Abstract
X-linked severe combined immunodeficiency (XSCID) is characterized by profound immunodeficiency and early mortality, the only potential cure being hematopoietic stem cell (HSC) transplantation or gene therapy. Current clinical gene therapy protocols targeting HSCs are based upon ex vivo gene transfer, potentially limited by the adequacy of HSC harvest, transduction efficiencies of repopulating HSCs, and the potential loss of their engraftment potential during ex vivo culture. We demonstrate an important proof of principle by showing achievement of durable immune reconstitution in XSCID dogs following intravenous injection of concentrated RD114-pseudotyped retrovirus vector encoding the corrective gene, the interleukin-2 receptor γ chain (γc). In 3 of 4 dogs treated, normalization of numbers and function of T cells were observed. Two long-term–surviving animals (16 and 18 months) showed significant marking of B lymphocytes and myeloid cells, normalization of IgG levels, and protective humoral immune response to immunization. There were no adverse effects from in vivo gene therapy, and in one dog that reached sexual maturity, sparing of gonadal tissue from gene transfer was demonstrated. This is the first demonstration that in vivo gene therapy targeting HSCs can restore both cellular and humoral immunity in a large-animal model of a fatal immunodeficiency.
Introduction
Gene therapy offers promise of clinical benefit and perhaps cure for many genetic diseases. The first successful gene therapy trial was in treatment of X-linked severe combined immunodeficiency (XSCID), a life-threatening primary immunodeficiency caused by mutations in the common gamma chain (γc) subunit of various interleukin (IL) receptors, thus presenting with absence of normal lymphocyte function and profound immunologic abnormalities.1,2 In this clinical study, XSCID infants received autologous hematopoietic stem cells (HSCs) transduced ex vivo with an oncoretroviral vector carrying the γc gene, resulting in immune reconstitution and reversal of the clinical phenotype in 10 of 11 treated boys.3,4 This trial documented that gene therapy alone can successfully provide long-term clinical benefit for a human disease. The immune reconstitution was similar to that observed in XSCID boys who successfully received bone marrow transplants, namely complete reconstitution of the T-cell population with γc+ (normal) T cells, with only approximately 1% of the B lymphocytes being derived from transduced cells. Unfortunately, 3 of the gene therapy–treated boys developed T-cell leukemias attributed to insertional mutagenesis approximately 3 years following treatment, indicating that gene therapy is not without potential serious side effects.4-6
To date, all human clinical gene therapy protocols targeting HSCs are based upon ex vivo gene transfer requiring that HSCs be mobilized, isolated, and cultured ex vivo for 4 to 5 days in the presence of multiple cytokines,3,7-10 which can cause differentiation and loss of homing abilities of the cells. Numerous animal studies have reported HSCs cultured ex vivo under similar conditions and cytokine exposure to be at a disadvantage compared with freshly isolated cells in terms of both their short-term and long-term engraftment potential that was evident as early as 3 to 6 days of culture.11-14 These studies raise the question of whether exhaustion of stem cells might result from growth factor stimulation ex vivo.15 Limitation of number of HSCs that can be isolated and potential further loss of cells during processing may ultimately lead to a low dose of transduced cells for transplantation.
An attractive alternative would be to transduce HSCs in vivo within their natural environment, thereby eliminating the necessity for ex vivo manipulation of the target cell. Low-level gene marking of blood lineages has previously been observed after intravenous administration of retrovirus vector to newborn dogs to treat mucopolysaccharidosis VII,16 suggesting the possibility that an in vivo approach to gene therapy could cure XSCID because of the potent selective growth advantage conferred on T lymphocytes by γc gene correction. Therefore, in this study, we explored this alternative approach in a canine XSCID model by direct intravenous injection of concentrated retroviral vector to evaluate in vivo transduction of HSCs and clinical benefit following in vivo gene therapy. Canine XSCID, unlike genetically engineered γc-deficient mice, has a clinical and immunologic phenotype identical to human XSCID,17-19 making it an ideal large-animal, preclinical model in which to evaluate new strategies for treating XSCID. It is also an ideal model to monitor for potential adverse effects with long latency following gene transfer into long-lived stem cells and/or committed progenitors.
Materials and methods
XSCID dogs
All experiments were performed in accordance with protocols approved by the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC no. 360600). The colony of XSCID dogs used in this study carry a 4–base pair deletion in the first exon of γc on the X-chromosome.20 The animals were intravenously injected with concentrated retroviral vector at day 3 of life.
Retroviral vector production
RD114-pseudotyped MFGS–canine-γc–IRES–GFP-MGMT* retrovirus vector was produced using a replication-incompetent murine leukemia virus (MLV)–based vector (MFGS) packaged by a cell line (FLYRD18) encoding the RD114 envelope that is derived from feline endogenous virus.21 The vector used is a bicistronic vector, meaning that a picornavirus internal ribosome entry site (IRES) element allows the translation of an upstream canine-γc gene and a downstream gene for a green fluorescent protein (GFP)–mutant methylguanine methyltransferase (MGMT*) fusion protein (GFP-MGMT*)22 from the same virus mRNA. Thus, marking can be evaluated by flow cytometric measurement of GFP expression. The mutant MGMT portion of the fusion protein confers resistance to cytotoxic effects of O6 -benzylguanine and a DNA-alkylating agent such as temozolomide. This can be used for either in vitro selection of gene-marked cells or treatment of dogs in vivo to achieve selective enrichment of marked marrow cells expressing the mutant MGMT-selective system. While such a selection regimen was not used in the current study, it will be exploited in future studies of animals treated with this vector. RD114 pseudotyping confers a physical stability, which allows vector concentration by tangential flow filtration (PXB300C50; Millipore, Bedford, MA) and ultracentrifugation (83 000g, 90 minutes, 4°C) without loss of titer. Activity of the concentrated vector preparation was established using a single transduction of 3-day–cultured CD34+ hematopoietic progenitor cells, where an optimum transduction of 80.1% GFP-expressing cells occurred at a multiplicity of infection (MOI) of 10 and virus concentration of 8 × 106 infectious units (IU)/mL.
Analysis of transgene expression by flow cytometry and confocal microscopy
Peripheral dog blood was lysed (ACK lysis buffer), washed with phosphate-buffered saline (PBS), and then analyzed for GFP and/or after staining with phycoerythrin (PE)– or biotin-labeled monoclonal antibodies specific against canine CD3 (CA17.2A12), CD21 (CA2.1D6), CD45RA (CA4.1D3), CD14 (TUG4), or neutrophils23 (Serotec, Raleigh, NC). Analysis was performed using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA) or confocal microscopy. Confocal images were collected on a Leica TCS-NT/SP confocal microscope (Leica Microsystems, Exton, PA) using a 63 ×/1.32 numeric aperture oil immersion objective, zoom 4 ×. Fluorochromes were excited using an argon laser at 488 nm for GFP, 568 nm for Alexa-Fluor 568 (Molecular Probes, Eugene, OR). Images were processed using Leica TCS-NT/SP software version 2.61 and Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA). The γc expression was determined by using a proprietary PE-labeled human γc antibody that cross-reacts with the canine γc.
In vitro functional assessment of PBMCs by evaluation of signaling and proliferative responses to cytokine stimulation
Stimulation of γc leads to phosphorylation and heterodimerization of Janus kinase 1 (Jak 1) and 3, which results in downstream tyrosine phosphorylation of signal transducer and activator of transcription 5 (STAT5). Peripheral blood mononuclear cells (PBMCs) obtained by centrifugation over a discontinuous density gradient of Hypaque-Ficoll were stimulated with IL-2 (50 000 units/mL) or IL-7 (10 ng/μL), followed by intracellular staining for phosphorylated STAT524,25 (ZYMED Lab, San Francisco, CA) or bromodeoxyuridine (BrdU; Becton Dickinson) for assaying cell proliferation as per manufacturer's instructions. Secondary staining was performed using an allophycocyanin (APC)–labeled anti–mouse IgG1 antibody for flow cytometric detection.
TCR Vβ variability analysis for clonality
We have recently cloned and sequenced 43 different canine T-cell receptor β (TCRβ) variable/diversity joining segment (VDJ) sequences and confirmed their specificity by sequence homology analysis with known TCRβ VDJ region sequences from other species (B.J.H., and P.J.F., manuscript in preparation). Twenty-three distinct TCR Vβ segments were identified that comprised 18 different families (> 80% homology at the nucleotide level). The TCR Vβ families were arbitrarily assigned numbers from BV1 to BV18, beginning with families that contained the greatest number of sequences, and numbered consecutively. Unique members of the same family were designated by assigning an additional consecutive number (eg, BV3.1, BV3.2, BV3.3). We are in the process of developing polymerase chain reaction (PCR) primers to specifically prime different TCR Vβ families and unique members of those families. In this study, spectratype analysis was performed on DNA sequences synthesized from PBMCs by PCR using 12 specific forward TCR Vβ primers and an IRD700-labeled reverse primer. Amplified products were loaded and run on a GeneReadIR 4200 sequencing machine (LiCor, Lincoln, NE).
Assessment of humoral immune function
Serum IgG was determined by radial immunodiffusion using canine IgG–specific plates (Bethyl Lab, Montgomery, TX). At normalization of serum IgG levels, the 2 long-term–surviving dogs (1547 and 1655) were immunized with a modified live canine parvovirus (CPV) and canine distemper virus (CDV) vaccine and serum levels of specific IgG were determined by either a passive hemagglutination (for CPV) or serum neutralization (for CDV) assay.
Multiplex quantitative PCR analysis of vector copy number
Genomic DNA for quantitative PCR (qPCR) was extracted from lysed whole blood or magnetic bead–isolated CD3+ T cells (Serotec) separated in GFP+ and GFP– cells by fluorescence-activated cell sorting. Sperm DNA was extracted as previously reported.26 Vector copy number was determined by real-time TaqMan PCR (Applied Biosystems, Foster City, CA). By using a multiplex method, sequences of both the retroviral vector (MFGS) and the housekeeping gene hypoxanthine-guanine phosphoribosyltransferase (HPRT) were amplified in the same well to allow normalization of DNA input most accurately. A primer/probe set located within the packaging signal of the MFGS vector was used: forward primer, 5′-CGCAACCCTGGGAGACGTCC-3′; probe, 5′-6-FAM-CCGTTTTTGTGGCCCGACCTGATAMRA; reverse primer, 5′-CGTCTCCTACCAGAACCACATATCC-3′. The primers and probe for detection of the HPRT gene were as follows: forward primer, 5′-GGATTTGAAATTCCAGACAAGTTTGTTG-3′; probe, 5′-VIC-GCCCTTGACTATAATGAATACTTCAGG-TAMRA-3′; reverse primer, 5′-GTGAGAAAAAGAAGCAATTACTTACATTC-3′. The following PCR parameters were used: 2 minutes at 50°C, 10 minutes at 95°C, 40 cycles of 15 seconds at 95°C, and 60 seconds at 60°C. Standard curves were established individually for both MFGS and HPRT by analyzing samples of serial dilutions of DNA from a K562 cell line with a single copy of MFGS vector per cell. All samples of interest were analyzed in triplicates, and copy number of MFGS vector was calculated using the standard curve and normalized using the results for HPRT.
Transgene integration site analysis using a modified LAM-PCR
Clonality assessment was performed using a modified protocol of previously described linear amplification-mediated (LAM)–PCR.27 In our modified assay, TasI was used as the frequent cutter restriction enzyme and PvuII to digest the 206-bp PCR product that derives from amplifying a vector sequence starting within the 3′ long terminal repeat (LTR). Insertion site sequences were aligned to canine genomic sequences in collaboration with The Institute for Genomic Research (TIGR) and to the recently assembled canine genome using the UCSC genome browser (BLAT search28 ).
Results
Intravenous administration of concentrated vector is well tolerated
Four 3-day-old XSCID puppies (weighing approximately 500 g each), dogs 1551, 1548, 1547, and 1655, were injected intravenously with escalating doses of concentrated RD1114-pseudotyped MFGS–canine-γc–IRES–GFP-MGMT* retrovirus vector every 3 hours. The total doses injected were 9 × 108, 16 × 108, 24 × 108, and 32 × 108 IU, respectively (Table 1). At time of treatment, all animals were T-cell lymphopenic with less than 0.5% of the peripheral blood lymphocytes being CD3+ as determined by flow cytometry. No acute adverse events were evident at any dosing schedule.
Doses of RD114-pseudotyped MFGS-canine-γc-IRES-GFP-MGMT* vector used for the 4 dogs treated
Dog no. . | Volume, mL . | Titer, × 108 IU/mL . | Final dose, × 108 IU . |
|---|---|---|---|
| 1551 | 5 | 1.8 | 9 |
| 1548 | 10 | 1.6 | 16 |
| 1547 | 15 | 1.6 | 24 |
| 1655 | 10 | 3.2 | 32 |
Dog no. . | Volume, mL . | Titer, × 108 IU/mL . | Final dose, × 108 IU . |
|---|---|---|---|
| 1551 | 5 | 1.8 | 9 |
| 1548 | 10 | 1.6 | 16 |
| 1547 | 15 | 1.6 | 24 |
| 1655 | 10 | 3.2 | 32 |
IU indicates infectious units.
Sustained transgene expression in both lymphoid and myeloid peripheral blood lineages following in vivo gene therapy
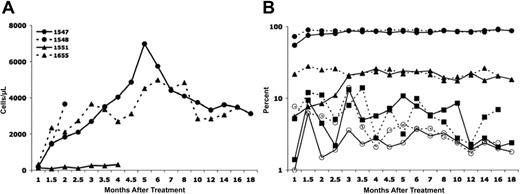
Following intravenous injection of the bicistronic vector, transduced cells could be detected easily by flow cytometric measurement of GFP expression. All 4 treated XSCID dogs had detectable GFP expression in peripheral blood lymphocytes (3% to 15.8%), monocytes (0.5% to 5.6%), and granulocytes (0.5% to 6.1%) at 3 weeks after treatment, the earliest time point evaluated in each of these animals. The percentage of lymphocytes and, to a lesser extent, of monocytes in the peripheral blood that were GFP+ continued to increase in all the dogs between 3 and 8 weeks after treatment (up to 80%-85% GFP+ T lymphocytes), whereas the percentage of GFP+ granulocytes remained relatively constant. Figure 1 illustrates the strategy used for the flow cytometric analysis for GFP+ cells in the various cell lineages following in vivo gene therapy. Because of the nuclear localization sequence found in MGMT*, the GFP-MGMT* fusion protein is localized to the cell nucleus of the different lineages of peripheral blood cells (Figure 2), making it unlikely that any of the observed GFP marking is due to passive cytoplasmic transfer between cells. By 8 weeks after treatment, the T-cell proportion of peripheral blood lymphocytes had normalized for dogs 1548, 1547, and 1655 with at least 78% expressing GFP. Dog 1551 showed an increased T-cell percentage but clearly below the normal range (49%, with 71.1% being GFP+; Table 2). The absolute number of gene-corrected (GFP+) peripheral T cells also attained normal age-matched levels by 8 weeks after treatment in all the dogs except for dog 1551, which remained T-cell lymphopenic (Figure 3A; Table 2). Dog 1548 died at 9 weeks after treatment where, at necropsy, the cause of death was attributed to a severe anemia of uncertain cause. Histopathologically, there were no indications of an infection, marrow failure, or a malignant process. Blood loss was another possible cause of anemia and, retrospectively, an excessive amount of blood was noted to have been drawn for monitoring purposes. Dog 1551 suffered from recurrent chronic infections and severe weight loss, necessitating that the dog be humanely killed at 5 months after gene therapy treatment. Long-term follow-up of dogs 1547 (18 months) and 1655 (16 months) showed sustained marking (gene-corrected cells) in all blood cell lineages (Figure 3B). The proportion of GFP+ T cells remained stable between 80% and 85% in both dogs. Of particular note is the sustained transgene marking of the peripheral B cells (up to 26%) and of the myeloid lineages represented by GFP+ granulocytes (2% to 6%) and monocytes (2% to 12%), demonstrating persistence of even the myeloid marking following in vivo gene therapy.
Gene-corrected (GFP+) peripheral blood cells in all 4 dogs 8 weeks following intravenous gene therapy as determined by flow cytometry
. | Neutrophils . | . | . | Monocytes . | . | . | Lymphocytes . | . | . | CD3+ . | . | . | . | CD21+ . | . | . | . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dog no. . | No.* . | % GFP+ . | No. GFP+* . | No.* . | % GFP+ . | No. GFP+* . | No.* . | % GFP+ . | No. GFP+* . | % . | No.* . | % GFP+ . | No. GFP+* . | % . | No.* . | % GFP+ . | No. GFP+* . | ||||||||||||
| 1551 | 5 420 | 1.0 | 54 | 340 | 0.6 | 2 | 560 | 38.7 | 216 | 49.1 | 274 | 71.1 | 195 | 28.7 | 160 | 8.4 | 14 | ||||||||||||
| 1548 | 11 000 | 3.2 | 352 | 770 | 5.9 | 45 | 7200 | 52.8 | 3802 | 64.8 | 4665 | 78.5 | 3662 | 7.9 | 568 | 26.5 | 151 | ||||||||||||
| 1547 | 5 900 | 1.5 | 88 | 860 | 4.5 | 39 | 3400 | 65.2 | 2217 | 69.5 | 2363 | 78.0 | 1843 | 4.9 | 167 | 6.1 | 10 | ||||||||||||
| 1655 | 6 320 | 3.8 | 242 | 930 | 11.1 | 102 | 3850 | 61.0 | 2348 | 65.3 | 2514 | 84.5 | 2120 | 20.3 | 782 | 24.2 | 189 | ||||||||||||
. | Neutrophils . | . | . | Monocytes . | . | . | Lymphocytes . | . | . | CD3+ . | . | . | . | CD21+ . | . | . | . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dog no. . | No.* . | % GFP+ . | No. GFP+* . | No.* . | % GFP+ . | No. GFP+* . | No.* . | % GFP+ . | No. GFP+* . | % . | No.* . | % GFP+ . | No. GFP+* . | % . | No.* . | % GFP+ . | No. GFP+* . | ||||||||||||
| 1551 | 5 420 | 1.0 | 54 | 340 | 0.6 | 2 | 560 | 38.7 | 216 | 49.1 | 274 | 71.1 | 195 | 28.7 | 160 | 8.4 | 14 | ||||||||||||
| 1548 | 11 000 | 3.2 | 352 | 770 | 5.9 | 45 | 7200 | 52.8 | 3802 | 64.8 | 4665 | 78.5 | 3662 | 7.9 | 568 | 26.5 | 151 | ||||||||||||
| 1547 | 5 900 | 1.5 | 88 | 860 | 4.5 | 39 | 3400 | 65.2 | 2217 | 69.5 | 2363 | 78.0 | 1843 | 4.9 | 167 | 6.1 | 10 | ||||||||||||
| 1655 | 6 320 | 3.8 | 242 | 930 | 11.1 | 102 | 3850 | 61.0 | 2348 | 65.3 | 2514 | 84.5 | 2120 | 20.3 | 782 | 24.2 | 189 | ||||||||||||
Normal ranges were determined in 8-week-old unaffected dogs and are as follows: neutrophils, 8500 ± 2900 cells/μL; monocytes, 1400 ± 700 cells/μL; lymphocytes, 5000 ± 1500 cells/μL; CD3+, 76.2% ± 14.4%; and CD21+, 12.9% ± 8.6%. The percentage of GFP+ cells in untreated control dogs (background signal) was less than 0.05%.
To convert neutrophil, monocyte, or lymphocyte counts to × 109 cells/L, divide cells/μL by 1000.
Cells/μL.
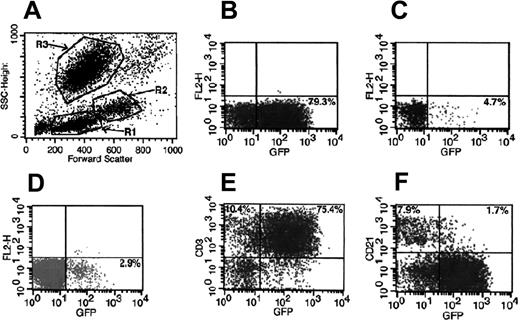
Strategy used for evaluation of transgene expression in different cell lineages by flow cytometry. A representative peripheral blood analysis of dog 1655 at 4 months after treatment is shown. (A) Scattergram of forward and side scatter (SSC) of peripheral blood showing gate R1, lymphocytes; gate R2, monocytes; and gate R3, granulocytes. (B) GFP+ cells in lymphocyte gate. (C) GFP+ cells in monocyte gate. (D) GFP+ cells in granulocyte gate. (E) GFP+ T (CD3+) cells in lymphocyte gate. (F) GFP+ B (CD21+) cells in lymphocyte gate.
Strategy used for evaluation of transgene expression in different cell lineages by flow cytometry. A representative peripheral blood analysis of dog 1655 at 4 months after treatment is shown. (A) Scattergram of forward and side scatter (SSC) of peripheral blood showing gate R1, lymphocytes; gate R2, monocytes; and gate R3, granulocytes. (B) GFP+ cells in lymphocyte gate. (C) GFP+ cells in monocyte gate. (D) GFP+ cells in granulocyte gate. (E) GFP+ T (CD3+) cells in lymphocyte gate. (F) GFP+ B (CD21+) cells in lymphocyte gate.
Nuclear localization effect of the GFP-MGMT* fusion protein in marked blood cells. Confocal microscopy image of GFP expression in peripheral blood (A) lymphocytes, (B) monocytes, and (C) granulocytes of in vivo–treated XSCID dog 1547. Note the green nuclei due to nuclear localization effect of MGMT* from the GFP-MGMT* fusion protein. Staining with an (A) anti-CD3 and (B-C) antineutrophil/antimonocyte monoclonal antibody with secondary red-Alexa Fluor–labeling confirms morphologic diagnosis.
Nuclear localization effect of the GFP-MGMT* fusion protein in marked blood cells. Confocal microscopy image of GFP expression in peripheral blood (A) lymphocytes, (B) monocytes, and (C) granulocytes of in vivo–treated XSCID dog 1547. Note the green nuclei due to nuclear localization effect of MGMT* from the GFP-MGMT* fusion protein. Staining with an (A) anti-CD3 and (B-C) antineutrophil/antimonocyte monoclonal antibody with secondary red-Alexa Fluor–labeling confirms morphologic diagnosis.
Gene-marked circulating T lymphocytes express a CD45RA+ naive phenotype consistent with restored thymic function
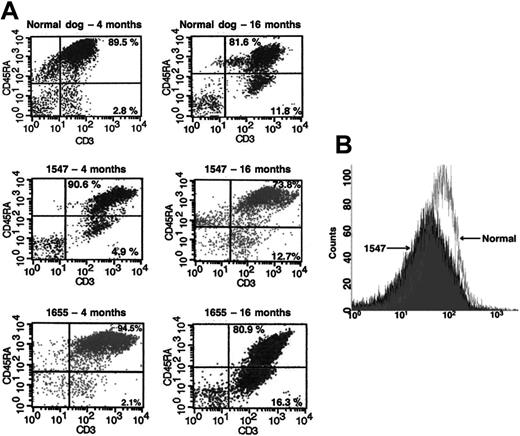
CD45RA+ T lymphocytes represent a population of unprimed (naive) T cells in contrast to the CD45RA– (CD45RO) primed/memory T cells. We observed that the majority of the newly emerging gene-marked T lymphocytes in the in vivo–treated dogs in this study expressed a CD45RA+ (naive) phenotype, suggesting that they were recent thymic emigrants. More than 90% of the GFP+ T cells in both long-term–surviving dogs, 1547 and 1655, at 4 months after treatment expressed a naive CD45RA+ phenotype compared with only 70% to 80% at 16 months after treatment (Figure 4A). This demonstrates a typical age-related decrease, similar to the pattern observed in normal, unaffected dogs at the same age.
Normal expression of the γc transgene in gene-marked peripheral blood lymphocytes
Transgene expression as determined by GFP expression has remained unchanged after the initial 2 to 3 months in both dogs 1547 and 1655. More importantly, there has not been any evidence of overexpression of the γc in the treated dogs. Using a limited supply of a proprietary human γc antibody that cross-reacts with the canine γc, we compared γc mean fluorescence intensity (MFI) of the treated dogs to an age-matched normal dog monthly from 4 months after treatment. The γc expression was stable, with ratios averaging 1.1 ± 0.5 (Figure 4B). It is of note that 10% to 15% of the cells that were γc marked were GFP–. This is most likely due to the γc gene being located upstream of the IRES element of our bicistronic vector, thus expressed at higher levels than the downstream GFP transgene.29 This also explains the 80% to 85% gene-corrected T cells based upon GFP expression instead of the close to 100% one would expect.
Longitudinal analysis of T-cell development and persistence of gene marking in different blood cell lineages. (A) Absolute numbers of gene-corrected (GFP+) T cells for all 4 dogs treated. The normal absolute number of peripheral T cells in age-matched unaffected dogs is greater than 750/μL. (B) Proportion of gene-corrected (GFP+) T cells (•), B cells (▴), monocytes (▪), and granulocytes (○) in dog 1547 (solid lines) and dog 1655 (broken lines).
Longitudinal analysis of T-cell development and persistence of gene marking in different blood cell lineages. (A) Absolute numbers of gene-corrected (GFP+) T cells for all 4 dogs treated. The normal absolute number of peripheral T cells in age-matched unaffected dogs is greater than 750/μL. (B) Proportion of gene-corrected (GFP+) T cells (•), B cells (▴), monocytes (▪), and granulocytes (○) in dog 1547 (solid lines) and dog 1655 (broken lines).
Gene-corrected lymphocytes exhibit normal function
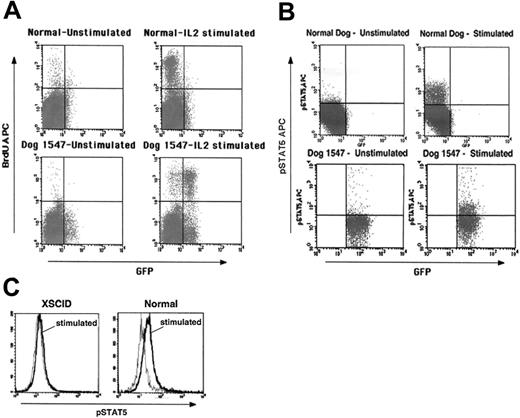
More detailed immunologic and molecular analyses have been possible with the long-term–surviving animals. The gene-corrected T cells in both dog 1547 and dog 1655 attained normal function as evidenced by normal in vitro proliferation following mitogenic stimulation (data not shown) and stimulation with IL-2 (Figure 5A), responses not evident in untreated XSCID cells. The few GFP– cells that exhibit BrdU incorporation most likely represent the γc+ GFP– T cells described before. Furthermore, in vitro IL-2 stimulation resulted in downstream signaling of the restored cytokine receptor as measured by STAT5 phosphorylation for PBMC samples of both dogs (Figure 5B). Cells from an untreated XSCID dog do not show such a response (Figure 5C). To analyze the diversity repertoire of the gene-corrected T cells, we performed TCR Vβ CDR3 spectratyping. By 6 months after treatment a diverse, polyclonal TCR repertoire was evident (Figure 6A) similar to that seen in normal dogs and XSCID dogs that successfully received bone marrow transplants (B.J.H. and P.J.F., unpublished data, January 2005). Serum IgG concentrations normalized in both dogs between 6 and 8 months after treatment (data not shown). Both animals were immunized with a modified live vaccine to canine parvovirus and canine distemper virus and, in contrast to untreated XSCID dogs, they developed protective levels of specific IgG antibody following immunization as summarized in Table 3. No adverse reaction to the vaccine was seen.
Specific IgG antibody titers following immunization in dogs 1547 and 1655 treated with intravenous gene therapy
. | Dog 1547 . | . | Dog 1655 . | . | ||
|---|---|---|---|---|---|---|
| Modified live vaccine . | Titer before imm . | Titer after imm . | Titer before imm . | Titer after imm . | ||
| CPV | < 10 | 640 | < 10 | 320 | ||
| CDV | < 4 | 1536 | < 4 | 1024 | ||
. | Dog 1547 . | . | Dog 1655 . | . | ||
|---|---|---|---|---|---|---|
| Modified live vaccine . | Titer before imm . | Titer after imm . | Titer before imm . | Titer after imm . | ||
| CPV | < 10 | 640 | < 10 | 320 | ||
| CDV | < 4 | 1536 | < 4 | 1024 | ||
Protective IgG titers: CPV, > 40; CDV, > 100.
Imm indicates immunization.
Single transgene copy number in peripheral blood GFP+ lymphocytes but sparing of germ line cells
qPCR analysis of purified GFP+ lymphocytes at 15 months after treatment showed that the proviral copy number in dog 1547 averaged 1.13 copies per GFP+ T cell and in dog 1655, 1.07 copies per cell. Interestingly, purified GFP– T cells showed marking at similar levels consistent with the flow cytometric finding of γc expression in GFP– cells as previously described. Since dog 1547 has reached sexual maturity, it was possible to evaluate the presence of vector insertions in purified sperm cells. Microscopy of isolated specimen confirmed absence of contaminating epithelial or blood cells. qPCR analysis of DNA from sperm did not detect any transgene at a detection limit of a single copy in 10 000 cells.
CD45RA and γc expression in treated dogs are reconstituted compared with normal controls. (A) Flow cytometric evaluation of CD45RA expression in CD3+ T lymphocytes in a normal dog (top 2 panels) and in both in vivo gene therapy–treated dogs 1547 and 1655 at 4 months (left) and at 16 months (right) of age. Naive T lymphocytes are denoted by presence of CD45RA+ (top right quadrants). (B) Overlay of γc expression in peripheral blood lymphocytes from dog 1547 with γc expression in an age-matched normal dog.
CD45RA and γc expression in treated dogs are reconstituted compared with normal controls. (A) Flow cytometric evaluation of CD45RA expression in CD3+ T lymphocytes in a normal dog (top 2 panels) and in both in vivo gene therapy–treated dogs 1547 and 1655 at 4 months (left) and at 16 months (right) of age. Naive T lymphocytes are denoted by presence of CD45RA+ (top right quadrants). (B) Overlay of γc expression in peripheral blood lymphocytes from dog 1547 with γc expression in an age-matched normal dog.
Polyclonal pattern of genomic vector insertion sites
Vector integration site analysis on purified T cells by modified LAM-PCR demonstrated a highly polyclonal pattern for the long-term–surviving dogs 1547 and 1655 compared with an oligoclonal pattern for the relatively short-lived dog 1551 (Figure 6B). Serial LAM-PCRs (approximately every 3 months) have been performed and, so far, around 500 sequences have been analyzed, of which 30 unique insertion sites have human homologs. While additional analysis is ongoing, no insertion appeared to be in proximity of the LMO2 gene, a known oncogene in humans and associated with leukemia development in at least 2 patients following ex vivo gene therapy for XSCID,4,6 or any other currently known oncogene. Of note, there are no clones that have appeared dominant or increasing in representation over time in either of the long-term–surviving dogs. However, this does not necessarily preclude potentially harmful integration sites, and potential adverse events can only be determined by long-term monitoring of the treated dogs.
Reconstitution of T-cell function following intravenous gene therapy. (A) Proliferation of PBMCs from in vivo–treated dog 1547 following stimulation with IL-2 as determined by BrdU incorporation by flow cytometry. (B) Flow cytometry analysis for STAT5 phosphorylation in PBMCs from dog 1547 following IL-2 stimulation. A proliferative response and phosphorylation of STAT5 are evident primarily in GFP+ cells. Controls from age-matched normal dogs are shown in the top panels. (C) Comparison of STAT5 phosphorylation in PBMCs from an untreated XSCID dog and a normal dog (gray line indicates prestimulation; black line, after IL-2 stimulation).
Reconstitution of T-cell function following intravenous gene therapy. (A) Proliferation of PBMCs from in vivo–treated dog 1547 following stimulation with IL-2 as determined by BrdU incorporation by flow cytometry. (B) Flow cytometry analysis for STAT5 phosphorylation in PBMCs from dog 1547 following IL-2 stimulation. A proliferative response and phosphorylation of STAT5 are evident primarily in GFP+ cells. Controls from age-matched normal dogs are shown in the top panels. (C) Comparison of STAT5 phosphorylation in PBMCs from an untreated XSCID dog and a normal dog (gray line indicates prestimulation; black line, after IL-2 stimulation).
Polyclonal T-cell reconstitution following intravenous gene therapy. (A) T-cell diversity in dog 1547 6 months following gene therapy as determined by TCR Vβ CDR3 spectratyping for 12 distinct TCR Vβ segments. (B) Assessment of vector integration sites by LAM-PCR from genomic DNA from CD3+ cells from dog 1547 (lanes 2-4) and dog 1551 (lanes 5-7) 5 months following gene therapy. Lane 1, DNA molecular weight (MW) marker; lane 8, H2O control. → indicates size of final vector control band with incomplete digestion by PvuII.
Polyclonal T-cell reconstitution following intravenous gene therapy. (A) T-cell diversity in dog 1547 6 months following gene therapy as determined by TCR Vβ CDR3 spectratyping for 12 distinct TCR Vβ segments. (B) Assessment of vector integration sites by LAM-PCR from genomic DNA from CD3+ cells from dog 1547 (lanes 2-4) and dog 1551 (lanes 5-7) 5 months following gene therapy. Lane 1, DNA molecular weight (MW) marker; lane 8, H2O control. → indicates size of final vector control band with incomplete digestion by PvuII.
Discussion
In this study we report for the first time immune reconstitution and reversal of the clinical phenotype of XSCID by direct intravenous injection of a retrovirus vector, encoding the normal canine γc, in XSCID dogs. XSCID dogs that do not receive immune-corrective allogeneic bone marrow transplants or are not sustained in a strictly isolated gnotobiotic environment usually succumb to infection by 3 months of age.
Three of the 4 in vivo gene therapy–treated dogs attained normal numbers of gene-corrected peripheral T cells by 8 weeks following treatment. This finding is analogous to the cell development observed in XSCID dogs following successful HSC transplantation with purified CD34+ bone marrow cells.30 In 2 dogs that have been followed for 16 and 18 months, respectively, the absolute numbers of gene-corrected T cells have remained in the normal to high normal range. The reconstituted peripheral T cells in the treated dogs were functional as assessed by their ability to proliferate and phosphorylate STAT5 in response to cytokine stimulation. They also exhibited a diverse, polyclonal repertoire as determined by TCR Vβ CDR3 spectratyping.
One dog that received the lowest vector dose remained severely T-cell lymphopenic, and insertion site analysis demonstrated that the T lymphocytes in this dog were derived from a very limited number of clones. Although the animal survived longer than untreated XSCID dogs, it succumbed to chronic infections, suggesting a failure to achieve the level of clinical benefit seen in the other dogs. This suggests that, despite the selective growth advantage for γc+ T lymphocytes,31 a certain threshold (dose) of vector and therefore gene-corrected cells appears necessary to trigger sufficient expansion of marked T lymphocytes. Moreover, there may be a need to reach a threshold level of corrected T-cell progenitors before robust immune reconstitution can occur. This is analogous to our HSC transplantation studies, demonstrating that a minimum dose of purified CD34+ bone marrow cells is required to achieve full T-cell immune reconstitution and correction of the clinical phenotype, whereas lower doses result in a state of T-cell lymphopenia with dogs rarely surviving past 5 to 6 months after transplantation.30 This hypothesis is also supported by recent findings in human ex vivo gene therapy studies for XSCID32 and adenosine deaminase (ADA)–deficient SCID,10 suggesting that the level of T-cell reconstitution may be related to the dose of transduced cells.
Although some XSCID dogs, like some XSCID boys, can develop phenotypically mature T cells with age because of peripheral expansion, these nonfunctional T cells have an activated (CD45RA–) phenotype, and the dogs remain T-cell lymphopenic and do not survive past 3 months of age.33,34 At the time of treatment all dogs studied had a peripheral blood T-cell lymphopenia, which accounted for less than 0.5% of lymphocytes, making it highly unlikely that observed T-lymphocyte reconstitution was solely based upon transduction of peripheral T cells. In addition, the majority of gene-corrected T lymphocytes exhibited a naive (CD45RA+) phenotype, suggesting that these cells developed through a thymic-dependent pathway.35,36
In contrast to the XSCID boys treated by gene therapy, in whom the proportion of gene-corrected B cells remained 1% or less,3,37 the XSCID dogs treated by intravenous retroviral gene therapy exhibited B-cell chimerism as early as 4 weeks after treatment. More importantly, the 2 dogs monitored long-term have maintained significant proportions of gene-corrected B cells ranging from 11% to 26%. These 2 animals developed normal to low normal concentrations of serum IgG and specific IgG antibody production following immunization. The clinical significance of the presence of gene-corrected B lymphocytes is that both dogs have survived longer than 16 months without any supportive treatment with intravenous immunoglobulin.
An interesting finding from this study was the sustained presence of gene-corrected monocytes and granulocytes, ranging between 2% and 12% and 2% and 6%, respectively. Persistent production of donor-derived myeloid cells is not observed following nonmyeloablative HSC transplantation in either XSCID boys or XSCID dogs, and in gene therapy–treated XSCID boys transgene was detected in 0.1% or less of myeloid cells.3 The presence of sustained levels of gene-marked myeloid cells for at least 18 months in our in vivo gene therapy XSCID dog study is highly suggestive of transduction of either multipotent HSCs or very primitive committed progenitors. The clinical significance of gene-corrected myeloid cells in XSCID is unclear; however, this level of marking may be significant for other diseases, where low levels of myeloid chimerism may be sufficient to correct the disease phenotype such as in the case of chronic granulomatous disease (CGD).
No adverse side effects have been observed in either of the 2 long-term–monitored dogs. In particular, no pathologic clonal expansions have been detected by serial LAM-PCR analyses and none of the integration sites thus far examined have been in the proximity of known oncogenes. However, it should be stressed that this does not necessarily preclude potentially harmful integration sites. Because of the lifespan of dogs and the number of dogs that can be treated, canine XSCID provides an ideal large-animal, preclinical model in which to evaluate new strategies for gene therapy of XSCID and the potential long-term sequelae following gene transfer into long-lived HSCs including genotoxicity (effect of transgene insertion), phenotoxicity (effect of transgene expression), and selection toxicity (effect of cell expansion).38
Some gene-marking studies have reported marking in hematopoietic stem cells following different approaches to in vivo gene therapy.39-41 One potential concern for a clinical application of a direct intravenous approach to gene therapy in humans is the half-life of the vector in the bloodstream, based upon the observation that MLV-based retroviral vectors can be inactivated in the presence of human serum.42 However, RD114-pseudotyped viral vectors, such as the vector used in this study, have been reported to have an enhanced stability in human and nonhuman primate sera.43,44 A recent report of a phase 1 dose escalation clinical trial evaluating the intravenous injection of a vesicular stomatitis virus G-protein (VSV-G)–pseudotyped MLV-based vector expressing human FVIII in 13 hemophilia A patients showed that vector-positive peripheral blood mononuclear cells persisted for up to 53 weeks in the patients, suggesting that the used vector was not inactivated following intravenous injection.45 Thus an in vivo gene therapy approach may be feasible for humans. We also demonstrated in the study reported here that use of an RD114-pseudotyped viral vector does not result in the transduction of germ line cells. This is consistent with the reported absence of RD114 receptors in gonadal tissue,46 an important safety consideration for any in vivo gene therapy.
In summary, our study demonstrates that in vivo retroviral gene therapy by direct intravenous administration of a γc-encoding vector is capable of transducing long-lived HSCs and/or committed progenitors in their natural environment, resulting in the correction of the XSCID phenotype. This method may be a viable alternative for gene therapy of XSCID and perhaps other genetic hematologic/immunologic diseases, thereby eliminating any potential detrimental effects of the ex vivo manipulation and culture of target cells that is required by current clinical gene therapy protocols.
Prepublished online as Blood First Edition Paper, December 29, 2005; DOI 10.1182/blood-2005-10-4057.
Supported by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases, and by NIH grant RO1 AI 43745 (P.J.F.).
H.L.M. and P.J.F. contributed equally to this study and share senior authorship.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Dr Patrick F. Kelly (Department of Hematology/Oncology, St Jude Children's Research Hospital, Memphis, TN) for his generous gift of the FLYRD18 producer cells. We thank Dr Margaret L. Casal and Patty O'Donnell (Veterinary Hospital, University of Pennsylvania, Philadelphia) for the veterinary care of the XSCID dogs. We would like to also thank Dr Linda Burkly of Biogen Idec for providing the γc monoclonal antibody used in this study, and Dr Meggan Czapiga of the National Institute of Allergy and Infectious Diseases, NIH, for collecting the confocal micrographs.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal