .04). These results indicate that DCs exposed to influenza virus attract PBMCs. To determine which cells were actually attracted, cells from lower wells were analyzed with a panel of monoclonal antibodies that identify monocytes, DC subsets, NK cells, CD4+ and CD8+ T cells and their subsets, as well as B cells. As shown in Figure 3B, CD14+ monocytes represented the largest number within migrating PBMCs (393 × 103 ± 150 × 103 for pDCs and 183 × 103 ± 64 × 103 for mDCs; P = .04 and .03, respectively; n = 3) as compared with control (44 × 103 ± 26 × 103). In 4 of 4 donors analyzed, mDC supernatants attracted significant numbers of CD3–CD16+CD56+ NK cells (4.9 × 103 ± 2.5 × 103 for mDCs and 1.4 × 103 ± 0.7 × 103 for control medium; P = .03; n = 4; Figure 3C). pDC supernatants significantly attracted NK cells in 3 donors (6.75 × 103 ± 1.4 × 103 for pDCs and 1.7 × 103 ± 0.6 × 103 for control medium; P = .03; n = 3, BC3, 4, and 6; Figure 3C). Finally, the analysis of T-cell subsets in migrating PBMCs revealed increased numbers of CD4+ T cells with effector memory phenotype (CD3+CD4+CD45RA–CCR7–; 19 × 103 ± 9 × 103 for pDCs, 22 × 103 ± 10 × 103 for mDCs, and 12 × 103 ± 7 × 103 for control medium; P = .04; n = 4; Figure 3D). These results suggest that early after exposure to virus, both mDCs and pDCs attract innate immune effectors and effector memory CD4+ T cells.
Migration of PBMCs toward supernatants of blood DC subsets exposed to influenza virus. Purified blood DCs from 4 donors were exposed to influenza virus for 24 hours and supernatants were used in a migration assay. Supernatant (30 μL) was added to the lower part of a migration chamber and 7.5 × 105 PBMCs were added on top of a 3-μm–diameter pore filter. After a 45-minute incubation at 37°C, with 5% CO2, migrating cells were harvested, counted, and stained for flow cytometry. Migration of each cell subset in the different conditions is expressed as absolute cell number ×103 in the lower well after a 45-minute incubation. Paired t test. Blue bars indicate medium + virus; red, pDC SN; and green, mDC SN. (A) Total cell counts by trypan blue exclusion. (B-D) Counts of CD14+ monocytes (B); CD16+CD56+ NK cells (C); and CD4+ effector memory T cells (D). BC = buffy coat.
Migration of PBMCs toward supernatants of blood DC subsets exposed to influenza virus. Purified blood DCs from 4 donors were exposed to influenza virus for 24 hours and supernatants were used in a migration assay. Supernatant (30 μL) was added to the lower part of a migration chamber and 7.5 × 105 PBMCs were added on top of a 3-μm–diameter pore filter. After a 45-minute incubation at 37°C, with 5% CO2, migrating cells were harvested, counted, and stained for flow cytometry. Migration of each cell subset in the different conditions is expressed as absolute cell number ×103 in the lower well after a 45-minute incubation. Paired t test. Blue bars indicate medium + virus; red, pDC SN; and green, mDC SN. (A) Total cell counts by trypan blue exclusion. (B-D) Counts of CD14+ monocytes (B); CD16+CD56+ NK cells (C); and CD4+ effector memory T cells (D). BC = buffy coat.
Discussion
Our results demonstrate that upon exposure to live influenza virus, freshly isolated blood mDCs and pDCs produce 3 waves of CKs, which allow attraction of immune effectors. Although pDCs and mDCs strikingly differ in their biology, they display a remarkably similar pattern of CK secretion despite some qualitative (CCL2, CCL8, and CXCL16 expressed in mDCs only) and quantitative (CCL3, CCL4, and CCL5 expressed at higher levels in pDCs) differences. Such a sequential CK secretion program might actually explain how the DCs coordinate immune responses as they mature and migrate toward lymphoid organs (Figure 4). Interestingly, this sequential CK program is reminiscent of the sequential CK secretion triggered by lipopolysaccharide (LPS) in cultured DCs.10 These results suggest that the various DC subsets share a common program to attract immune effectors.
Thus, the first-produced CKs are those attracting innate effectors and cytotoxic cells, which might limit the spread of infection. The next wave involves CKs that attract memory T cells and those that attract monocytes, which could replenish the pool of DCs or tissue macrophages. Finally, as mature DCs land in the secondary lymphoid organs they secrete CKs that attract B cells and naive T cells, allowing their priming. Attraction of T cells with regulatory/suppressor function might finally permit the termination of immune response. Accordingly, CXCL13 has been shown to attract B cells as well as CXCR5+CD4+ memory T cells that can enhance IgG and IgA production.32 Furthermore, CXCL13-producing cells are located uniquely in the follicles and germinal centers.31 Thus, influenza virus–activated blood DCs that migrate to B-cell areas of peripheral lymphoid organs could launch humoral immunity via CXCL13. Influenza-activated DCs that migrate to the T-cell zone could launch T-cell immunity via CCL19 and CCL22. Indeed, both chemokines are expressed by mature DCs in the T-cell areas.26,33 CCL19 is chemotactic for CCR7 expressing naive and central memory T cells,26,27 whereas CCL22 is chemoattractant for Th2 effector memory T cells.30
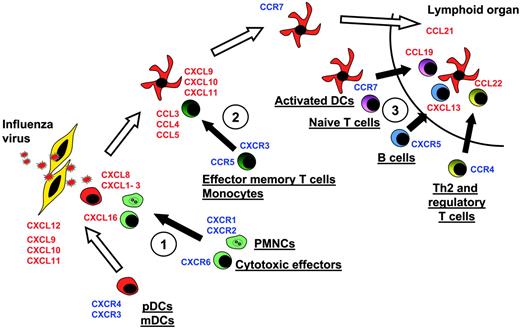
Model of DC migration and sequential effector-cell attraction. Blood DCs expressing, at the steady state, CXCR4 and CXCR3, can migrate through virally infected tissues, expressing CXCL12, CXCL9, CXCL10, and CXCL11. There, they can penetrate the tissues. Upon encountering the virus, they start to release at the first step CXCL16, CXCL1, CXCL2, CXCL3, CXCL7, and CXCL8. These CKs attract Th1 effectors cells expressing CXCR6 and neutrophils expressing CXCR2 (no. 1 in figure). Later, activated DCs secrete CCL2, CCL3, CCL4, CCL5, and CCL8, which essentially attract CCR5-expressing memory T lymphocytes and monocytes (no. 2). Upon maturation, DCs up-regulate CCR7 and down-regulate CXCR4, allowing, with l-selectin expression, their migration to high endothelial venules expressing CCL21 in lymphoid organs. In the T-cell area, activated pDCs secrete CCL19 and CXCL13, which respectively attract CCR7-expressing naive T cells and CXCR5-expressing naive B cells (no. 3). They also secrete CCL22, attracting CCR4-expressing Th2 and CD4+CD25+ regulatory cells (no. 3).
Model of DC migration and sequential effector-cell attraction. Blood DCs expressing, at the steady state, CXCR4 and CXCR3, can migrate through virally infected tissues, expressing CXCL12, CXCL9, CXCL10, and CXCL11. There, they can penetrate the tissues. Upon encountering the virus, they start to release at the first step CXCL16, CXCL1, CXCL2, CXCL3, CXCL7, and CXCL8. These CKs attract Th1 effectors cells expressing CXCR6 and neutrophils expressing CXCR2 (no. 1 in figure). Later, activated DCs secrete CCL2, CCL3, CCL4, CCL5, and CCL8, which essentially attract CCR5-expressing memory T lymphocytes and monocytes (no. 2). Upon maturation, DCs up-regulate CCR7 and down-regulate CXCR4, allowing, with l-selectin expression, their migration to high endothelial venules expressing CCL21 in lymphoid organs. In the T-cell area, activated pDCs secrete CCL19 and CXCL13, which respectively attract CCR7-expressing naive T cells and CXCR5-expressing naive B cells (no. 3). They also secrete CCL22, attracting CCR4-expressing Th2 and CD4+CD25+ regulatory cells (no. 3).
It is intriguing that each wave is composed of several CKs with similar functions. Recent studies have demonstrated that CKs act in synergy, where one CK would sensitize the cells to another, thereby amplifying the attraction of immune effectors.34,35 Furthermore, the production of several CKs able to attract the same type of immune cells might represent a mechanism to compensate for the CK antagonists that viruses have created during their evolutionary fight against the immune system. For example, the poxviruses and herpes viruses evolved several means to compromise the CK network.36,37 These include secretion of CK binding proteins that compete with CKs secreted by host cells (reviewed in Seet and McFadden36 ). Immature/nonactivated DCs patrolling via blood could be attracted to the site of pathogen entry in a process mediated via their expression of CXCR3 and CXCR4. The respective ligands are secreted by epithelial cells upon virus entry. Indeed, McWilliam et al38 found that DCs are the first to arrive at the site of pathogen entry, preceding even neutrophils, a finding consistent with our model. Furthermore, we have found that both mDCs and pDCs are attracted in vivo to the respiratory tract in children with acute viral infections triggered by influenza virus or respiratory syncytial virus (RSV). Upon influenza exposure, the down-regulation of CXCR4 expression and up-regulation of CCR7 expression (data not shown) would allow the migration of pDCs and lnDCs into lymphoid organs where, by production of appropriate CKs, they would launch immune response. This coordinated program of successive waves of CK production triggered by viral exposure in blood DCs has been observed on purified cells. Therefore, influenza virus infection triggers an autonomous program of CK production in these cells, independently of any cell-to-cell interaction. This capacity of blood DCs could thus have been evolutionally selected to allow a rapid development of protective immune response.
Prepublished online as Blood First Edition Paper, November 29, 2005; DOI 10.1182/blood-2005-07-2965.
Supported by Baylor Health Care Systems (BHCS) foundation, DANA foundation, and the National Institutes of Health (U19 AIO57 234 and CA78 846). J.B. holds the Caruth Chair for Transplantation Immunology Research. A.K.P. holds the Ramsay Chair for Cancer Immunology Research.
J.B. and A.K.P. codirected this work.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Elisabeth Kraus and Sebastien Coquery for their help in DC sorting, and Dr Christophe Combadiere, PhD (Unite INSERM 543, Paris, France) for advice in migration assays.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal