Treatment of adult Philadelphia chromosome-positive lymphocytic leukemia is rarely successful. We report here the effects of TZD18, a novel dual ligand specific for peroxisome proliferator-activated receptor α and γ (PPARα/γ) on Ph+ lymphocytic leukemia cell lines BV173, SD1, and SupB-15. Exposure of these cells to TZD18 resulted in growth inhibition in a dose- and time-dependent manner that was associated with G1 cell cycle arrest. This effect was much stronger than that mediated by the PPARγ ligand pioglitazone (PGZ), which also belongs to the thiazolidinediones (TZD) class of ligands. However, it may not be mediated through PPARγ or PPARα activation because antagonists of PPARγ and PPARα cannot reverse it. Study of the key regulators of cell cycle progression by Western blot analysis showed that the expression of the cyclin-dependent kinase inhibitor (CDKI) p27kip1, but not that of p21cip1, was enhanced, whereas that of c-Myc, cyclin E, cyclin D2, and cyclin-dependent kinases 2 and 4 (CDK-2 and CDK-4) was decreased when these cells were treated with TZD18 (10 or 20 μM). Therefore, the up-regulation of p27kip1 and the down-regulation of CDK-2 and CDK-4 may, at least in part, account for the G1 cell cycle arrest. Furthermore, a remarkable induction of apoptosis was observed in the cells treated with this dual ligand. No obvious alteration of bcl-2 protein level occurred, but bax was up-regulated in these TZD18-treated cells. Activation of caspase 8 and caspase 9 by TZD18 was also observed. Importantly, NF-κB DNA-binding activity was markedly decreased by the TZD18 treatment. In addition, TZD18 enhanced the growth inhibitory effect of imatinib, a specific tyrosine kinase inhibitor therapeutically used in the treatment of Ph+ leukemia. Overall, our findings strongly suggest that TZD18 may offer a new therapeutic approach to aid in the treatment of Ph+ lymphocytic leukemia.
Introduction
Although significant progress has been made in the treatment of acute lymphocytic leukemia (ALL), the prognosis for patients with Philadelphia chromosome-positive (Ph+)/Bcr-Abl+ adult ALL is still very poor. After first remission, allogeneic stem cell transplantation is the treatment of choice. However, most patients are not eligible for this therapy because of advanced age or lack of a suitable stem cell donor (for reviews, see Redaelli et al1 and Bassan et al2 ). Imatinib (Gleevec, previously known as STI571 and CGP57148), the selective tyrosine kinase inhibitor, displayed pronounced antileukemic activity in Ph+ chronic myeloid leukemia (CML) and ALL. It is used after allogeneic stem cell transplantation and is recommended for relapsed or refractory Ph+ ALL as salvage therapy to facilitate subsequent transplantation.3 However, quick emergence of resistance to this agent is a major problem in the treatment of patients with Ph+ leukemia. Another major obstacle to imatinib-based therapies is the persistence of Ph+ cells despite the application of imatinib. Based on these arguments, the development of novel therapeutic agents for human ALL, especially for Ph+ ALL, is necessary.
Peroxisome proliferator-activated receptors (PPARs) belong to the family of nuclear hormone receptors that include receptors for estrogen, thyroid hormone, retinoic acid, 1,25-dihydroxy vitamin D3, and retinoid X. To date, 3 subtypes of PPARs (α, β/δ, and γ) have been identified that exhibit distinct tissue distribution and are associated with selective ligands. PPARγ can be activated by synthetic ligands such as those belonging to the antidiabetic thiazolidinedione (TZD) class of compounds.4,5 A naturally occurring arachidonic acid metabolite, 15-deoxy-delta (12,14-prostaglandin J2 (15d-PGJ2),6 and certain nonsteroidal anti-inflammatory drugs5 are also identified as its ligands.
PPARγ was initially noted to be highly expressed in adipose tissue and was found to have a key function in adipocyte differentiation, insulin sensitization, and lipid metabolism.7,8 We and others9-13 have demonstrated that PPARγ is also widely expressed in a variety of types of tumor cells and that it has crucial roles in suppressing cell growth and invasion and in promoting differentiation and apoptosis. Interestingly, Braissant et al14 found that the expression of this receptor in immune system was just as high as its expression in the adipose tissue. We demonstrated that PPARγ was expressed in ALL cells and its ligands, such as the synthetic TZD-class ligand pioglitazone (PGZ), and that 15d-PGJ2 also potently inhibited growth and induced apoptosis of human ALL cells, including Ph+ ALL cells.12
PPARα is another subtype of the PPAR family. The important physiologic role of PPARα is to modulate lipid metabolism by, for instance, lowering serum triglyceride levels and raising high-density lipoprotein cholesterol levels through increased clearance and decreased synthesis of triglyceride-rich, very low-density lipoprotein.15 Fibrates are PPARα synthetic ligands that have been used clinically for several metabolic syndromes, such as dyslipidemia.16 This class of drugs inhibited the IL-1-stimulated release of IL-6 and inflammatory prostaglandins in vascular smooth muscle cells,17 possibly through negative regulation of NF-κB and AP-1, thus contributing to the treatment of coronary diseases.
The role of PPARα in tumor cell growth is unclear. PPARα ligand fibrates may lead to peroxisome proliferation in the liver of rodents that could ultimately result in hepatocellular carcinoma.18 However, some evidence also suggests that fibrates can inhibit hepatoma cell growth.19-21 Furthermore, fibrates cause growth arrest by regulating cell cycle-related factors and induce the monocytic differentiation of HL-60 cells.22,23 These data indicate that this subtype of the PPAR family could also have anticancer activities.
TZD18 (Figure 1A) is one of a series of compounds synthesized by Merck (Rahway, NJ) that can specifically bind and activate PPARα and PPARγ. In addition to its specificity to both these receptors, this compound has pharmacokinetic parameters superior to those of other members of this series: high bioavailability, high-dose normalized AUC, and relatively low clearance.24,25 It also exhibits favorable effects on lipid homeostasis.26
Based on the previously observed antileukemia activity of PPARα and PPARγ ligands in human leukemia cell lines, we hypothesized that a dual ligand specific for these 2 subtypes of PPAR might be even more effective than ligands for either PPARγ or PPARα alone. Therefore, we investigated the antiproliferative and proapoptotic activity of TZD18 against human Ph+ lymphocytic leukemia cells in the anticipation that it may have potential as a therapeutic agent for this poorly responsive disease.
Materials and methods
Compounds, reagents, and plasmids
TZD18 was kindly provided by Merck and was dissolved in dimethyl sulfoxide (DMSO) at 10-2 M as a stock solution. Imatinib was kindly provided by Novartis (Basel, Switzerland) and was dissolved in DMSO at 10-3 M. PPARγ antagonist GW9662 was purchased from GlaxoSmithKline (Hertfordshire, United Kingdom). The PPARα ligand WY14,643 and the antagonist MK886 were from Alexis Biochemicals (Berlin, Germany). The pan-caspase inhibitor benzylocarbonylvalylalanylaspartyl fluoromethylketone (Z-VAD-FMK) was obtained from R&D (Wiesbaden-Nordenstadt, Germany). All stock solutions were stored at -70°C and were further diluted to appropriate concentrations with medium before use.
The luciferase reporter vector PPREx3-tk-luc with 3 × human PPRE consensus sequence was kindly provided by R. M. Evans (Salk Institute for Biological Studies, La Jolla, CA), the full-length human PPARγ1 expression plasmid pcDNAFlag-PPARγ1 was provided by Prof K. K. Chatterjee (Department of Medicine, University of Cambridge, Cambridge, United Kingdom), and the PPARα expression vector pCMX-mPPARα was provided by Dr P. Tontonoz (Howard Hughes Medical Institute, University of California, Los Angeles). The Renilla luciferase reporter vector was a gift from Dr C. Mueller-Tidow (Division of Hematology and Oncology, University of Muenster, Muenster, Germany).
Cell lines
Human Ph+ lymphocytic leukemia cell lines BV173 and SD1 were purchased from Deutsche Sammlung für Mikroorganismen und Zellkulturen (Braunschweig, Germany) and were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS). The Sup B-15 cell line, which was established from the bone marrow of a 10-year-old boy with Ph+ ALL (B cell precursor ALL) in second relapse,27 was also purchased from Deutsche Sammlung für Mikroorganismen und Zellkulturen and was cultured under the same conditions but with 20% FCS. Human mesenchymal stem cells were obtained from Dr Markus Rojewski (Institut für Klinische Transfusionsmedizin und Immungenetik, University Hospital Ulm, Germany). Cells in logarithmic growth phase were used for further experiments.
Assessment of cell proliferation
A previously described method was used to measure cell proliferation.12 Briefly, cells at 2 × 105/mL were treated with various agents in 96-well plates. After incubation, 10 μL of 3-4,5-dimetylthiazol-2-yl-2,5-diphenyltetrazoliumnbromide (MTT; Sigma-Aldrich, Taufkirchen, Germany) solution was added to each well, and cells were incubated for another 4 hours. Water-insoluble formazan was formed during incubation and was solubilized by the addition of 100 μL solubilization solution to each well. The formazan dye was quantified using an HTII ELISA reader (Anthos Mikrosysteme, Krefeld, Germany).
Cell cycle analysis
Cells were treated with different concentrations of TZD18 for different durations, washed with PBS, and fixed with ice-cold 70% ethanol. These samples were treated with RNase, stained with propidium iodide, and analyzed with the FACSCalibur flow cytometer (Becton Dickinson, Heidelberg, Germany).
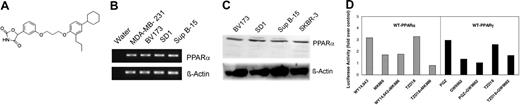
Analysis of expression and function of PPARα and PPARγ in Ph+ ALL cells. (A) Structure of compound TZD18. (B) RT-PCR analysis of the expression of PPARα. Total RNA was isolated, 0.2 μg each RNA sample was applied to RT-PCR, and specific amplification products for PPARα were visualized by electrophoresis and subsequent ethidium bromide staining. Total RNA extracted from the breast cancer cell line MDA-MB-231 was used as a positive control. (C) PPARα expression was determined by Western blot, as described in “Materials and methods.” Total protein isolated from the breast cancer cell line SKBR-3 served as a positive control for PPARα expression. (D) Functional analysis of several PPAR agonists and antagonists. SD1 cells were cotransfected with either a PPARα or a PPARγ expression vector (wt-PPARα or wt-PPARγ), a PPRE reporter vector (PPREx3-tk-luciferase), and a Renilla luciferase vector as an internal control for transfection efficiency. Eighteen hours after transfection, the cells were incubated with PPARα ligand (WY14,643), PPARγ ligand (PGZ), TZD18, PPARα antagonist (MK886), PPARγ antagonist (GW9662), or their combinations for another 24 hours. All drugs were used at 10 μM. After incubation, firefly and Renilla luciferase activities were determined. The firefly luciferase activity was normalized with Renilla luciferase activity and is shown as the fold increase over vehicle-treated controls. The result is a representative of 3 independent experiments.
Analysis of expression and function of PPARα and PPARγ in Ph+ ALL cells. (A) Structure of compound TZD18. (B) RT-PCR analysis of the expression of PPARα. Total RNA was isolated, 0.2 μg each RNA sample was applied to RT-PCR, and specific amplification products for PPARα were visualized by electrophoresis and subsequent ethidium bromide staining. Total RNA extracted from the breast cancer cell line MDA-MB-231 was used as a positive control. (C) PPARα expression was determined by Western blot, as described in “Materials and methods.” Total protein isolated from the breast cancer cell line SKBR-3 served as a positive control for PPARα expression. (D) Functional analysis of several PPAR agonists and antagonists. SD1 cells were cotransfected with either a PPARα or a PPARγ expression vector (wt-PPARα or wt-PPARγ), a PPRE reporter vector (PPREx3-tk-luciferase), and a Renilla luciferase vector as an internal control for transfection efficiency. Eighteen hours after transfection, the cells were incubated with PPARα ligand (WY14,643), PPARγ ligand (PGZ), TZD18, PPARα antagonist (MK886), PPARγ antagonist (GW9662), or their combinations for another 24 hours. All drugs were used at 10 μM. After incubation, firefly and Renilla luciferase activities were determined. The firefly luciferase activity was normalized with Renilla luciferase activity and is shown as the fold increase over vehicle-treated controls. The result is a representative of 3 independent experiments.
RT-PCR
Reverse transcription-polymerase chain reaction (RT-PCR) reactions were carried out with GeneAmp PCR System 2700 (Applied Biosystems, Weiterstadt, Germany). RNA extraction and cDNA preparation were as described previously.12 After reverse transcription, 1 μL cDNA was used for further PCR analysis. Primer sequences for PPARα were: forward, 5′-acttatcctgtggtccccgg; reverse, 5′-ccgacagaaaggcacttgtga. A touch-down PCR protocol was used as described by Suchanek et al28 with the following modifications: samples were amplified for 32 cycles, and the annealing temperature ranged between 65°C and 49°C, decreasing 0.5°C after each cycle, and then amplified for another 10 cycles at the annealing temperature of 49°C.
Western blot analysis
Protein concentrations of whole cell lysates and cytoplasmic or nuclear extracts were measured using a BCA protein assay kit (Pierce, Bonn, Germany). Western blot analysis was performed as described previously.12 The following antibodies from Santa Cruz Biotechnology (Heidelberg, Germany) were used in this study: anti-PPARα (sc-9000), anti-bax (sc-493), anti-p27 (sc-528), anti-cyclin D2 (sc-754), anti-cyclin E (sc-247), anti-cyclin-dependent kinase (CDK-2 (sc-6248), anti-CDK-4 (sc-260), anti-c-Myc (sc-40), anti-NF-κB p65 (sc-372), anti-IκBα (sc-371), and anti-β-actin (sc-1616). Anti-phospho-IκBα (Ser32) (no. 9241) was from Cell Signaling (Beverly, MA). Primary antibodies were diluted 1:200 to 1:500.
Transient transfection assay
Transient transfections were carried out in SD1 cells by using electroporation with an EPI2500 electroporator (Dr L. Fischer Lab Instruments, Heidelberg, Germany). Transfection mixes included 7.5 μg PPREx3-tk-luc, 7.5 μg wild-type human PPARγ expression vector (pcDNAFlag-PPARγ1) or wild-type PPARα expression vector (pCMX-mPPARα), and 100 ng Renilla luciferase reporter plasmid (served as an internal transfection efficiency control). After transfection, cells were cultured and exposed to various agents. Luciferase activities were determined using the dual luciferase reporter assay system (Promega, Madison, WI) according to the instructions of manufacturer.
Measurement of apoptosis by cell-death ELISA
Cells were cultured in the presence or absence of TZD18. Apoptotic cell death was determined with the cell death detection ELISAplus (Roche Diagnostics, Heidelberg, Germany) according to the manufacturer's recommendations with modifications. Briefly, after lysis and centrifugation, cell lysates were incubated with the biotin-labeled antihistone and peroxidase-labeled anti-DNA antibodies in a streptavidin-coated microtiter plate for 2 hours at room temperature. After incubation, the peroxidase substrate ABTS was added, the plate was incubated at room temperature for approximately 10 minutes, and the peroxidase activity was determined by an HTII ELISA reader (Anthos) at 405 nm. Data were presented as fold increase of optical density (OD) and compared with untreated control.
Measurement of apoptosis by TUNEL
Cells were treated with either PGZ (20 μM) or TZD18 (20 μM). After incubation, apoptosis was determined by the terminal deoxynucleotidyl-transferase-mediated UTP nick end labeling (TUNEL) technique using an in situ cell death detection kit (Roche Applied Biosystems, Heidelberg, Germany) by following the instructions of the manufacturer. Briefly, cells were cytospun on sides, permeabilized using 0.1% Triton-PBS for 60 minutes, and stained with fluorescein-dUTP and dNTP. Mounting was performed using Immersol 518F (Zeiss, Jena, Germany). Images were obtained using a Zeiss Axioskop 2 microscope equipped with a Plan Neofluar 40 ×/1.4 objective lens (total magnification, × 400). Oil was used as the imaging medium (Zeiss). For documentation of the images, an MRm cooled grayscale camera and AxioVision 4.0 software (Zeiss) were used.
Measurement of caspase 8 and caspase 9 activities
Apoptosis was induced by incubation of cells with TZD18. The activities of caspase 8 and caspase 9 were determined with caspase 8 and caspase 9 colorimetric activity assay kits (Chemicon, Temecula, CA) according to the manufacturer's recommendations with modifications. Briefly, 2 × 106 cells were lysed in lysis buffer, and the protein concentration for each sample was measured using the BCA protein assay kit. Incubation of cell lysates with caspase 8-specific substrate IETD-p-nitroaniline or caspase 9-specific substrate LEHD-p-nitroaniline led to the cleavage of these substrates by caspase 8 or caspase 9, respectively, and the release of chromophore p-nitroaniline. Caspase activities were detected by measuring this chromophore using the HTII ELISA reader (Anthos) at 405 nm. OD values were then normalized to protein concentrations of the samples. Fold-increase in either caspase 8 or caspase 9 activities before and after TZD18 treatment was determined by comparing the OD value of the apoptotic sample with that of untreated control.
Measurement of NF-κB activity by EMSA and ELISA
SD1 cells were treated with either vehicle or 20 μM TZD18 for 1 to 4 days, respectively. Cells were harvested, and nuclear and cytoplasmic protein was extracted by the use of NE-PER nuclear and cytoplasmic extraction kit (Pierce). For determination of NF-κB-binding activities, a double-stranded oligonucleotide containing the NF-κB-binding site (5′-AGT TGA GGG GAC TTT CCC AGG C-3′) was used.29 This oligonucleotide was synthesized and 3′-end labeled with biotin by TIB Molbiol (Berlin, Germany). Electrophoretic mobility shift assay (EMSA) was performed using a LightShift chemiluminescence EMSA kit (Pierce) according to the instructions of the manufacturer with modifications. Equal amounts of nuclear protein (5 μg per sample) were incubated with biotin-labeled NF-κB DNA probe in binding buffer for 30 minutes at room temperature. DNA-protein complexes were separated by nondenaturing 6% polyacrylamide gel electrophoresis. The specificity of the shifted bands was verified by including 100-fold unlabeled double-stranded oligonucleotide in the reaction complexes. Identities of the NF-κB subunits were determined by adding antibodies specific for p50 (sc-114), p65 (sc-109), p52 (sc-7386), c-Rel (sc-71), and RelB (sc-226) (Santa Cruz Biotechnology) to the reactions and observing the supershifted bands. Detection of biotin-labeled DNA by chemiluminescence was performed according to the instructions of manufacturer. The NF-κB DNA-binding activities of cells before and after treatment with TZD18 were also quantitatively analyzed by using a TransAM NFκB Family Transcription Factor ELISA Assay Kit (Active Motif, Rixensart, Belgium) according to the instructions of the manufacturer.
Statistical analysis
Results were presented as a mean plus or minus standard deviation (SD) of at least 3 independent experiments. Synergistic effects of the combination of TZD18 and imatinib on cell proliferation were assessed using the Chou-Talalay method30 and Calcusyn software (Biosoft, Ferguson, MO). Briefly, the dose-effect curve for each drug alone was determined based on the experimental observations using the median-effect principle; the combination index (CI) for each experimental combination was then calculated according to the following equation:
where (D)1 and (D)2 are the doses of drug 1 and drug 2, respectively, that have x effect when used in combination and (Dx)1 and (Dx)2 are the doses of drug 1 and drug 2 that have the same x effect when used alone. CI values equaling 1 indicate additive effects; CI values less than 1 indicate a greater than expected additive effect (synergism).
Results
PPARα and PPARγ were expressed in lymphocytic leukemia cells
PPARγ has previously been shown to be expressed in human Ph+ lymphocytic leukemia cell lines.12 Direct evidence for the expression of PPARα in ALL cell lines is still lacking. We showed here, with the use of RT-PCR, the expression of PPARα in all 3 lymphocytic leukemia cell lines (Figure 1B). RNA isolated from the breast cancer cell line MDA-MB-231 was used as a positive control.28 Western blot analysis confirmed these results (Figure 1C). Total protein isolated from the breast cancer cell line SKBR-3 served as a positive control because PPARα protein was previously identified in this cell line.31 Next, we tested whether all the agonists and antagonists used in this study were functional in our cell system by cotransfection of a WT-PPARα or a WT-PPARγ expression vector and a PPRE-luciferase reporter vector into SD1 cells. PPARα ligand WY14,64332 and PPARγ ligand PGZ12 individually resulted in approximately a 3-fold increase of luciferase activity, whereas PPARα antagonist MK88633 and PPARγ antagonist GW9662,34 respectively, significantly reversed this effect. As a dual ligand for these 2 subtypes of PPARs, the increased luciferase activities by TZD18 could be reversed by the addition of appropriate antagonists (Figure 1D). These results clearly indicated that all ligands used in this study were able to activate PPARγ or PPARα, whereas the antagonists were able to inhibit the activation of PPARγ or PPARα under our conditions.
TZD18 caused growth inhibition of Ph+ lymphocytic leukemia cells
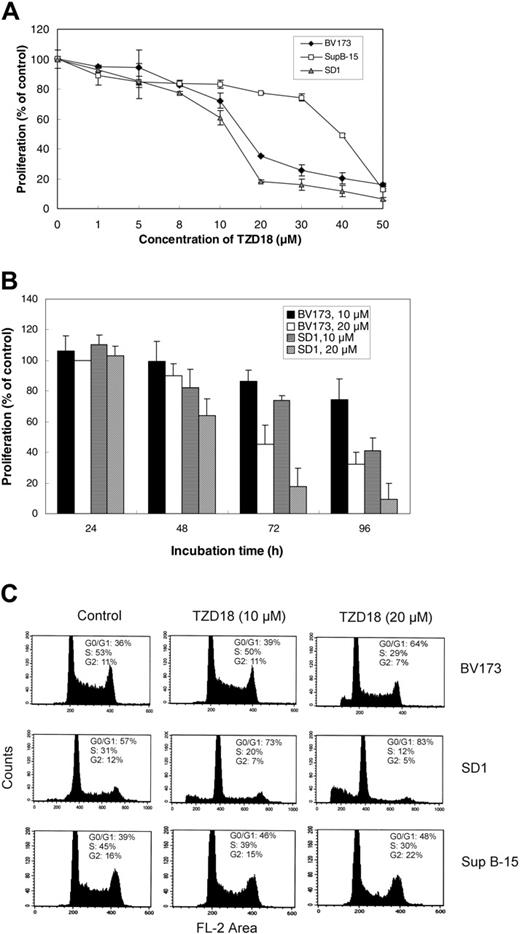
We studied the effect of TZD18 on cell growth in Ph+ lymphocytic leukemia cell lines BV173, SD1, and Sup B-15. This dual ligand inhibited the proliferation of all of these cell lines in a dose- and time-dependent manner (Figure 2A-B). The rank order of sensitivity of these cell lines to TZD18 was SD1 > BV173 > Sup B-15. BV173 and SD1 were more sensitive to the treatment, with growth inhibition of approximately 70% and 80% at the concentration of 20 μM after 4 days, respectively, whereas Sup B-15 was relatively resistant.
The growth inhibitory effect of TZD18 was much stronger than that of PGZ. At the concentration of 20 μM in cell lines BV173 and SD1, PGZ showed only 10% to 20% growth inhibition (data not shown). We tested TZD18 in different types of tumors and normal cells and found that different types of cells reacted to the drug differently. In the human glioblastoma cell line GMS-10, up to 40 μM TZD18 resulted in approximately 10% growth increase after 4 days of treatment (data not shown), whereas human mesenchymal stem cells isolated from human bone marrow and normal stromal cells from breast cancer samples showed no significant change in proliferation after 4-day culture with up to 50 μM TZD18 (data not shown). These results suggest that the inhibitory effects on human lymphocytic leukemia cells might not have been caused by the nonspecific cytotoxicity of TZD18.
We have previously shown that the antiproliferative effect of PPARγ ligands was attributable to their inhibition of cell cycle progression.12,35 Therefore, we investigated the possible effect of TZD18 on the cell cycle distribution of the lymphocytic cell lines. As indicated in Figure 2C, cell cycle progression after exposure to TZD18 (10 and 20 μM) for 3 days was inhibited in a dose-dependent fashion for all cell lines. In all cases, G0/G1-phase arrest was observed. The S phase in BV173 and SD1 after treatment with 20 μM TZD18 was decreased from 52% and 31% to 29% and 13%, respectively. The data clearly indicate that cell cycle arrest, at least in part, might have contributed to the inhibitory effects of this ligand on the growth of lymphocytic leukemia cell lines.
Effect of the PPARα/γ ligand TZD18 on the proliferation of tested cell lines. TZD18 inhibits proliferation of tested cell lines in a dose-dependent (A) and a time-dependent (B) manner. Cells (2 × 105/mL) were incubated in the presence of TZD18 at various concentrations for different numbers of days (measured in hours). Cell proliferation was measured by MTT test; results were expressed as a percentage of control (without treatment). Values are mean ± SD of 6 individual experiments. (C) TZD18 alters cell cycle progression. Cells (2 × 105/mL) were incubated in the presence or absence of TZD18 (10 or 20 μM, 3 days), fixed, treated with RNase, and stained for DNA with PI. Cell cycle distribution was determined by FACS analysis. Results represent the percentage of the total cell population. Figure is representative of 3 independent experiments.
Effect of the PPARα/γ ligand TZD18 on the proliferation of tested cell lines. TZD18 inhibits proliferation of tested cell lines in a dose-dependent (A) and a time-dependent (B) manner. Cells (2 × 105/mL) were incubated in the presence of TZD18 at various concentrations for different numbers of days (measured in hours). Cell proliferation was measured by MTT test; results were expressed as a percentage of control (without treatment). Values are mean ± SD of 6 individual experiments. (C) TZD18 alters cell cycle progression. Cells (2 × 105/mL) were incubated in the presence or absence of TZD18 (10 or 20 μM, 3 days), fixed, treated with RNase, and stained for DNA with PI. Cell cycle distribution was determined by FACS analysis. Results represent the percentage of the total cell population. Figure is representative of 3 independent experiments.
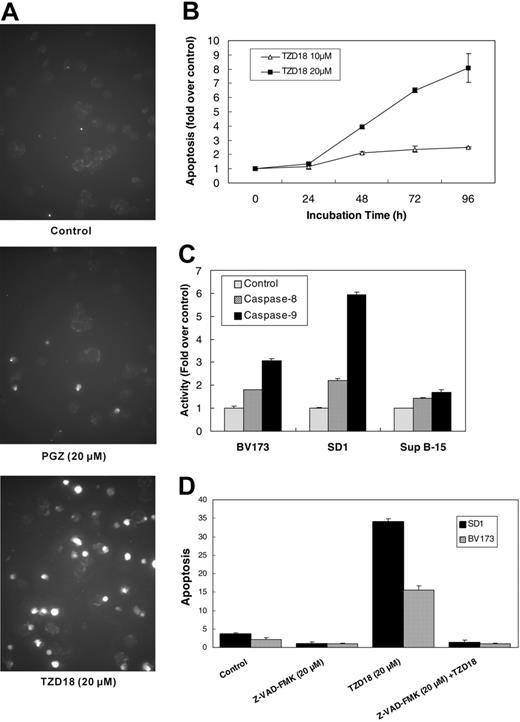
TZD18-induced apoptosis of Ph+ lymphocytic leukemia cells
As did the PPARγ ligands PGZ and 15d-PGJ2,12 TZD18 caused significant apoptosis of each cell line, as determined by TUNEL and cell death ELISA. Sup B-15 cells were least sensitive to TZD18-induced apoptosis, consistent with data from the proliferation assay. In contrast, a great part of BV173 and SD1 cells underwent apoptosis when they were exposed to TZD18 (10 or 20 μM) (Figure 3A; Table 1). In comparison, PGZ (20 μM, 4 days) induced only a 2-fold increase of apoptosis in BV173 and none in SD1 cells (Table 1). Similar to the growth inhibitory effects, apoptosis was also induced by TZD18 (10 and 20 μM) in a time-dependent fashion in SD1 cells (Figure 3B). In addition, TZD18 remarkably activated caspase 9 and, to a lesser extent, caspase 8 in SD1 and BV173 cells (Figure 3C). Again, only minimal activation of caspase 8 and caspase 9 was observed in Sup B-15 cells after their exposure to TZD18 (data not shown). Furthermore, ligand-induced apoptosis was totally reversed by addition of the pan-caspase inhibitor Z-VAD-FMK to the BV173 and SD1 cell lines (Figure 3D), indicating that apoptosis induced by TZD18 was caspase dependent.
Induction of apoptosis by PGZ and TZD18 in Ph+ ALL cell lines
Cell line and treatment (dose, μM) . | TUNEL, % positive, mean ± SD . | Enrichment factor, mean ± SD* . |
|---|---|---|
| BV173 | ||
| Control | 4.4 ± 0.17 | 1.00 ± 0.05 |
| PGZ (20) | 10.2 ± 0.47 | 1.96 ± 0.11 |
| TZD18 (10) | 7.6 ± 0.76 | 2.15 ± 0.06 |
| TZD18 (20) | 29.2 ± 6.5 | 9.46 ± 0.27 |
| SD1 | ||
| Control | 3.4 ± 0.85 | 1.00 ± 0.06 |
| PGZ (20) | 4.8 ± 1.56 | 1.01 ± 0.06 |
| TZD18 (10) | 9.2 ± 1.84 | 1.53 ± 0.09 |
| TZD18 (20) | 21.7 ± 1.89 | 8.24 ± 0.25 |
| Sup B-15 | ||
| Control | ND | 1.00 ± 0.01 |
| PGZ (20) | ND | ND |
| TZD18 (10) | ND | 1.91 ± 0.14 |
| TZD18 (30) | ND | 2.36 ± 0.07 |
Cell line and treatment (dose, μM) . | TUNEL, % positive, mean ± SD . | Enrichment factor, mean ± SD* . |
|---|---|---|
| BV173 | ||
| Control | 4.4 ± 0.17 | 1.00 ± 0.05 |
| PGZ (20) | 10.2 ± 0.47 | 1.96 ± 0.11 |
| TZD18 (10) | 7.6 ± 0.76 | 2.15 ± 0.06 |
| TZD18 (20) | 29.2 ± 6.5 | 9.46 ± 0.27 |
| SD1 | ||
| Control | 3.4 ± 0.85 | 1.00 ± 0.06 |
| PGZ (20) | 4.8 ± 1.56 | 1.01 ± 0.06 |
| TZD18 (10) | 9.2 ± 1.84 | 1.53 ± 0.09 |
| TZD18 (20) | 21.7 ± 1.89 | 8.24 ± 0.25 |
| Sup B-15 | ||
| Control | ND | 1.00 ± 0.01 |
| PGZ (20) | ND | ND |
| TZD18 (10) | ND | 1.91 ± 0.14 |
| TZD18 (30) | ND | 2.36 ± 0.07 |
BV173, SD1, and Sup B-15 cells were treated with PGZ (20 μM) or TZD18 (10 μM and 20 μM) for 4 days. Apoptosis was determined by TUNEL assay and cell death ELISAplus kit, as described in “Materials and methods.” TUNEL assay results are expressed as percentage of positive cells. Cell death ELISA results are expressed as fold change of enrichment of nucleosomes in the cytoplasm of cells treated with drugs compared with controls (without treatment). Data represent the mean ± SD of triplicate experiments.
ND indicates not done.
Enrichment factor is the enrichment of nucleosomes in the cytoplasm measured by cell death ELISA and is an indicator of apoptosis. Values represent fold change over control
Effect of TZD18 on apoptosis of tested cell lines. (A) Apoptotic cells in BV173 after treatment with PGZ (20 μM) or TZD18 (20 μM) were measured by TUNEL assay, as described in “Materials and methods.” (B) SD1 cells were cultured in the presence of TZD18 (10 or 20 μM) for different hours, washed, and lysed. Apoptosis was measured by cell death ELISA, as described in “Materials and methods.” Results are expressed as the fold change of enrichment of nucleosomes in the cytoplasm of cells treated with drugs compared with controls (without treatment). Data represent the mean ± SD of triplicate experiments. (C) Cells were cultured in the presence of TZD18 (20 μM) for 4 days, washed, and lysed in lysis buffer. Caspase 8 and caspase 9 activities in the cell lysates were measured as described in “Materials and methods.” Results were expressed as the fold increase of OD values compared with control (without treatment). Figure is representative of the results of 3 independent experiments. (D) Cells were incubated in the presence of TZD18 (20 μM), Z-VAD-FMK (20 μM), or both for 4 days. Cell apoptosis was examined as described in “Materials and methods.” Apoptosis was expressed as fold change in enrichment of nucleosomes in the cytoplasm of cells treated with drugs compared with the sample with the lowest value (cells treated with Z-VAD-FMK). Results are the mean ± SD of 3 individual experiments.
Effect of TZD18 on apoptosis of tested cell lines. (A) Apoptotic cells in BV173 after treatment with PGZ (20 μM) or TZD18 (20 μM) were measured by TUNEL assay, as described in “Materials and methods.” (B) SD1 cells were cultured in the presence of TZD18 (10 or 20 μM) for different hours, washed, and lysed. Apoptosis was measured by cell death ELISA, as described in “Materials and methods.” Results are expressed as the fold change of enrichment of nucleosomes in the cytoplasm of cells treated with drugs compared with controls (without treatment). Data represent the mean ± SD of triplicate experiments. (C) Cells were cultured in the presence of TZD18 (20 μM) for 4 days, washed, and lysed in lysis buffer. Caspase 8 and caspase 9 activities in the cell lysates were measured as described in “Materials and methods.” Results were expressed as the fold increase of OD values compared with control (without treatment). Figure is representative of the results of 3 independent experiments. (D) Cells were incubated in the presence of TZD18 (20 μM), Z-VAD-FMK (20 μM), or both for 4 days. Cell apoptosis was examined as described in “Materials and methods.” Apoptosis was expressed as fold change in enrichment of nucleosomes in the cytoplasm of cells treated with drugs compared with the sample with the lowest value (cells treated with Z-VAD-FMK). Results are the mean ± SD of 3 individual experiments.
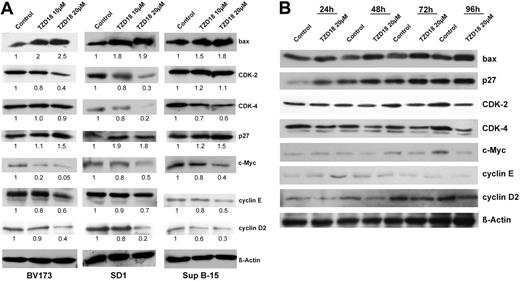
TZD18-regulated cell-cycle- and apoptosis-related proteins
Given that cyclins D and E, as well as CDK-2 and -4 and their inhibitors p21cip1 and p27kip1, are key regulators of G1- to S-phase cell cycle progression, we investigated the effects of TZD18 on the expression of these molecules. The pro-growth molecules cyclin D2, CDK-2, CDK-4, and cyclin E were down-regulated, whereas anti-growth molecule CDK inhibitor (CDKI) p27kip1, but not p21 (data not shown), was up-regulated by TZD18 (Figure 4A). We found previously that alteration of the bcl-2-related apoptotic pathways is associated with PPARγ-induced apoptosis in human breast cancer cell lines.10 Therefore, we assessed the regulation of bcl-2 and bax after exposure to TZD18. The antiapoptotic factor bcl-2 was unchanged (data not shown), whereas the proapoptotic factor bax was dramatically up-regulated (Figure 4A). In addition, the oncoprotein c-Myc, which is a master regulator of the G1/S transition cascade and apoptosis, was prominently down-regulated in all 3 cell lines (Figure 4A). We also analyzed the changes of these cell cycle- and apoptosis-related factors at different time points after exposure to TZD 18 (20 μM) in SD1 cells. As shown in Figure 4B, expression change could be observed as early as 24 hours after culture with TZD18.
TZD18-induced cell growth inhibition was independent of PPARα and PPARγ
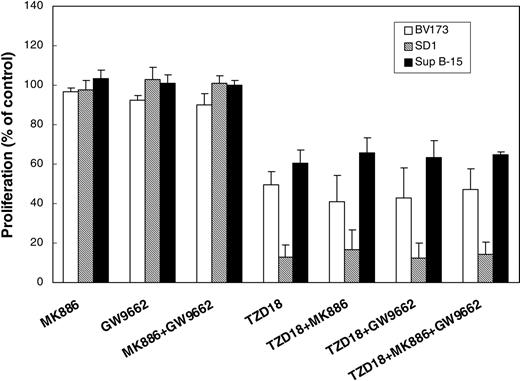
We have previously demonstrated that the PPARγ antagonists could not reverse the PPARγ agonist-induced cell growth inhibition of human lymphocytic leukemia.12 Similarly, although all the antagonists used in this study were able to inhibit PPARα or PPARγ activities (Figure 1D), the PPARγ antagonist GW9662 and the PPARα antagonist MK886, either alone or in combination, could not reverse the growth inhibitory effect of TZD18 (Figure 5), suggesting that this dual ligand may exert its effect on the proliferation and apoptosis of lymphocytic leukemia cells independently of its activation of PPARα and PPARγ.
NF-κB-binding activities were decreased by TZD18
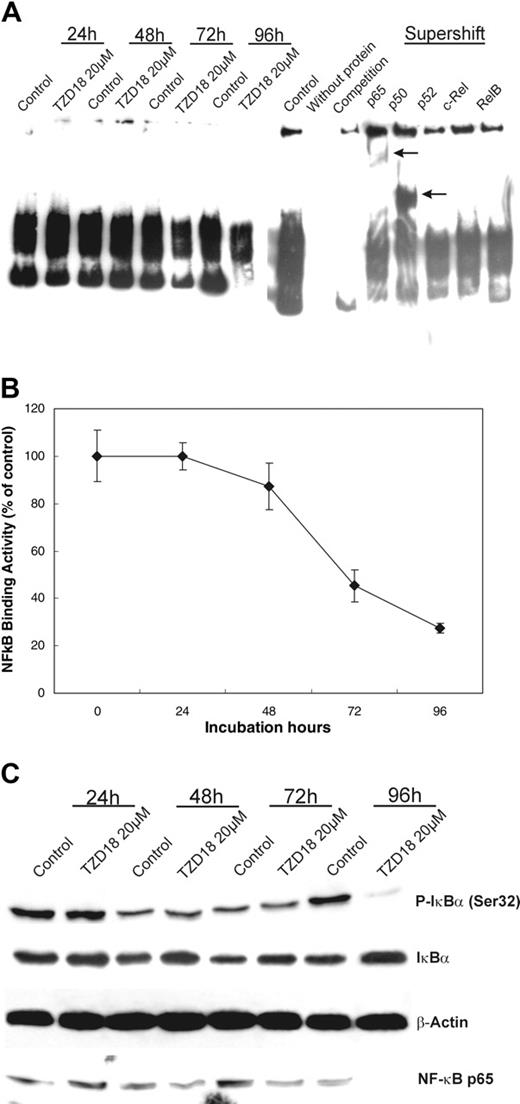
Increasing data suggest that PPARγ ligands exert their effects through interaction with the transcription factor NF-κB. Therefore, the influence of TZD18 on the NF-κB-DNA-binding activities was first investigated with EMSA. Treatment with TZD18 resulted in a time-dependent decrease in the NF-κB-DNA-binding activities in cell line SD1. The specificity of the NF-κB bands was confirmed by competition assay and supershift analysis (Figure 6A). We quantitatively measured the NF-κB activities in the same cell nuclear protein used in EMSA (Figure 6B). The obtained results confirmed the EMSA data. For further analysis of the mechanism responsible for the decreased NF-κB activities, Western blotting was performed with nuclear and cytoplasmic extracts prepared from SD1 cells at various time points after treatment with TZD18. Nuclear p65 levels decreased after TZD18 treatment (Figure 6C), whereas the p65 band in whole cell lysates was similar (data not shown), suggesting that p65 translocation to the nucleus was blocked. Because IκBα phosphorylation on serine residues 32 and 36 with subsequent ubiquitination and degradation plays an important role in NF-κB translocation and activation,36 serine phosphorylation of IκBα (Ser P-IκBα) was also analyzed. Decreased Ser P-IκBα was detected in cytoplasmic lysate isolated from TZD18-treated SD1 cells, whereas the total IκBα level was increased (Figure 6C). These findings indicate that increased expression and decreased degradation of IκBα may inhibit the translocation of NF-κB to the nucleus and, subsequently, contribute to the decrease of NF-κB DNA-binding activities.
Expression of cell cycle- and apoptosis-related proteins in tested cell lines cultured with TZD18. (A) BV173, SD1, and Sup B-15 cells were incubated with or without TZD18 (10 or 20 μM, 4 days), and cell lysates were examined for cell cycle- and apoptosis-related proteins by Western blot, as described in the “Materials and methods.” A representative blot of 3 independent experiments is shown. The relative intensity of each band was quantified using Phoretix ID Quantifier software and was expressed as percentage of control. (B) Equal amounts of protein prepared from SD1 cells exposed to TZD18 (20 μM) for different durations were examined for the same cell cycle- and apoptosis-related proteins as in panel A.
Expression of cell cycle- and apoptosis-related proteins in tested cell lines cultured with TZD18. (A) BV173, SD1, and Sup B-15 cells were incubated with or without TZD18 (10 or 20 μM, 4 days), and cell lysates were examined for cell cycle- and apoptosis-related proteins by Western blot, as described in the “Materials and methods.” A representative blot of 3 independent experiments is shown. The relative intensity of each band was quantified using Phoretix ID Quantifier software and was expressed as percentage of control. (B) Equal amounts of protein prepared from SD1 cells exposed to TZD18 (20 μM) for different durations were examined for the same cell cycle- and apoptosis-related proteins as in panel A.
TZD18 effect on cell proliferation is independent of PPARγ and PPARα. Cells (2 × 105/mL) were incubated with antagonists for either PPARγ (GW9662, 2 μM) or PPARα (MK886, 2 μM) in the absence or presence of TZD18 (20 μM) for 4 days. Cell proliferation was measured by the MTT assay, as described in “Materials and methods.” Results are expressed as a percentage of control (without treatment). Values are mean ± SD of at least 3 experiments.
TZD18 effect on cell proliferation is independent of PPARγ and PPARα. Cells (2 × 105/mL) were incubated with antagonists for either PPARγ (GW9662, 2 μM) or PPARα (MK886, 2 μM) in the absence or presence of TZD18 (20 μM) for 4 days. Cell proliferation was measured by the MTT assay, as described in “Materials and methods.” Results are expressed as a percentage of control (without treatment). Values are mean ± SD of at least 3 experiments.
Combination of TZD18 and imatinib markedly inhibited cell growth of Ph+ ALL cell lines
BV173 cells were extremely sensitive to imatinib. For example, 0.5 μM imatinib inhibited the cell proliferation of BV173 cells by 68%, and 2.5 μM imatinib resulted in 98% inhibition (data not shown). Sup B-15 cells were less sensitive; nevertheless, 2.5 μM imatinib caused more than 60% growth inhibition of these cells. In contrast to these 2 lymphocytic leukemia cell lines, SD1 cells were relatively resistant to imatinib, with 2.5 μM of the drug producing only approximately 12% growth inhibition. Interestingly, TZD18 (10 μM) significantly enhanced the inhibitory effect of imatinib on cell proliferation in all 3 cell lines (Figure 7A). We further analyzed the pharmacologic interactions between TZD18 and imatinib using a nonconstant ratio combination design with Calcusyn (Biosoft) software. The obtained isobologram suggested that the combination of the 2 drugs resulted in the synergistic inhibition (CI, less than 1.0) of cell proliferation in all 3 cell lines (Figure 7B).
Discussion
We have shown that the PPARγ ligands PGZ and 15d-PGJ2 inhibit the cell proliferation of lymphocytic leukemia cell lines, which is associated with G1-phase cell cycle arrest and apoptosis.12 Furthermore, cell cycle- and apoptosis-related genes, such as cyclin D1, cyclin D2, and p27kip1, are altered by these ligands. In this study, we demonstrated that a novel PPARα and PPARγ dual ligand, TZD18, which belongs structurally to the TZD class, also showed anti-growth effects on Ph+ lymphocytic leukemia cell lines that were stronger that those of PGZ. Our studies further revealed that the activation of PPARα and PPARγ seemed not to be required for TZD18 to exert its effects. Of more clinical relevance, this compound enhanced the ability of imatinib to inhibit cell proliferation of imatinib-sensitive and -resistant Ph+ lymphocytic leukemia cell lines.
PPARγ is expressed in human lymphocytic leukemia cell lines, as observed by our and other groups.12,37,38 However, to date, reports of PPARα expression in hematopoietic cells are limited to human myeloblastic leukemia cell line HL-6023,39 and murine lymphocytes.40 We have shown in this study that human Ph+ ALL cell lines also express PPARα.
Compared with PGZ, TZD18 more potently inhibits the growth of all Ph+ lymphocytic leukemia cell lines tested through G0/G1 cell cycle arrest. In contrast, the percentage of cells in the G2/M phase changed only minimally. This result suggests that the effect of the PPARα/γ ligand on cell cycle progression was similar to what we have previously reported for the PPARγ ligands. Interestingly, we have found that several tumor and normal cell lines did not respond to exposure to this substance, strongly suggesting that the effects of TZD18 observed on human Ph+ ALL cell lines are not attributed to the nonspecific cytotoxicity of this agent.
TZD18 inhibits NF-κB DNA-binding activities. Nuclear and cytoplasmic extracts were prepared from untreated and TZD18-treated (20 μm) SD1 cells after 24-, 48-, 72-, and 96-hour cultures. Equal amounts of protein were used for all assays. (A) EMSA analysis was performed to determine the DNA-binding capacity of NF-κB with the use of nuclear extracts. Competition analysis was performed by using a 100-fold excess of unlabeled NF-κB consensus-binding site. Antibodies specific for p50, p65, p52, cRel, and RelB were used in supershift analysis. Arrows indicate the positions of the super-shifted bands. The result is representative of 3 independent experiments. (B) Quantitative measurement of NF-κB activity was performed with nuclear proteins, as described in “Materials and methods.” NF-κB activity of TZD18-treated cells was expressed as a percentage of untreated control. Data represent mean ± SD of triplicate experiments. (C) NF-κB expression in nuclear extracts and phosphorylated I-κBα expression in cytoplasmic extracts were analyzed by Western blot. Membranes were blotted with antibodies specific for NF-κB subunit p65 and I-κBα phosphorylated at serine 32.
TZD18 inhibits NF-κB DNA-binding activities. Nuclear and cytoplasmic extracts were prepared from untreated and TZD18-treated (20 μm) SD1 cells after 24-, 48-, 72-, and 96-hour cultures. Equal amounts of protein were used for all assays. (A) EMSA analysis was performed to determine the DNA-binding capacity of NF-κB with the use of nuclear extracts. Competition analysis was performed by using a 100-fold excess of unlabeled NF-κB consensus-binding site. Antibodies specific for p50, p65, p52, cRel, and RelB were used in supershift analysis. Arrows indicate the positions of the super-shifted bands. The result is representative of 3 independent experiments. (B) Quantitative measurement of NF-κB activity was performed with nuclear proteins, as described in “Materials and methods.” NF-κB activity of TZD18-treated cells was expressed as a percentage of untreated control. Data represent mean ± SD of triplicate experiments. (C) NF-κB expression in nuclear extracts and phosphorylated I-κBα expression in cytoplasmic extracts were analyzed by Western blot. Membranes were blotted with antibodies specific for NF-κB subunit p65 and I-κBα phosphorylated at serine 32.
PPARγ ligands have been shown to inhibit G1/S-phase progression of colon,41,42 pancreatic,43,44 and breast cancers45 and of glioblastomas35 and leukemias.12,46 Targeting the key regulators of the G1/S transition is probably the reason for the observed cell cycle arrest. For example, PPARγ ligands result in the down-regulation of D-type cyclins47-50 and CDKs50 and the up-regulation of CDKIs such as p21cip1,44,47,49,51 , p27kip1,12 and p16ink4a.48 A similar effect was also observed with the PPARα ligand clofibrate using the human leukemia cell line HL-60. This hypolipidemia drug caused growth inhibition and cell cycle blockade related to the down-regulation of cyclin D2.52 Congruent with these reports, we showed here that TZD18 also modulated the key regulators of G1/S progression in such a way that these alterations might lead to cell blockage in G0/G1—that is, it reduced the expression of cyclin D2, cyclin E, CDK-2, and CDK-4 and enhanced the expression of CDKI p27kip.1 We have further noted that c-Myc protein was markedly down-regulated by TZD18. This oncoprotein stimulates G1 progression through multiple mechanisms such as repressing, directly or indirectly, p21cip1 and p27kip1 expression, increasing CDK-4, and inducing cyclin D1 and cyclin D2 synthesis (for a review, see Nasi et al53 ). Although our studies did not show whether the alterations of these cell cycle regulators are a direct or an indirect consequence of the down-regulation of c-Myc, we believe that these alterations may account for the observed cell cycle arrest induced by TZD18.
Down-regulation of cyclin D1 by PPARγ ligands has been described in studies of human vascular smooth muscle cells54 and non-small lung cancer cells47 and our previous studies with human lymphocytic leukemia cells.12 The reduced expression of cyclin D1 was demonstrated to be responsible for troglitazone (a PPARγ ligand of the TZD class)-mediated cell cycle arrest in MCF-7 cells.50 However, we did not observe changes of cyclin D1 in Ph+ lymphocytic leukemia cell lines after their exposure to TZD18. On the contrary, we found in this study that cyclin D2 was highly expressed in these cells, and its expression was markedly reduced by TZD18 treatment. Cyclin D2 is constitutively expressed at high levels in lymphocytic malignancies, especially in BCR-ABL-transformed cells, whereas its expression is either low or absent in Ph-negative lymphocytic leukemia cell lines.55-57 The link between BCR-ABL transformation and cyclin D2 overexpression was further elucidated by experiments showing that ectopic overexpression of cyclin D2 rescued the inhibition of Bcr-Abl protein tyrosine kinase activities caused by imatinib.56 Furthermore, bone marrow cells from cyclin D2-deficient strains of mice failed to proliferate in response to transfection of BCR-ABL expression vectors.57 We have also observed that growth inhibition caused by the PPARγ ligands PGZ and 15d-PGJ2 was associated with the down-regulation of cyclin D2 in lymphocytic leukemia cells, including Ph+ lymphocytic leukemia.12 Hence, cyclin D2 may be one of the key regulators of the growth and maintenance of Ph+ lymphocytic leukemia cells. Down-regulation of cyclin D2 by the PPARα ligands clofibrate and ciprofibrate has also been described in HL-60 cells.52 Therefore, we speculate that the repression of cyclin D2 by PPAR ligands may represent a crucial mechanism underlying the PPAR ligand-induced growth inhibition of Ph+ lymphocytic leukemia cells.
TZD18 enhances growth inhibition of imatinib. (A) Cells (2 × 105/mL) were incubated with different concentrations of imatinib, as indicated in the figure, in the presence or absence of TZD18 (10 μM, 4 days). Cell proliferation was measured by the MTT assay, as described in “Materials and methods.” Results are expressed as proliferation percentage of control (without treatment). Values are mean ± SD of 6 parallel experiments. (B) Statistical analysis of the effects of the combination of TZD18 and imatinib. BV173, SD1, and Sup B-15 cells were cultured in the presence of escalating doses of imatinib (0.25, 0.5, 1, 2.5 μM) or TZD18 (5, 10, 15, 20 μM) and the combination of 0.5 μM imatinib with varied concentrations of TZD18 (5, 10, 15, 20 μM) or the combination of 10 μM TZD18 with varied concentrations of imatinib (0.25, 0.5, 1, 2.5 μM). After 4 days, cell proliferation was measured with MTT assay. CI values for each data point of this nonconstant ratio design were calculated using Calcusyn software. All CI values were less than 1. Representative diagnosis-normalized isobolograms obtained from SD1 cells are shown (top, TZD18 concentration fixed at 10 μM; bottom, imatinib concentration fixed at 0.5 μM).
TZD18 enhances growth inhibition of imatinib. (A) Cells (2 × 105/mL) were incubated with different concentrations of imatinib, as indicated in the figure, in the presence or absence of TZD18 (10 μM, 4 days). Cell proliferation was measured by the MTT assay, as described in “Materials and methods.” Results are expressed as proliferation percentage of control (without treatment). Values are mean ± SD of 6 parallel experiments. (B) Statistical analysis of the effects of the combination of TZD18 and imatinib. BV173, SD1, and Sup B-15 cells were cultured in the presence of escalating doses of imatinib (0.25, 0.5, 1, 2.5 μM) or TZD18 (5, 10, 15, 20 μM) and the combination of 0.5 μM imatinib with varied concentrations of TZD18 (5, 10, 15, 20 μM) or the combination of 10 μM TZD18 with varied concentrations of imatinib (0.25, 0.5, 1, 2.5 μM). After 4 days, cell proliferation was measured with MTT assay. CI values for each data point of this nonconstant ratio design were calculated using Calcusyn software. All CI values were less than 1. Representative diagnosis-normalized isobolograms obtained from SD1 cells are shown (top, TZD18 concentration fixed at 10 μM; bottom, imatinib concentration fixed at 0.5 μM).
Cell cycle arrest can be followed by cell differentiation, apoptosis, or both.58 The TZD class of PPARγ ligands has been reported to induce apoptosis in a variety of human solid tumors and hematologic malignancies (for reviews, see Koeffler59 and Michalik et al60 ). By using TUNEL and cell death ELISA, we have demonstrated that TZD18 induced apoptosis in all 3 Ph+ lymphocytic leukemia cell lines in a time- and dose-dependent manner. We showed that a pan-caspase inhibitor, Z-VAD-FMK, completely abolished TZD18-induced apoptosis, indicating that apoptosis caused by this substance is caspase dependent. Normally, caspase-dependent apoptosis is initiated through 2 partially interdependent routes. One is the extrinsic pathway, triggered by the ligation of cell death receptors and leading to the activation of caspase 8. The other is the intrinsic pathway in which the bcl-2 superfamily and mitochondria are involved. Changes of expression of antiapoptotic and proapoptotic proteins of the bcl-2 family disrupt the mitochondrial membranes and result in the release of proapoptotic proteins and the activation of caspase 9. Both caspase 8 and 9 in turn activate apoptosis executers such as the caspase 3 launching apoptosis program. Although we did not observe any change of the antiapoptotic protein bcl-2, we noticed that the proapoptotic protein bax was significantly up-regulated after the Ph+ lymphocytic leukemia cells were cultured with TZD18. This resulted in a reduced bcl-2/bax ratio that might have resulted in increased mitochondrial permeability and apoptosis. Numerous reports demonstrated that the bcl-2/bax ratio, rather than the level of bcl-2 alone, was important for drug-induced apoptosis of leukemic cells.61 At the same time, we observed a dramatic increase in activities of caspase 9 by TZD18 treatment. These data strongly suggest apoptosis induced by TZD 18 may occur through the intrinsic pathway. This is congruent with recent reports that 15d-PGJ262 and the PPARγ ligand CDDO63 induced apoptosis of lymphocytes by activating the mitochondrial pathway. Interestingly, a modest increase in caspase 8 was also observed in this study. However, we did not observe any changes in the expression of cell death receptors (fas, DR4) or their ligands, such as Fas ligands and TRAIL. Neither TNFα, activating antibodies of Fas, nor recombinant TRAIL enhanced the apoptosis produced by TZD18 (data not shown). We hypothesized that the cell death receptor pathway is not important for apoptosis triggered by TZD18. Caspase 8 activation probably is caused by a mechanism independent of cell death receptor.64
We previously showed that the antiproliferative effect of PGZ was independent of activation of PPARγ in lymphocytic leukemia cells.12 In this study, we explored the ability of the PPARγ antagonist GW9662 and the PPARα antagonist MK886 to reverse the effects of TZD18. Although both antagonists could inhibit the activities of PPARα and PPARγ under our conditions—as shown by PPRE-luciferase assay—GW9662 and MK886, either alone or in combination, failed to reverse the antiproliferative effects of TZD18, indicating that this PPAR ligand may act through PPARα- and γ-independent mechanisms. Furthermore, the fact that a classic PPARα ligand, WY14643, did not inhibit but instead slightly enhanced proliferation of the 3 Ph+ lymphocytic leukemia cell lines (data not shown) further supports this speculation.
NF-κB is a collective term that refers to a small class of closely related dimeric transcription factors for a number of target genes, including growth factors, modulators of angiogenesis, cell-adhesion molecules, and antiapoptotic factors.65 Activation of NF-κB is tightly controlled by inhibitory IκB proteins. Increased IκB degradation by phosphorylation and subsequent ubiquitination leads to increased nuclear translocation of NF-κB and its DNA-binding activities. It is believed that NF-κB is a positive regulator of cell cycle progression and that it activates target genes such as cyclin-D1, -D2, -D3, and -E.66-68 It is also an important regulatory factor for c-Myc protein.69,70 Furthermore, activated NF-κB in tumor cells has been shown to increase the transcription of target genes whose products block apoptosis. These genes included members of the TNF family (TRAF1 and TRAF2), members of the BCL-2 family, cellular inhibitors of apoptosis (cIAPs), and others.71 Recently, activation of NF-κB/Rel by Bcr-Abl was demonstrated in Ph+ leukemia in vitro and in vivo.72-75 NF-κB/Rel has been shown to be required for Bcr-Abl-mediated tumorigenicity and for the transformation of Bcr-Abl-positive myeloid cells in vitro and in a murine model.74 Numerous reports have shown that PPARγ and PPARα ligands exert their antitumor or anti-inflammation effects by inhibiting multiple steps of the NF-κB signaling pathway.76-79 We have shown in this study that TZD18 exerted an inhibitory effect on NF-κB DNA-binding activities similar to that of other PPARγ and PPARα ligands. Furthermore, we showed that the expression of NF-κB inhibitor IκBα increased and that the phosphorylation of IκBα decreased after TZD18 exposure. All these changes might have resulted in an increased level of IκBα and, subsequently, in a blocked translocation of NF-κB to the nucleus. Indeed, we showed in this study that the level of p65, one partner of NF-κB dimer, was decreased in the nucleus after TZD18 treatment. Most likely, TZD18 exerted its inhibitory effect on NF-κB DNA-binding activities by increasing the level of NF-κB inhibitors and blocking the nuclear translocations of NF-κB. It is not surprising that the expression of several NF-κB target genes, such as c-MYC, CYCLIN D, and BAX, were also regulated by TZD18. Therefore, interference of the NF-κB signaling pathway by TZD18 may represent a major mechanism for the observed inhibition of proliferation and apoptosis in Ph+ lymphocytic leukemia cells.
The tyrosine kinase inhibitor imatinib binds competitively to the ATP binding site of Abl tyrosine kinase of the Bcr-Abl fusion protein, thereby inhibiting signaling by this oncogenic protein and resulting in significant growth inhibition of Ph+ ALL cells.80 This agent frequently induces remission in patients with Ph+ ALL (for reviews, see von Bubnoff et al81 and Ottmann et al82 ), but, unfortunately, drug resistance and disease relapse occur after several months. Alternative strategies include combination therapy with imatinib and a cytotoxic agent such as cytarabine or daunorubicin, or both, and a novel signal transduction modulator, such as farnesyl transferase inhibitor.83-86 In this study, TZD18 enhanced synergistically the growth-inhibitory activities of imatinib in these Ph+ lymphocytic leukemia cell lines. More interestingly, TZD18, either alone or in combination with imatinib, was extremely effective in the Ph+ ALL cell line SD1, which was relatively resistant to imatinib therapy. Although the use of combination therapy with TZD18 and imatinib for Ph+ ALL requires further evaluation, our data suggest that TZD18 may represent a promising therapeutic agent to aid in the treatment of this extremely aggressive disease.
We have evaluated the anticancer effects of the novel PPARα/γ dual agonist TZD18, for the first time, in 3 Ph+ ALL cell lines. This compound was able to inhibit the proliferation and to induce the apoptosis of these cells more effectively than was the conventional TZD PPAR ligand PGZ. Of potential clinical importance is that this compound enhanced the cytotoxic effects of imatinib. Hence, our data indicate the potential usefulness of TZD18 in the treatment of Ph+ lymphocytic leukemia.
Prepublished online as Blood First Edition Paper, January 10, 2006; DOI 10.1182/blood-2005-05-2103.
Supported by grants from Deutsche José Carreras Leukämie-Stiftung and Deutsche Forschungsgemeinschaft (E.E.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr M. Ruthardt for the cell line Sup B-15, Ms Q. Guo from Merck (USA) for the compound TZD18, Prof R. M. Evans for PPRE vector, Prof K. K. Chatterjee for PPARγ vector, Dr P. Tontonoz for PPARα vector, Mr H. Krebbel for phosphorylated IκBα antibody, Dr C. Mueller-Tidow for Renilla vector and advice in transfection assays, Ms S. Liu and T. Tian for excellent technical assistance, Mr G. Adie for help in the preparation of this manuscript, and Dr A. C. Hocke for help in microscopic imaging. E.E. thanks Deutsche J. Carreras Leukämie-Stiftung and Deutsche Forschungsgemeinschaft for their generous support.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal