We investigated the relative prognostic significance of cytogenetics in 635 adult acute myeloid leukemia (AML) patients 60 years of age or older treated on front-line protocols. Classification trees and tree-structured survival analysis (TSSA) were used to identify important cytogenetic groups, and their prognostic significance was then assessed in multivariable analysis (MVA). Overall, 48.5% achieved complete remission (CR); 6.6% survived at 5 years. Complex karyotypes with at least 3 abnormalities (complex ≥ 3) and a group including “rare aberrations” predicted lower CR rates (25% and 30%) versus other patients (56%). Compared with complex ≥ 3, the odds of CR were significantly higher for noncomplex karyotypes without rare aberrations on MVA. Cytogenetically, complex ≥ 5 predicted inferior disease-free survival on TSSA, remaining significant on MVA together with white blood cell count (WBC), sex, and age. For survival, complex ≥ 5, rare aberrations, and core-binding factor (CBF) abnormalities were prognostic (P < .001), with 5-year survivals of 0%, 0%, and 19.4%, respectively, and 7.5% for remaining patients. Together with WBC, marrow blasts, sex, and age, the cytogenetic groups remained significant on MVA. In conclusion, pretreatment cytogenetics adds to other prognostic factors in older AML patients. Patients with complex ≥ 5 appear to benefit minimally from current treatment and are better suited for investigational therapy or supportive care. (Blood. 2006;108:63-73)
Introduction
Despite advances in supportive care, the prognosis of patients aged 60 years or older with acute myeloid leukemia (AML) remains extremely poor, with 5% to 15% or less patients alive beyond 5 years.1,2 Although a poor performance status associated with advanced age is contributory, it is also clear that AML in older patients is associated with many adverse features, including increased incidence of primary drug resistance due to higher multi-drug resistance protein expression, antecedent myelodysplasia, and karyotypic abnormalities that are known to be associated with poor outcome.3-5 Despite the generally bad prognosis, however, a small number of long-term survivors exist among patients treated with current standard chemotherapy. The ability to accurately identify this small subset of older AML patients who benefit from current treatment approaches, as well as those who do not, is of critical importance. For the latter patients, novel investigational therapy or supportive care can be offered.
Cytogenetic findings at presentation are among the most important prognostic factors in predicting initial response to therapy, remission duration, and overall survival in AML.6-8 A number of studies have successfully identified 3 broad cytogenetic prognostic groups in patients younger than 55 or 60 years of age.9-11 In the United Kingdom Medical Research Council (MRC) studies,10,11 prognostic groups were identified by comparing response to induction therapy and long-term outcome for patients with recurring karyotypic abnormalities, using those with normal karyotype as the reference group. If survival was better than that for cytogenetically normal patients, the associated karyotypic abnormalities were considered to confer a “favorable risk.” Conversely, if survival was worse than for the cytogenetically normal, the abnormalities were considered to bestow an “adverse risk.” A slightly modified cytogenetic classification system has also been shown to be of prognostic use in patients older than 55 years.12
This study explores the prognostic significance of cytogenetic abnormalities in AML patients 60 years of age or older enrolled in the prospective Cancer and Leukemia Group B (CALGB) cytogenetic study 8461 and treated with standard AML therapy, using classification trees and tree-structured survival analysis (TSSA) to identify prognostic groups.13,14 These methods do not require the selection of a reference group, such as cytogenetically normal, against which other cytogenetic abnormalities are individually compared. Rather, intensive search procedures are used to identify subsets that are most homogeneous with respect to the outcome being modeled, such as achievement of complete remission or survival.
Patients, materials, and methods
Patients
Patients included in this analysis were newly diagnosed AML patients 60 years of age or older, with evaluable karyotype, enrolled on all CALGB treatment protocols that included older untreated AML patients between July 1984 and June 1999 and on a prospective cytogenetic companion study, CALGB 8461. All protocols were approved by the Institutional Review Board of each CALGB participating institution, and written informed consent was obtained from all patients prior to enrollment on study. All patients had AML defined by the French-American-British (FAB) Cooperative Group criteria.15 Patients with AML-M3 (n = 14) were excluded from this analysis. Pathologic diagnoses were centrally reviewed in all cases.
Cytogenetic analysis
Chromosomal analysis was performed on pretreatment bone marrow, or, rarely, blood when marrow was unavailable, from all patients at diagnosis.8 All cytogenetic results were centrally reviewed. At least 20 bone marrow metaphases were analyzed in patients designated as having normal cytogenetics. Karyotypes were interpreted according to the International System for Human Cytogenetic Nomenclature.16 Patients with t(15;17)(q22; q12∼21) were excluded from analysis. It was sufficient that an aberration occurred in only 1 of 2 or more clones identified in the karyotype for the patient to be included in a given cytogenetic group. Patients with deletions of the chromosome arm 5q [eg, del(5)(q13q33)], those with the loss of the whole chromosome 5 [ie, monosomy 5 (-5)], and those with various structural aberrations resulting in the loss of material from 5q {eg, unbalanced translocations between chromosome 5 and a known partner chromosome, eg, der(5)t(5;17)(q11.2;q11.2), the presence of additional material of unknown origin replacing part of 5q [add(5)(q15)], isochromosome of 5p [i(5)(p10)], etc} were grouped together and designated as having “-5/5q-” abnormalities. Similar patient groups were created for cases with either the whole chromosome loss or structural aberrations leading to the loss of material from 7q, 9q, 11q, 12p, 13q, 17p, and 20q. Complex karyotypes ≥ 3 and ≥ 5 were defined as the presence of, respectively, 3 or more and 5 or more chromosome abnormalities in at least 1 clone in the absence of t(8;21)(q22;q22) and inv(16)(p13q22). Karyotypes containing 1 or 2 abnormalities that were unrelated to abnormalities included in the other karyotype groups and occurred in fewer than 5 cases were considered “rare.” Patients with these abnormalities were combined in a single “rare aberrations” group for analysis.
Treatment
Six hundred thirty-five AML patients included in this analysis were registered on front-line CALGB treatment protocols 8221 (n = 24), 8321 (n = 15), 8525 (n = 196), 8923 (n = 218), 9420 (n = 64), and 9720 (n = 118). These patients, who had adequate cytogenetics, comprised 76% of the total number of 837 patients with AML [other than FAB M3 or t(15;17)], were aged 60 years or older, and were registered both on the CALGB 8461 companion study and one of the aforementioned treatment protocols. Patients treated on protocols 8221, 8321, 8525, and 8923 received induction therapy with cytarabine and daunorubicin.17-19 Etoposide with or without the multi-drug resistance protein modulator PSC-833 was also included together with cytarabine and daunorubicin in protocols 9420 and 9720.20,21 In all protocols, a second induction cycle using the same drugs was administered if marrow aplasia (≤ 5% blasts, ≤ 15% cellularity) was not achieved on day 14 with the first course. All patients achieving complete remission (CR) after 1 or 2 cycles of induction chemotherapy were assigned to postremission therapy according to the specific protocol. This comprised repeated cycles of cytarabine in standard, intermediate, or high dose, followed by 4 cycles of maintenance therapy with cytarabine and daunorubicin on CALGB 8221 and 852517,18 ; standard-dose cytarabine or mitoxantrone plus intermediate-dose cytarabine on CALGB 892319 ; and cytarabine, daunorubicin, and etoposide, with or without maintenance treatment with interleukin-2, on CALGB 9420 and 9720,20,21 as previously reported.
Definition of response and survival outcomes
CR was defined as the achievement of a morphologically normal marrow, a granulocyte count of 1.5 × 109/L or greater, and a platelet count of 100 × 109/L or greater. Relapse was defined by greater than 5% blasts in marrow aspirates or the development of extramedullary leukemia in patients with previously documented CR, according to the National Cancer Institute criteria.22 Disease-free survival (DFS) was defined only for patients who achieved CR and was measured from the date of CR until relapse or death, regardless of cause. Overall survival (OS) was measured from the protocol on-study date until death, regardless of cause, censoring for patients alive.
Statistical analysis
To determine the prognostic significance of pretreatment cytogenetics in predicting achievement of CR, DFS, and OS, classification trees and TSSA were used initially. Once prognostic cytogenetic abnormalities were identified, multivariable models were constructed to adjust for other pretreatment variables.
In using classification trees to identify prognostic cytogenetic aberrations predictive of CR, the splitting criterion for each node was selected as that partition of the covariate space that maximized homogeneity within the terminal nodes of the tree. The Gini index was used as the impurity measure that partitioned the covariate space.14 Pruning was used to select the best subtree. The pruning method used for the classification tree was to prune all branches based on the one-standard error cost complexity rule.23 The final pruned tree is displayed in the customary format such that ovals are used to indicate an internal branch in the tree whereas rectangles are used to indicate final subsets.
Univariable estimates of DFS and OS were calculated using the Kaplan-Meier method,24 and the log-rank test was used to compare differences between survival curves. In TSSA, used to model DFS and OS, the log-rank test was used as the splitting criterion to select between prognostic cytogenetic aberrations.13 For each node, the splitting criterion was selected as that partition of the covariate space that maximized the value of the log-rank test statistic. Splitting according to this rule continued until there were fewer than 20 subjects observed in a terminal node or until censoring became excessive. Pruning was used to select the best subtree.13
In multivariable analyses, covariates included in the full models were the continuous variables (age, pretreatment white blood cell count [WBC], hemoglobin level, platelet count, and percent blasts in bone marrow) and the categoric variables (sex, race [white versus nonwhite], type of AML [de novo versus secondary]), and indicators representing the prognostic cytogenetic groups as determined by the classification and survival tree analyses. Additionally, age was categorized as 60 to 69 years, 70 to 79 years, and 80 years and older and was included in the full model with its continuous version excluded to identify the better-fitting form of this covariate. Using backward selection, variables with a P value greater than .05 by the likelihood ratio test were removed from the model one at a time, and the change in coefficients was sequentially assessed to ensure there was no confounding. A multivariable logistic regression model predicting achievement of CR was derived to determine whether the prognostic cytogenetic groups identified by the classification tree remained significant once other variables adjusted the model. After the final model was determined, the Hosmer and Lemeshow goodness-of-fit test was performed.25 For DFS and OS, a Cox proportional hazards model was developed using a backward selection procedure. After the final model was determined, the proportional hazards assumption was assessed globally as well as for the individual variables. Functions developed for S-Plus (Insightful Corporation, Seattle, WA), TSSA for tree-structured survival analysis,26 and Recursive Partitioning and Regression Trees (RPART) for classification trees27 were used. All analyses were performed by the CALGB Statistical Center.
Results
Patient characteristics
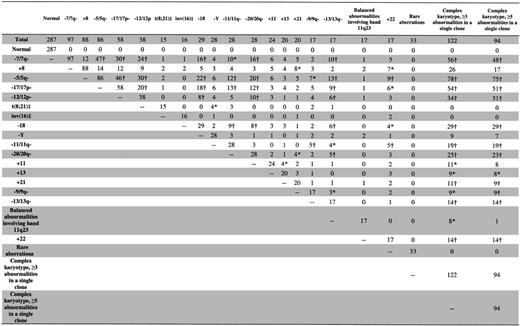
The pretreatment clinical characteristics of the 635 AML patients are shown in Table 1. The median age was 68 years (range, 60-86 years), with 233 patients aged 70 to 79 and 29 patients aged 80 years or older. The majority (97.5%) had de novo AML, with FAB-M1,-M2, and -M4 predominating. Figure 1 shows the number of patients in each karyotype group. The most common karyotype groups were normal (seen in 45% of patients), complex karyotype with at least 3 abnormalities (19%), complex karyotype with at least 5 abnormalities (15%), and abnormalities of chromosomes 7 (-7/7q-; 15%) and 5 (-5/5q-; 14%) and trisomy 8 (+8; 14%). Abnormalities characteristic of core-binding factor (CBF) AML, t(8;21) and inv(16)/t(16;16), which are associated with a favorable prognosis in younger patients, were detected in only 5% of all patients studied. A variety of other recurrent abnormalities, trisomies, deletions, and balanced and unbalanced chromosomal translocations were also detected (Figure 1). Thirty-three cases with 1 or 2 rare chromosomal abnormalities unrelated to the other karyotype groups were placed into a rare-aberrations group and are summarized in Table 2.
Pretreatment clinical characteristics (N = 635 patients)
Data | |
| Age, median (range), y | 68 (60-86) |
| Age distribution, no. (%) | |
| 60-69 y | 373 (58.7) |
| 70-79 y | 233 (36.7) |
| 80 y or older | 29 (4.6) |
| Sex, male, no. (%) | 346 (54.5) |
| Race, no. (%) | |
| White | 568 (89.5) |
| Black | 49 (7.7) |
| Hispanic | 11 (1.7) |
| Other | 7 (1.1) |
| FAB classification, no. (%) | |
| M0 | 14 (2.2) |
| M1 | 134 (21.1) |
| M2 | 234 (36.9) |
| M4 | 126 (19.8) |
| M5 | 70 (11.0) |
| M6 | 18 (2.8) |
| M7 | 4 (0.6) |
| Unclassified | 35 (5.5) |
| De novo AML, no. (%) | 619 (97.5) |
| WBC, median (range), × 109/L | 13.8 (0.5-450.0) |
| Hemoglobin level, median (range), g/dL | 9.4 (3.0-15.4) |
| Platelet count, median (range), × 109/L | 64 (2-1200) |
| Percentage myeloblasts in marrow, median (range) | 65 (20-98) |
| Percentage myeloblasts in blood, median (range) | 48 (0-99) |
| CNS involvement, % | 1 |
| Splenomegaly, % | 7 |
| Hepatomegaly, % | 9 |
| Gum hypertrophy, % | 7 |
| Lymphadenopathy, % | 7 |
| Skin infiltration, % | 7 |
| Mediastinal mass, % | 1 |
Data | |
| Age, median (range), y | 68 (60-86) |
| Age distribution, no. (%) | |
| 60-69 y | 373 (58.7) |
| 70-79 y | 233 (36.7) |
| 80 y or older | 29 (4.6) |
| Sex, male, no. (%) | 346 (54.5) |
| Race, no. (%) | |
| White | 568 (89.5) |
| Black | 49 (7.7) |
| Hispanic | 11 (1.7) |
| Other | 7 (1.1) |
| FAB classification, no. (%) | |
| M0 | 14 (2.2) |
| M1 | 134 (21.1) |
| M2 | 234 (36.9) |
| M4 | 126 (19.8) |
| M5 | 70 (11.0) |
| M6 | 18 (2.8) |
| M7 | 4 (0.6) |
| Unclassified | 35 (5.5) |
| De novo AML, no. (%) | 619 (97.5) |
| WBC, median (range), × 109/L | 13.8 (0.5-450.0) |
| Hemoglobin level, median (range), g/dL | 9.4 (3.0-15.4) |
| Platelet count, median (range), × 109/L | 64 (2-1200) |
| Percentage myeloblasts in marrow, median (range) | 65 (20-98) |
| Percentage myeloblasts in blood, median (range) | 48 (0-99) |
| CNS involvement, % | 1 |
| Splenomegaly, % | 7 |
| Hepatomegaly, % | 9 |
| Gum hypertrophy, % | 7 |
| Lymphadenopathy, % | 7 |
| Skin infiltration, % | 7 |
| Mediastinal mass, % | 1 |
FAB indicates French-American-British; WBC, white blood cell count; and CNS, central nervous system.
*Percentage of myeloblasts may be less than 30% in cases of AML-M6.
Characteristics of rare-aberrations group
No. . | Karyotype . | Sex . | Age, y . | WBC, × 109/L . | % BM blasts . | Response to induction . | DFS, mo . | OS, mo . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Abnormalities reported previously to confer adverse prognosis in AML | ||||||||||
| 1 | 46,XX,t(6;9)(p23;q34)[19]/46,XX[1] | F | 62 | 25 | 56 | CR | 3 | 11 | ||
| 2 | 46,XY,t(9;22)(q34;q11.2)[29]/46,XY[2] | M | 60 | 106 | 65 | R | NA | 12 | ||
| 3 | 46,XX,t(9;22)(q34;q11.2)[29]/46,XX[1] | F | 63 | 25 | 84 | CR | 2 | 8 | ||
| 4 | 46,XX,t(5;9;22)(p31;q34;q11.2)[20] | F | 72 | 112 | 48 | R | NA | 3 | ||
| 5 | 46,XX,t(3;21)(q26;q22)[3] | F | 79 | — | 36 | CR | 8 | 12 | ||
| 6 | 46,XX,t(2;3)(p21;q27)[28] | F | 66 | 110 | 79 | R | NA | 5 | ||
| 7 | 46,XY,del(3)(q13.2q27)[15]/46,XY[7] | M | 67 | 17 | 40 | R | NA | 25 | ||
| 8 | 46,XY,add(3)(q12),del(16)(q22q24)[19]/46,XY[1] | M | 83 | 1 | — | D | NA | <1 | ||
| 9 | 47,XX,+4[13]/46,XX[27] | F | 65 | 11 | 72 | D | NA | <1 | ||
| 10 | 47,XX,+4[26]/46,XX[6] | F | 68 | 2 | 65 | D | NA | <1 | ||
| 11 | 47,XX,+4[5]/46,XX[15] | F | 79 | 9 | 55 | CR | 7 | 10 | ||
| 12 | 47,XY,+i(11)(q10)[20] | M | 68 | 47 | 89 | R | NA | 21 | ||
| 13 | 46,XY,i(13)(q10)[8]/46,XY[12] | M | 77 | 188 | 87 | R | NA | 1 | ||
| 14 | 46,XX,+2-17dmin[23]/47,XX,+r,+3-24dmin[4] | F | 76 | 9 | 77 | R | NA | 15 | ||
| Abnormalities with thus far unknown prognostic significance in AML | ||||||||||
| 15 | 45,XY,−10[5]/46,XY[24] | M | 67 | 46 | 88 | D | NA | <1 | ||
| 16 | 47,XX,+15[14]/46,XX[14] | F | 62 | 1 | 45 | CR | 5 | 14 | ||
| 17 | 47,XY,+20[12]/46,XY[18] | M | 73 | 30 | 79 | R | NA | 2 | ||
| 18 | 46,XY,del(2)(p21)[13]/46,XY[7] | M | 62 | 4 | 76 | D | NA | <1 | ||
| 19 | 46,XY,add(2)(p23)[3]/46,XY[22] | M | 70 | 6 | 56 | CR | 18 | 51 | ||
| 20 | 46,XX,del(14)(q?13q?22)[3]/46,XX[50] | F | 73 | 1 | 54 | CR | 19 | 24 | ||
| 21 | 46,XY,t(4;14;8)(p16;q1?1.2;p21)[37] | M | 66 | 53 | 80 | D | NA | <1 | ||
| 22 | 46,XX,der(15)t(1;15)(q11;p13)[44]/46,XX[1] | F | 62 | 81 | 96 | D | NA | <1 | ||
| 23 | 46,XX,der(18)t(1;18)(q25;q23)[20]/46,XX[1] | F | 66 | 133 | 97 | D | NA | <1 | ||
| 24 | 47,XY,+der(1;19)(q10;p10)[20] | M | 68 | 106 | 60 | R | NA | 4 | ||
| 25 | 46,XX,t(9;13)(q21.1;q33)[23] | F | 65 | 5 | 31 | D | NA | <1 | ||
| 26 | 46,XX,?dup(9)(q13q22)[20] | F | 71 | 89 | 86 | D | NA | <1 | ||
| 27 | 46,X,del(Y)(q12)[20]/46,XY[6] | M | 64 | 1 | 69 | D | NA | <1 | ||
| 28 | 46,XY,t(6;12)(q11;p11.2)[20] | M | 71 | 51 | 84 | R | NA | 1 | ||
| 29 | 46,XY,del(8)(q2?2q2?4.1)[20] | M | 78 | 6 | 50 | CR | 9 | 12 | ||
| 30 | 46,XY,add(13)(p11)[19]/46,XY[1] | M | 66 | 2 | 76 | R | NA | 11 | ||
| 31 | 46,XX,del(16)(q12.1)[16]/46,XX[14] | F | 67 | 3 | 62 | CR | 17 | 28 | ||
| 32 | 47,XX,+r[7]/48,XX,+2r[23] | F | 70 | 49 | 76 | CR | 16 | 22 | ||
| 33 | 47,XY,+mar[5]/46,XY[35] | M | 67 | 4 | 35 | R | NA | 27 | ||
No. . | Karyotype . | Sex . | Age, y . | WBC, × 109/L . | % BM blasts . | Response to induction . | DFS, mo . | OS, mo . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Abnormalities reported previously to confer adverse prognosis in AML | ||||||||||
| 1 | 46,XX,t(6;9)(p23;q34)[19]/46,XX[1] | F | 62 | 25 | 56 | CR | 3 | 11 | ||
| 2 | 46,XY,t(9;22)(q34;q11.2)[29]/46,XY[2] | M | 60 | 106 | 65 | R | NA | 12 | ||
| 3 | 46,XX,t(9;22)(q34;q11.2)[29]/46,XX[1] | F | 63 | 25 | 84 | CR | 2 | 8 | ||
| 4 | 46,XX,t(5;9;22)(p31;q34;q11.2)[20] | F | 72 | 112 | 48 | R | NA | 3 | ||
| 5 | 46,XX,t(3;21)(q26;q22)[3] | F | 79 | — | 36 | CR | 8 | 12 | ||
| 6 | 46,XX,t(2;3)(p21;q27)[28] | F | 66 | 110 | 79 | R | NA | 5 | ||
| 7 | 46,XY,del(3)(q13.2q27)[15]/46,XY[7] | M | 67 | 17 | 40 | R | NA | 25 | ||
| 8 | 46,XY,add(3)(q12),del(16)(q22q24)[19]/46,XY[1] | M | 83 | 1 | — | D | NA | <1 | ||
| 9 | 47,XX,+4[13]/46,XX[27] | F | 65 | 11 | 72 | D | NA | <1 | ||
| 10 | 47,XX,+4[26]/46,XX[6] | F | 68 | 2 | 65 | D | NA | <1 | ||
| 11 | 47,XX,+4[5]/46,XX[15] | F | 79 | 9 | 55 | CR | 7 | 10 | ||
| 12 | 47,XY,+i(11)(q10)[20] | M | 68 | 47 | 89 | R | NA | 21 | ||
| 13 | 46,XY,i(13)(q10)[8]/46,XY[12] | M | 77 | 188 | 87 | R | NA | 1 | ||
| 14 | 46,XX,+2-17dmin[23]/47,XX,+r,+3-24dmin[4] | F | 76 | 9 | 77 | R | NA | 15 | ||
| Abnormalities with thus far unknown prognostic significance in AML | ||||||||||
| 15 | 45,XY,−10[5]/46,XY[24] | M | 67 | 46 | 88 | D | NA | <1 | ||
| 16 | 47,XX,+15[14]/46,XX[14] | F | 62 | 1 | 45 | CR | 5 | 14 | ||
| 17 | 47,XY,+20[12]/46,XY[18] | M | 73 | 30 | 79 | R | NA | 2 | ||
| 18 | 46,XY,del(2)(p21)[13]/46,XY[7] | M | 62 | 4 | 76 | D | NA | <1 | ||
| 19 | 46,XY,add(2)(p23)[3]/46,XY[22] | M | 70 | 6 | 56 | CR | 18 | 51 | ||
| 20 | 46,XX,del(14)(q?13q?22)[3]/46,XX[50] | F | 73 | 1 | 54 | CR | 19 | 24 | ||
| 21 | 46,XY,t(4;14;8)(p16;q1?1.2;p21)[37] | M | 66 | 53 | 80 | D | NA | <1 | ||
| 22 | 46,XX,der(15)t(1;15)(q11;p13)[44]/46,XX[1] | F | 62 | 81 | 96 | D | NA | <1 | ||
| 23 | 46,XX,der(18)t(1;18)(q25;q23)[20]/46,XX[1] | F | 66 | 133 | 97 | D | NA | <1 | ||
| 24 | 47,XY,+der(1;19)(q10;p10)[20] | M | 68 | 106 | 60 | R | NA | 4 | ||
| 25 | 46,XX,t(9;13)(q21.1;q33)[23] | F | 65 | 5 | 31 | D | NA | <1 | ||
| 26 | 46,XX,?dup(9)(q13q22)[20] | F | 71 | 89 | 86 | D | NA | <1 | ||
| 27 | 46,X,del(Y)(q12)[20]/46,XY[6] | M | 64 | 1 | 69 | D | NA | <1 | ||
| 28 | 46,XY,t(6;12)(q11;p11.2)[20] | M | 71 | 51 | 84 | R | NA | 1 | ||
| 29 | 46,XY,del(8)(q2?2q2?4.1)[20] | M | 78 | 6 | 50 | CR | 9 | 12 | ||
| 30 | 46,XY,add(13)(p11)[19]/46,XY[1] | M | 66 | 2 | 76 | R | NA | 11 | ||
| 31 | 46,XX,del(16)(q12.1)[16]/46,XX[14] | F | 67 | 3 | 62 | CR | 17 | 28 | ||
| 32 | 47,XX,+r[7]/48,XX,+2r[23] | F | 70 | 49 | 76 | CR | 16 | 22 | ||
| 33 | 47,XY,+mar[5]/46,XY[35] | M | 67 | 4 | 35 | R | NA | 27 | ||
WBC indicates white blood cell count; BM, bone marrow; F, female; CR, complete remission; M, male; R, primary resistant disease; NA, not applicable; —, not available; and D, induction death.
Overall outcome
Of the 635 patients, 308 (48.5%) achieved CR following initial induction chemotherapy. Two hundred (31.5%) patients had primary chemotherapy-resistant disease, and 127 (20%) died during induction. There was no difference in CR rates between patients receiving induction chemotherapy that included etoposide and those receiving only daunorubicin and cytarabine (47% versus 49%; P = .69). There was also no difference in CR rates between patients aged 60 to 69 years and those aged 70 to 79 years (51% versus 48%; P = .43). However, patients aged 80 years and older had a significantly inferior CR rate (24%) compared with patients aged 60 to 69 (P = .005) and patients aged 70 to 79 years (P = .02). Of the 22 patients 80 years or older who did not achieve CR, 13 had chemotherapy-resistant disease and 9 died during induction, which was similar in distribution to induction failures due to chemotherapy-resistant disease and induction deaths, respectively, among patients aged 60 to 69 and 70 to 79 years not achieving CR (P = .65 across all 3 age groups; results not shown).
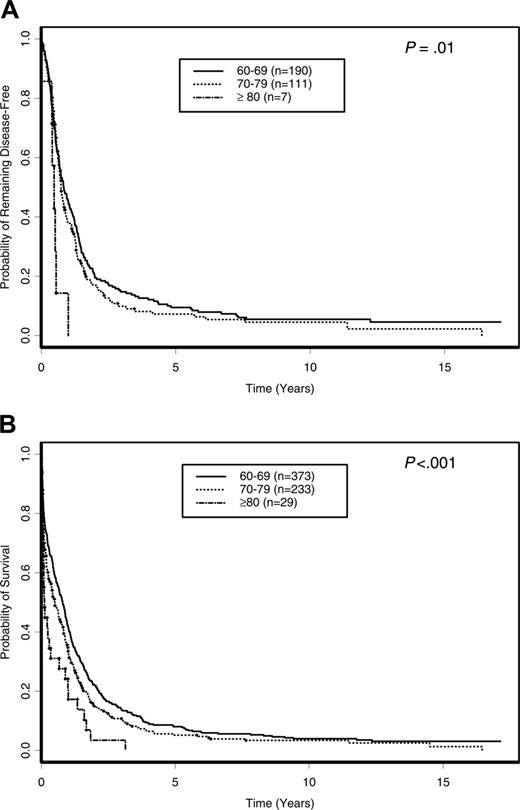
With a median follow-up of 10.9 years (range, 6.1-17.1 years) for surviving patients, the OS of all 635 patients was 6.6% (95% confidence interval [CI], 4.9%-8.7%) at 5 years. There were no significant differences in the DFS and OS of patients treated on different protocols (P = .47 and P = .57, respectively) and between protocols assigning repeated cycles of cytarabine after remission (CALGB 8221, 8321, 8525, and 8923) and protocols where only a single cycle of postremission cytarabine-based therapy was assigned (CALGB 9420 and 9720; P = .18 and P = .50, respectively; data not shown). Further, among patients who were assigned repeated cycles of cytarabine postremission therapy (CALGB 8221 and 8525), there were no differences in DFS (P = .71) or OS (P = .90) among patients receiving standarddose (100 mg/m2/d × 5 days), intermediate-dose (400 mg/m2/d × 5 days), or high-dose (3 g/m2 every 12 hours on days 1, 3, and 5) cytarabine (data not shown). Patients 80 years or older had the shortest DFS and OS. The 5-year DFS for patients 80 years or older was 0% compared with 9.5% (95% CI, 5.8%-14.1%) and 7.2% (95% CI, 3.4%-13.0%) for those aged 60 to 69 and 70 to 79 years, respectively (P = .01; Figure 2A). Similarly, the 5-year OS for patients 80 years or older was 0% compared with 8.0% (95% CI, 5.6%-11.1%) and 5.2% (95% CI, 2.8%-8.5%) for patients aged 60 to 69 and 70 to 79 years, respectively (P < .001; Figure 2B). The outcomes of induction treatment, relapse rate, 5-year DFS, and 5-year OS for individual cytogenetic groups are summarized in Table 3.
Outcome of defined karyotype groups
. | . | Outcome of induction therapy . | . | . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|
. | No.(%) . | CR, no.(%) . | Resistant disease, no.(%) . | Induction death, no.(%) . | Relapsed, no.(%) . | 5-y DFS, % (95% CI) . | 5-y OS, % (95% CI) . | ||
| Normal | 287 (45) | 163 (57) | 75 (26) | 49 (17) | 135 (83) | 9 (5-14) | 8 (5-12) | ||
| −7/7q− | 97 (15) | 37 (38) | 44 (45) | 16 (16) | 30 (81) | 3 (0.2-12) | 2 (0.4-7) | ||
| +8 | 88 (14) | 35 (40) | 31 (35) | 22 (25) | 29 (83) | 9 (2-21) | 7 (3-13) | ||
| −5/5q− | 86 (14) | 19 (22) | 44 (51) | 23 (27) | 18 (95) | 0 | 0 | ||
| −17/17p− | 58 (9) | 16 (28) | 25 (43) | 17 (29) | 16 (100) | 6 (0.4-25) | 2 (0.1-8) | ||
| −12/12p− | 38 (6) | 7 (18) | 23 (61) | 8 (21) | 7 (100) | 0 | 0 | ||
| CBF AML | 31 (5) | 24 (77) | 3 (10) | 4 (13) | 18 (75) | 17 (5-34) | 19 (8-35) | ||
| t(8;21) | 15 (2) | 11 (73) | 2 (13) | 2 (13) | 7 (64) | 27 (7-54) | 20 (5-42) | ||
| inv(16) | 16 (3) | 13 (81) | 1 (6) | 2 (13) | 11 (85) | 8 (0.5-29) | 19 (5-40) | ||
| −18 | 29 (5) | 6 (21) | 13 (45) | 10 (34) | 6 (100) | 0 | 0 | ||
| −Y | 28 (4) | 14 (50) | 8 (29) | 6 (21) | 11 (79) | 14 (2-37) | 11 (3-25) | ||
| −11/11q− | 28 (4) | 10 (36) | 14 (50) | 4 (14) | 9 (90) | 10 (0.6-36) | 4 (0.3-15) | ||
| −20/20q− | 28 (4) | 5 (18) | 18 (64) | 5 (18) | 4 (80) | 0 | 0 | ||
| +11 | 24 (4) | 10 (42) | 9 (38) | 5 (21) | 10 (100) | 0 | 0 | ||
| +13 | 20 (3) | 12 (60) | 2 (10) | 6 (30) | 9 (75) | 8 (0.5-31) | 5 (0.3-21) | ||
| +21 | 20 (3) | 8 (40) | 8 (40) | 4 (20) | 7 (88) | 0 | 0 | ||
| −9/9q− | 17 (3) | 10 (59) | 2 (12) | 5 (29) | 9 (90) | 10 (0.6-36) | 6 (0.4-24) | ||
| −13/13q− | 17 (3) | 5 (29) | 9 (53) | 3 (18) | 4 (80) | 20 (0.8-58) | 6 (0.4-24) | ||
| Balanced abnormalities involving band 11 q23 | 17 (3) | 10 (59) | 4 (24) | 3 (18) | 7 (70) | 10 (0.6-36) | 12 (2-31) | ||
| +22 | 17 (3) | 7 (41) | 6 (35) | 4 (24) | 6 (86) | 14 (0.7-46) | 6 (0.4-24) | ||
| Rare aberrations | 33 (5) | 10 (30) | 12 (36) | 11 (33) | 10 (100) | 0 | 0 | ||
| Complex karyotype, at least 3 abnormalities in a single clone | 122 (19) | 30 (25) | 61 (50) | 31 (25) | 27 (90) | 3 (0.2-15) | 2 (0.3-5) | ||
| Complex karyotype, at least 5 abnormalities in a single clone | 94 (15) | 22 (23) | 46 (49) | 26 (28) | 21 (95) | 0 | 0 | ||
. | . | Outcome of induction therapy . | . | . | . | . | . | ||
|---|---|---|---|---|---|---|---|---|---|
. | No.(%) . | CR, no.(%) . | Resistant disease, no.(%) . | Induction death, no.(%) . | Relapsed, no.(%) . | 5-y DFS, % (95% CI) . | 5-y OS, % (95% CI) . | ||
| Normal | 287 (45) | 163 (57) | 75 (26) | 49 (17) | 135 (83) | 9 (5-14) | 8 (5-12) | ||
| −7/7q− | 97 (15) | 37 (38) | 44 (45) | 16 (16) | 30 (81) | 3 (0.2-12) | 2 (0.4-7) | ||
| +8 | 88 (14) | 35 (40) | 31 (35) | 22 (25) | 29 (83) | 9 (2-21) | 7 (3-13) | ||
| −5/5q− | 86 (14) | 19 (22) | 44 (51) | 23 (27) | 18 (95) | 0 | 0 | ||
| −17/17p− | 58 (9) | 16 (28) | 25 (43) | 17 (29) | 16 (100) | 6 (0.4-25) | 2 (0.1-8) | ||
| −12/12p− | 38 (6) | 7 (18) | 23 (61) | 8 (21) | 7 (100) | 0 | 0 | ||
| CBF AML | 31 (5) | 24 (77) | 3 (10) | 4 (13) | 18 (75) | 17 (5-34) | 19 (8-35) | ||
| t(8;21) | 15 (2) | 11 (73) | 2 (13) | 2 (13) | 7 (64) | 27 (7-54) | 20 (5-42) | ||
| inv(16) | 16 (3) | 13 (81) | 1 (6) | 2 (13) | 11 (85) | 8 (0.5-29) | 19 (5-40) | ||
| −18 | 29 (5) | 6 (21) | 13 (45) | 10 (34) | 6 (100) | 0 | 0 | ||
| −Y | 28 (4) | 14 (50) | 8 (29) | 6 (21) | 11 (79) | 14 (2-37) | 11 (3-25) | ||
| −11/11q− | 28 (4) | 10 (36) | 14 (50) | 4 (14) | 9 (90) | 10 (0.6-36) | 4 (0.3-15) | ||
| −20/20q− | 28 (4) | 5 (18) | 18 (64) | 5 (18) | 4 (80) | 0 | 0 | ||
| +11 | 24 (4) | 10 (42) | 9 (38) | 5 (21) | 10 (100) | 0 | 0 | ||
| +13 | 20 (3) | 12 (60) | 2 (10) | 6 (30) | 9 (75) | 8 (0.5-31) | 5 (0.3-21) | ||
| +21 | 20 (3) | 8 (40) | 8 (40) | 4 (20) | 7 (88) | 0 | 0 | ||
| −9/9q− | 17 (3) | 10 (59) | 2 (12) | 5 (29) | 9 (90) | 10 (0.6-36) | 6 (0.4-24) | ||
| −13/13q− | 17 (3) | 5 (29) | 9 (53) | 3 (18) | 4 (80) | 20 (0.8-58) | 6 (0.4-24) | ||
| Balanced abnormalities involving band 11 q23 | 17 (3) | 10 (59) | 4 (24) | 3 (18) | 7 (70) | 10 (0.6-36) | 12 (2-31) | ||
| +22 | 17 (3) | 7 (41) | 6 (35) | 4 (24) | 6 (86) | 14 (0.7-46) | 6 (0.4-24) | ||
| Rare aberrations | 33 (5) | 10 (30) | 12 (36) | 11 (33) | 10 (100) | 0 | 0 | ||
| Complex karyotype, at least 3 abnormalities in a single clone | 122 (19) | 30 (25) | 61 (50) | 31 (25) | 27 (90) | 3 (0.2-15) | 2 (0.3-5) | ||
| Complex karyotype, at least 5 abnormalities in a single clone | 94 (15) | 22 (23) | 46 (49) | 26 (28) | 21 (95) | 0 | 0 | ||
Prognostic value of cytogenetics for CR attainment
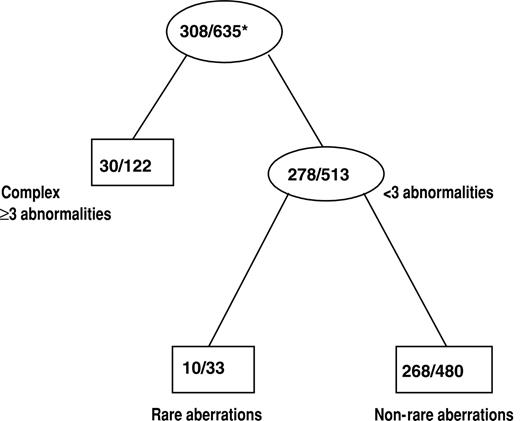
The effect of cytogenetics on achievement of CR was assessed by classification tree analysis using all cytogenetic groups defined in Figure 1. In addition, t(8;21) and inv(16) patients were combined together to form one cytogenetic group, CBF AML, which was also used in the tree analysis. The final classification tree for CR after pruning is shown in Figure 3. The tree predicting CR first identified complex karyotype ≥ 3 as a significant prognostic group. Among the remaining patients, those with rare aberrations had a significantly lower CR rate than other patients with noncomplex karyotypes. The CR rates for the 3 prognostic cytogenetic risk groups identified by the classification tree are shown in Table 4. The CR rates for patients with complex karyotype ≥ 3 (25%) and those with rare aberrations (30%) were lower than the CR rate for the remaining patients with noncomplex karyotype (56%; P < .001 and P = .006, respectively). The CR rate between patients with a complex karyotype of at least 3 and those with rare aberrations was not significantly different (P = .51).
Significant prognostic cytogenetic risk groups identified by tree analysis for CR, DFS, and OS
Cytogenetic risk groups . | No. . | Outcome, % (95% CI)* . |
|---|---|---|
| Complete remission | ||
| Complex ≥3 | 122 | 25 (17-32) |
| Rare aberrations | 33 | 30 (15-46) |
| Noncomplex ≥3, nonrare aberrations | 480 | 56 (51-60) |
| Disease-free survival | ||
| Complex >5 | 22 | 0 |
| Fewer than 5 aberrations | 286 | 9.1 (6.1-12.8) |
| Overall survival | ||
| Complex ≥5 | 94 | 0 |
| CBF AML | 31 | 19.4 (7.9-34.6) |
| Rare aberrations | 33 | 0 |
| Fewer than 5 non-CBF AML, nonrare aberrations | 477 | 7.5 (5.4-10.1) |
Cytogenetic risk groups . | No. . | Outcome, % (95% CI)* . |
|---|---|---|
| Complete remission | ||
| Complex ≥3 | 122 | 25 (17-32) |
| Rare aberrations | 33 | 30 (15-46) |
| Noncomplex ≥3, nonrare aberrations | 480 | 56 (51-60) |
| Disease-free survival | ||
| Complex >5 | 22 | 0 |
| Fewer than 5 aberrations | 286 | 9.1 (6.1-12.8) |
| Overall survival | ||
| Complex ≥5 | 94 | 0 |
| CBF AML | 31 | 19.4 (7.9-34.6) |
| Rare aberrations | 33 | 0 |
| Fewer than 5 non-CBF AML, nonrare aberrations | 477 | 7.5 (5.4-10.1) |
The outcome data show the complete remission rate, the 5-year DFS, and the 5-year OS.
The results of the multivariable logistic regression model developed to investigate the independent prognostic value of the identified cytogenetic groups after adjusting for other baseline variables are shown in Table 5. The cytogenetic indicator variables (P < .001), WBC (P = .001), sex (P = .002), and age categorized by decade (P = .043) emerged as independent significant covariates predictive of CR. The likelihood of achieving CR was less for patients with an increasing WBC as well as increasing patient age and lower among males compared with females. Relative to patients with complex karyotypes ≥3, the odds of achieving CR were significantly higher for patients with noncomplex karyotype without rare aberrations (odds ratio [OR] 4.80; 95% CI, 2.91-7.92) but not significantly different for patients with rare aberrations (OR 1.41; 95% CI, 0.56-3.54).
Significant variables predictive of outcome on multivariable analysis
. | OR/HR . | 95% CI . | P . |
|---|---|---|---|
| Complete remission | |||
| WBC | 0.90* | 0.85-0.96 | .001 |
| Sex, male | 0.58 | 0.41-0.81 | .002 |
| Age | .043 | ||
| 70-79 y | 0.93† | 0.65-1.34 | |
| 80 y or older | 0.32† | 0.13-0.82 | |
| Cytogenetics | <.001 | ||
| Rare aberrations | 1.41‡ | 0.56-3.54 | |
| 1 or 2 nonrare aberrations | 4.80‡ | 2.91-7.92 | |
| Disease-free survival | |||
| WBC | 1.06* | 1.02-1.11 | .013 |
| Sex, male | 1.29 | 1.01-1.64 | .040 |
| Age | .014 | ||
| 70-79 y | 1.17† | 0.91-1.50 | |
| 80 y or older | 3.60† | 1.66-7.78 | |
| Cytogenetics | <.001 | ||
| Complex ≥5 | 3.46§ | 2.07-5.78 | |
| Overall survival | |||
| WBC | 1.05* | 1.02-1.08 | .002 |
| Sex, male | 1.39 | 1.18-1.65 | .001 |
| % marrow blasts | 1.06∥ | 1.01-1.11 | .012 |
| Age | .001 | ||
| 70-79 y | 1.23† | 1.04-1.47 | |
| 80 y or older | 2.04† | 1.37-3.06 | |
| Cytogenetics | <.001 | ||
| Complex ≥5 | 4.49¶ | 2.85-7.06 | |
| Rare aberrations | 2.23¶ | 1.31-3.79 | |
| Fewer than 5 non-CBF AML, nonrare aberrations | 1.52¶ | 1.03-2.25 |
. | OR/HR . | 95% CI . | P . |
|---|---|---|---|
| Complete remission | |||
| WBC | 0.90* | 0.85-0.96 | .001 |
| Sex, male | 0.58 | 0.41-0.81 | .002 |
| Age | .043 | ||
| 70-79 y | 0.93† | 0.65-1.34 | |
| 80 y or older | 0.32† | 0.13-0.82 | |
| Cytogenetics | <.001 | ||
| Rare aberrations | 1.41‡ | 0.56-3.54 | |
| 1 or 2 nonrare aberrations | 4.80‡ | 2.91-7.92 | |
| Disease-free survival | |||
| WBC | 1.06* | 1.02-1.11 | .013 |
| Sex, male | 1.29 | 1.01-1.64 | .040 |
| Age | .014 | ||
| 70-79 y | 1.17† | 0.91-1.50 | |
| 80 y or older | 3.60† | 1.66-7.78 | |
| Cytogenetics | <.001 | ||
| Complex ≥5 | 3.46§ | 2.07-5.78 | |
| Overall survival | |||
| WBC | 1.05* | 1.02-1.08 | .002 |
| Sex, male | 1.39 | 1.18-1.65 | .001 |
| % marrow blasts | 1.06∥ | 1.01-1.11 | .012 |
| Age | .001 | ||
| 70-79 y | 1.23† | 1.04-1.47 | |
| 80 y or older | 2.04† | 1.37-3.06 | |
| Cytogenetics | <.001 | ||
| Complex ≥5 | 4.49¶ | 2.85-7.06 | |
| Rare aberrations | 2.23¶ | 1.31-3.79 | |
| Fewer than 5 non-CBF AML, nonrare aberrations | 1.52¶ | 1.03-2.25 |
OR indicates odds ratio for complete remission; HR, hazard ratio of relapse and/or death for DFS and OS; and WBC, white blood cell count.
OR/HR for a change in WBC of 20 × 109/L.
OR/HR are relative to age 60 to 69 years.
OR relative to complex karyotype ≥ 3.
HR relative to all other karyotypes.
HR for an increase in bone marrow blasts of 10%.
HR relative to CBF AML group.
Prognostic value of cytogenetics for DFS
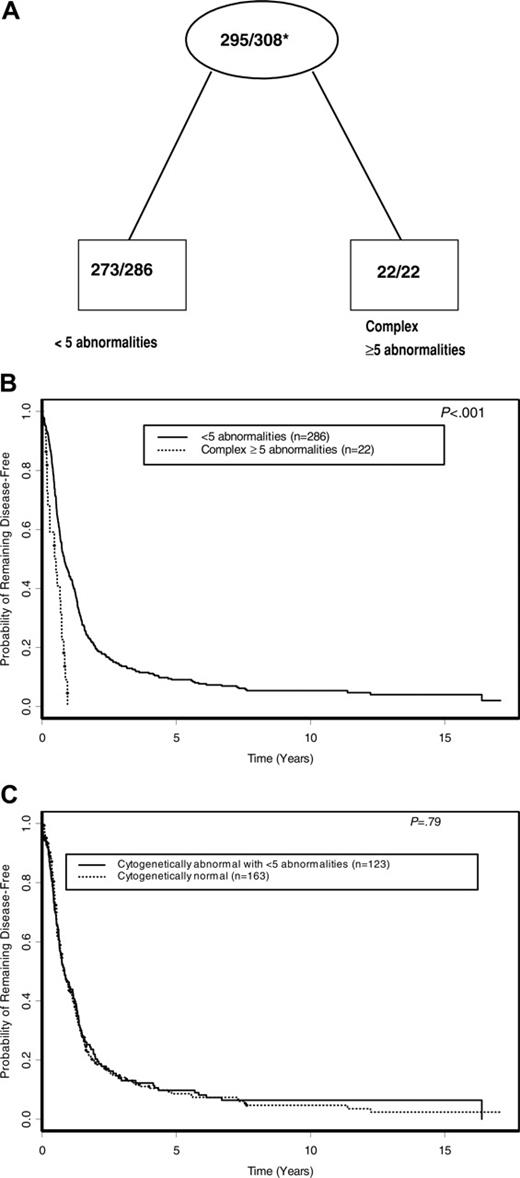
Using TSSA, the tree initially identified complex karyotype ≥ 5as a significant predictor of DFS, with no further significant splitting of the tree by other karyotypes used in the analysis (Figure 1). Therefore, as shown in Figure 4A and summarized in Table 4, complex karyotype ≥ 5 (versus all other karyotypes) was the only prognostic cytogenetic characteristic identified on pruning. The 5-year DFS for patients with complex karyotype ≥ 5 was 0% compared with 9.1% (95% CI, 6.1%-12.8%) for patients with other karyotypes (P < .001; Figure 4B). Patients with normal karyotype, who comprised the single largest karyotypic group, had a DFS not significantly different from that of patients with karyotypic abnormalities other than complex karyotype ≥ 5(P = .79; Figure 4C). On multivariable analysis, complex karyotype ≥ 5 remained significantly predictive of inferior DFS (P < .001), together with WBC (P = .013), sex (P = .040), and age (P = .014; Table 5). The risk of relapse or death was higher with an increasing WBC and patient age and higher among males compared with females. Relative to all other karyotypes (including normal karyotype), the hazard ratio (HR) of relapse or death for complex karyotype ≥ 5 patients was 3.46 (95% CI, 2.07-5.78).
Prognostic value of cytogenetics for OS
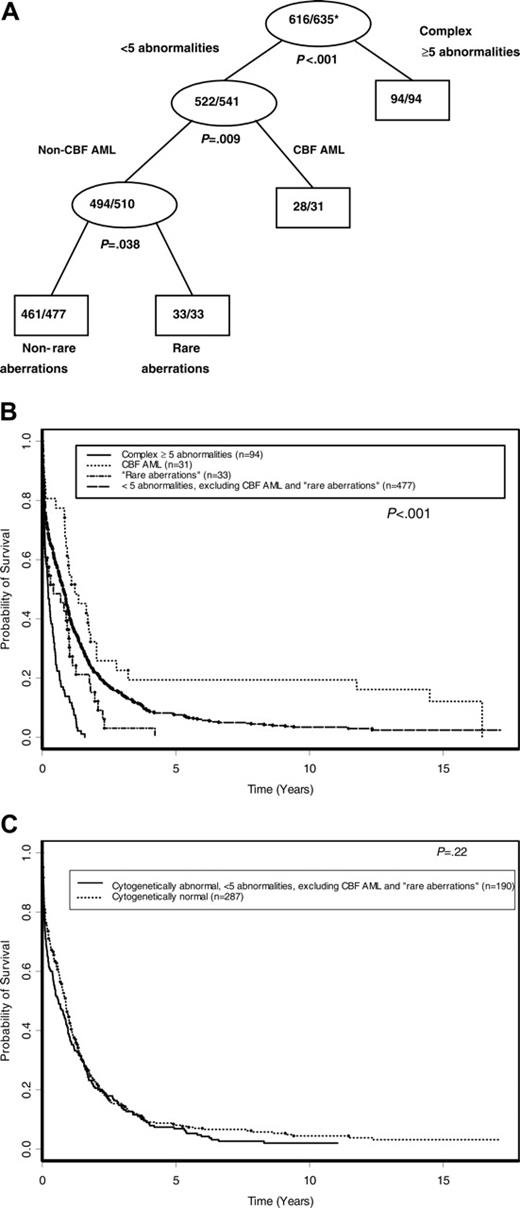
Figure 5A shows the tree developed for OS following pruning, using all karyotype groups shown in Figure 1 and the CBF AML group. Initially, complex karyotype ≥ 5 split the tree into 2 daughter nodes (P < .001). Then, among patients without complex karyotype ≥ 5, CBF AML-associated abnormalities emerged as the next best split (P = .009). The remaining patients without complex karyotype ≥ 5 or CBF AML-associated abnormalities were then further partitioned by the rare-aberrations group (P = .038). No further significant splitting occurred. As shown in Figure 5B and Table 4, the 5-year OS was 19.4% (95% CI, 7.9%-34.6%) for patients with CBF AML-associated abnormalities, 7.5% (95% CI, 5.4%-10.1%) for those with fewer than 5 abnormalities that did not include CBF AML-associated or rare aberrations, and 0% for both those with rare aberrations and those with complex karyotype ≥ 5(P < .001). The OS of patients with normal karyotype was not significantly different than that of patients with 1 to 4 abnormalities that did not include CBF AML-associated or rare aberrations (P = .22; Figure 5C).
On multivariable analysis, sex (P < .001), age (P = .001), WBC (P = .002), percentage of marrow blasts (P = .012), and the cytogenetic indicator variables (P < .001) emerged as the only significant variables predictive of survival (Table 5). The risk of death was higher with an increasing patient age, WBC, and percent marrow blasts and was higher among males compared with females. Compared with patients harboring CBF AML-associated abnormalities, the risk of death was significantly higher for patients with complex karyotype ≥ 5 (HR 4.49; 95% CI, 2.85-7.06), rare aberrations (HR 2.23; 95% CI, 1.31-3.79), and those with fewer than 5 abnormalities other than CBF AML-associated abnormalities and rare aberrations (HR 1.52; 95% CI, 1.03-2.25).
Discussion
Our results confirm the generally poor outcome of older AML patients treated with induction and postremission chemotherapy in doses commonly used in these patients for curative intent. The 5-year OS of 6.6% observed in our large series of older patients is similar to that previously reported for this population.28-31 Because of the generally poor outcome, the point might be made that supportive care represents the best option for many patients 60 years of age or older in terms of quality of life and reduced need for hospitalization. However, given that a small proportion of older AML patients can enjoy several years free from disease and/or be cured with intensive therapy, it is crucial to distinguish patients who might benefit from treatment from those for whom supportive or investigational therapy is more appropriate. The objective of this study was to investigate the significance of cytogenetics for identifying such patients, particularly those who derive little or no benefit from more intensive treatment beyond palliative intent.
The value of pretreatment cytogenetics in predicting response and long-term survival in AML is well established for younger patients6-10,32 and has recently been confirmed in the older population.12,33 Compared with the survival of patients with normal cytogenetics, abnormalities associated with a better outcome [eg, t(8;21), inv(16), and t(15;17)] were considered to be of favorable risk, whereas those associated with a worse outcome were considered be associated with adverse risk. As an alternative strategy, we used tree analysis to investigate the value of cytogenetic groups in predicting CR and long-term outcome in older AML patients. This strategy identifies distinguishing cytogenetic variables based on an exhaustive search of all possibilities and does not rely on an arbitrarily chosen reference group for definition of risk category.14
We identified 2 groups of older AML patients whose pretreatment cytogenetics predict an extremely poor outcome with no long-term survivorship. In the first group, defined by the presence of at least 5 abnormalities in a single clone, no patient is alive at 5 years despite relatively intensive AML therapy. In addition to being incurable with current treatments, patients with complex karyotype ≥ 5 have a very short survival and may be better suited for investigational therapy or supportive care, a conclusion also reached by others.12,34 Although this group overlaps significantly with an adverse risk category in another study,33 the 2 groups are not identical; patients with complex karyotype ≥ 5 are only a subset of the adverse-risk group defined by Wahlin et al.33 The second cytogenetic group associated with a very poor long-term outcome in our study comprised patients with 1 or 2 rare karyotypic aberrations. Although the median survival of this group was slightly better than that of patients with complex karyotype ≥ 5, no patient remained disease-free beyond 19 months or alive at 5 years (Table 2). While the majority of rare abnormalities (except for abnormalities involving 3q) have been previously grouped with normal karyotype in the same intermediate-risk group,12 our data suggest that, as a group, these rare aberrations predict an adverse outcome. Indeed, the first 14 of the 33 cases in the rare-aberrations group shown in Table 2 have chromosome aberrations that have been previously suggested to confer poor prognosis including t(6;9)(p23;q34)9,32 ; t(9;22)(q34;q11.2) and complex variants9,35,36 ; abnormalities of 3q,9,10,37 +4,38,39 i(11)(q10),39,40 and i(13)(q10)39,41-43 ; and double minutes (dmin).44,45 While to our knowledge the prognostic relevance of abnormalities detected in the remaining 19 cases in this group has not yet been established, the survival of these patients was as poor as that of the aforementioned 14 patients (P = .55; data not shown), suggesting that these aberrations may also confer a poor outcome, although no definitive conclusions can be drawn because of small patient numbers. A collaborative study pooling large datasets is required to investigate prognostic impact of each rare aberration separately.
Outcome of patients 60 years of age or older according to age decade. (A) DFS. (B) OS.
Outcome of patients 60 years of age or older according to age decade. (A) DFS. (B) OS.
Some differences between our results and those of others are noteworthy. In our analysis, karyotype complexity, defined by the presence of at least 5 abnormalities in a single clone, was highly predictive of DFS and OS. Although a similar definition for complex karyotype has been used to include patients in the unfavorable risk category in large UK MRC studies,10,12 other studies that were restricted to younger patients,9 or included patients both younger and older than 60 years,34 have included patients with at least 3 abnormalities in the definition of poor-risk patients. While complex karyotype ≥ 3 predicted a lower chance of achieving CR, it was not the best predictor of long-term outcome. Our analysis indicates that, at least in older patients, the definition of at least 5 abnormalities is more discriminating in identifying individuals with an extremely poor outcome with short median DFS (5.9 months) and OS (2.6 months). Patients with complex karyotype ≥ 5 had a significantly worse DFS (P = .02) and OS (P = .01) compared with those with 3 or 4 cytogenetic abnormalities in a single clone (data not shown). Furthermore, while none of the patients with at least 5 abnormalities were alive at 5 years, 7.1% (95% CI, 1.3%-20.4%) of patients with 3 or 4 abnormalities remained alive. In addition, while other studies have included patients with -5/5q-abnormalities in an unfavorable risk category,9,12,33 the presence of -5/5q-did not significantly add to our ability to identify patients who were incurable in our analysis, likely because most -5/5q-abnormalities are part of complex karyotype. Indeed, 75 of our 86 cases with -5/5q-were also part of a complex karyotype ≥ 5 (data not shown).
A limitation of our analysis is that we have not been able to meaningfully divide the patient population into learning and testing samples because of the relatively small number of patients carrying some of the recurring karyotypic abnormalities. Therefore, it will be important to apply our results to a large independent dataset for validation. An additional limitation is that the cytogenetically normal patients, who comprised 45% of the study population, are not further distinguishable from other patients without complex karyotype ≥ 5, CBF AML, and the rare aberrations with respect to OS. In particular, it is becoming apparent that cytogenetically normal AML is a molecularly heterogeneous disease, with different outcomes associated with molecular genetic rearrangements of critical genes.3,46-55 Future studies investigating risk stratification of AML using molecular analysis together with cytogenetics will likely add important prognostic information.
The prognostic significance of pretreatment clinical variables, independent of cytogenetics, has not been extensively studied in older AML patients. Although elevated lactate dehydrogenase level,56-58 secondary AML,59,60 high WBC,60 bone marrow blasts greater than 80%,56 and age older than 70 years56,61,62 have been reported as predictive of lower CR rate or worse survival, none of the aforementioned studies investigated the role of these variables once adjusted for cytogenetic risk groups. In the only analysis confined to older AML patients, and where detailed cytogenetics was also considered, increasing age, high WBC, and secondary AML were significant independent pretreatment variables associated with a reduced chance of achieving CR and poor survival.33 In this study, however, a significant number of patients received treatment of low intensity such as low-dose cytarabine, hydroxyurea, or thioguanine, often as single agents with palliative intent. In agreement with some of the published reports,56,60-62 we have also demonstrated that increasing age, higher WBC, and high percentage of marrow blasts are independent prognostic variables, in addition to sex (male) and cytogenetics, in predicting poorer survival. As only 2.5% of our patients had secondary AML, we could not meaningfully assess the significance of this variable. Importantly, while earlier studies indicated that patients older than 70 years have the poorest outcome,56,62 we suggest that a cutoff of 80 years might be more meaningful. Although relative to patients aged 60 to 69 years, the DFS and OS for patients aged 70 to 79 years were slightly (but statistically significantly) worse after adjusting for other significant variables. No long-term survivors were observed among patients aged 80 years or older in our series, even in the absence of complex karyotype ≥ 5 (data not shown). However, the number of patients aged 80 years or older was very small, and thus the question of whether such patients may be treated more appropriately using other therapies rather than intensive chemotherapy remains open.
Classification tree for CR after pruning. Internal leaves of the classification tree are displayed as ovals. Rectangles represent terminal leaves. The numbers inside each internal and terminal leaf represent the number of patients achieving CR out of the total number of patients included in the leaf. *Proportion of patients achieving CR.
Classification tree for CR after pruning. Internal leaves of the classification tree are displayed as ovals. Rectangles represent terminal leaves. The numbers inside each internal and terminal leaf represent the number of patients achieving CR out of the total number of patients included in the leaf. *Proportion of patients achieving CR.
It is important to note that the conclusions from our data relate only to the outcome of older AML patients treated with conventional cytotoxic agents used in conventional doses. None of our patients received high-dose chemotherapy with hematopoietic stem cell (HSC) support,63,64 novel molecular targeting agents,65-67 or allogeneic HSC transplantation.68-70 The relative benefit of these therapies on the outcome of older AML patients is currently uncertain. In a large series investigating the feasibility of intensive therapy that included consolidation with autologous HSC transplantation in 258 de novo AML patients older than 60 years, only 135 patients (none aged ≥ 80 years) qualified for intensive therapy, of whom only 16 actually received a transplant, suggesting that this approach is unlikely to have a major impact on the disease in this population.63 Furthermore, the prognosis is unlikely to be improved in patients with unfavorable cytogenetics.62,63 Recently, while the feasibility of nonmyeloablative allogeneic HSC transplantation in selected older AML patients has been demonstrated,68-70 the true impact of this treatment remains to be defined, particularly in patients with adverse cytogenetics.
DFS according to cytogenetic risk groups. (A) Final tree by TSSA after pruning. Internal leaves are shown by ovals and terminal leaves by rectangles. The numbers inside each internal and terminal leaf represent the number of patients suffering relapse or death out of the total number of patients at risk. *Proportion of patients suffering relapse or death. (B) DFS curves comparing cytogenetic risk groups identified by TSSA. (C) DFS curves comparing patients with normal karyotype with patients who have an abnormal karyotype with fewer than 5 abnormalities.
DFS according to cytogenetic risk groups. (A) Final tree by TSSA after pruning. Internal leaves are shown by ovals and terminal leaves by rectangles. The numbers inside each internal and terminal leaf represent the number of patients suffering relapse or death out of the total number of patients at risk. *Proportion of patients suffering relapse or death. (B) DFS curves comparing cytogenetic risk groups identified by TSSA. (C) DFS curves comparing patients with normal karyotype with patients who have an abnormal karyotype with fewer than 5 abnormalities.
In summary, we have confirmed that pretreatment cytogenetic analysis is also useful in predicting the outcome of older AML patients. Importantly, cytogenetic analysis can accurately predict a group of older patients who are not suited for intensive chemotherapy, especially those with complex karyotype, defined by the presence of at least 5 unrelated abnormalities in a single clone. Other pretreatment variables may also add independent prognostic information that could assist in treatment decisions. Despite the generally poor outcome of the remaining patients, however, it seems prudent to continue to recommend intensive chemotherapy because a proportion of these patients may enjoy prolonged disease-free survival and still be cured. It is possible that future molecular analyses may provide better means of identifying the latter patients and also potentially define appropriate pathways for targeted therapies.
OS according to cytogenetic risk groups. (A) Final tree by TSSA after pruning. Internal leaves are shown by ovals and terminal leaves by rectangles. The numbers inside each internal and terminal leaf represent the number of patients dead out of the total number of patients at risk. *Proportion dead. (B) OS curves comparing cytogenetic risk groups identified by TSSA. (C) OS curves comparing patients with normal karyotype with patients who have an abnormal karyotype with fewer than 5 abnormalities, excluding CBF AML and rare aberrations.
OS according to cytogenetic risk groups. (A) Final tree by TSSA after pruning. Internal leaves are shown by ovals and terminal leaves by rectangles. The numbers inside each internal and terminal leaf represent the number of patients dead out of the total number of patients at risk. *Proportion dead. (B) OS curves comparing cytogenetic risk groups identified by TSSA. (C) OS curves comparing patients with normal karyotype with patients who have an abnormal karyotype with fewer than 5 abnormalities, excluding CBF AML and rare aberrations.
Appendix
The following Cancer and Leukemia Group B institutions, principal investigators, and cytogeneticists participated in this study. Wake Forest University School of Medicine, Winston-Salem, NC: David D. Hurd, Harold O. Goodman, P. Nagesh Rao, Wendy L. Flejter, and Mark J. Pettenati (grant no. CA03927); Duke University Medical Center, Durham, NC: Jeffrey Crawford, Sandra H. Bigner, and Mazin B. Qumsiyeh (grant no. CA47577); North Shore-Long Island Jewish Health System, Manhasset, NY: Daniel R. Budman and Prasad R. K. Koduru (grant no. CA35279); University of Maryland Cancer Center, Baltimore, MD: Martin Edelman, Joseph R. Testa, Deana Hallman, Stuart Schwartz, Maimon M. Cohen, and Judith Stamberg (grant no. CA31983); University of Alabama at Birmingham, Birmingham, AL: Robert Diasio and Andrew J. Carroll (grant no. CA47545); Dana-Farber Cancer Institute, Boston, MA: George P. Canellos, Ramana Tantravahi, Cynthia C. Morton, and Leonard L. Atkins (grant no. CA32291); Christiana Care Health Services, Newark, DE: Stephen S. Grubbs, Digamber S. Borgaonkar, and Jeanne M. Meck (grant no. CA45418); University of Iowa Hospitals, Iowa City, IA: Gerald H. Clamon and Shivanand R. Patil (grant no. CA47642); Dartmouth Medical School, Lebanon, NH: Marc S. Ernstoff, Doris H. Wurster-Hill, and Thuluvancheri K. Mohandas (grant no. CA04326); Roswell Park Cancer Institute, Buffalo, NY: Ellis G. Levine and Anne Marie W. Block (grant no. CA02599); University of Missouri/Ellis Fischel Cancer Center, Columbia, MO: Michael C. Perry, Judith H. Miles, Jeffrey R. Sawyer, and Tim H. Huang (grant no. CA12046); University of Minnesota, Minneapolis, MN: Bruce A. Peterson, Betsy A. Hirsch, and Diane C. Arthur (grant no. CA16450); University of Tennessee Cancer Center, Memphis, TN: Harvey B. Niell and Sugandhi A. Tharapel (grant no. CA47555); Weill Medical College of Cornell University, New York, NY: Scott Wadler, Ram S. Verma, and Prasad R. K. Koduru (grant no. CA07968); Rhode Island Hospital, Providence, RI: William Sikov, Teresita Padre-Mendoza, Jennifer A. Ahearn, and Hon Fong L. Mark (grant no. CA08025); Long Island Jewish Medical Center, Lake Success, NY: Marc Citron, Alan L. Shanske, and Prasad R. K. Koduru (grant no. CA11028); University of North Carolina, Chapel Hill, NC: Thomas Shea and Kathleen W. Rao (grant no. CA47559); University of Chicago Medical Center, Chicago, IL: Gini Fleming, Diane Roulston, and Michelle M. Le Beau (grant no. CA41287); SUNY Upstate Medical University, Syracuse, NY: Stephen L. Graziano, Navnit S. Mitter, Edward J. Hallinan, Lawrence P. Gordon, and Constance K. Stein (grant no. CA21060); Eastern Maine Medical Center, Bangor, ME: Philip L. Brooks and Laurent J. Beauregard (grant no. CA35406); The Ohio State University, Columbus, OH: Clara D. Bloomfield and Karl S. Theil (grant no. CA77658); University of California, San Diego, CA: Stephen L. Seagren, Renée Bernstein, and Marie L. Dell'Aquila (grant no. CA11789); Washington University School of Medicine, St Louis, MO: Nancy L. Bartlett and Michael S. Watson (grant no. CA77440); Massachusetts General Hospital, Boston, MA: Michael L. Grossbard, Cynthia C. Morton, and Leonard L. Atkins (grant no. CA 12449); Walter Reed Army Medical Center, Washington, DC: Joseph J. Drabick and Rawatmal B. Surana (grant no. CA26806); University of Massachusetts Medical Center, Worcester, MA: Pankaj Bhargava, Philip L. Townes, and Vikram Jaswaney (grant no. CA37135); SUNY Maimonides Medical Center, Brooklyn, NY: Sameer Rafla and Ram S. Verma (grant no. CA25119); Medical University of South Carolina, Charleston, SC: Mark R. Green, Eduardo S. Cantu, G. Shashidhar Pai, and Daynna J. Wolff (grant no. CA03927); Parkview Hospital, Ft Wayne, IN: Sreenivasa Nattam and Patricia I. Bader; Columbia-Presbyterian Medical Center, New York, NY: Rose R. Ellison and Dorothy Warburton; Finsen Institute, Copenhagen, Denmark: Nis I. Nissen and Preben Philip; Georgetown University Medical Center, Washington, DC: Edward P. Gelmann and Jeanne M. Meck (grant no. CA77597); McGill Department of Oncology, Montreal, QC, Canada: J. L. Hutchison and Jacqueline Emond (grant no. CA31809); Mount Sinai School of Medicine, New York, NY: Lewis R. Silverman and Vesna Najfeld (grant no. CA04457); Vermont Cancer Center, Burlington, VT: Hyman B. Muss and Elizabeth F. Allen (grant no. CA77406); University of Illinois, Chicago, IL: Jeffrey A. Sosman and Maureen M. McCorquodale (grant CA74811); University of Puerto Rico, San Juan, PR: Enrique Velez-Garcia; University of Nebraska Medical Center, Omaha, NE: Margaret A. Kessinger Wegner and Warren G. Sanger (grant no. CA77298); Virginia Commonwealth University MB CCOP, Richmond, VA: John D. Roberts and Colleen Jackson-Cook (grant no. CA52784); University of Cincinnati Medical Center, Cincinnati, OH: Orlando J. Martelo and Ashok K. Srivastava; and Southern Nevada Cancer Research Foundation CCOP, Las Vegas, NV: John Ellerton (grant no. CA35421).
Prepublished online as Blood First Edition Paper, March 7, 2006; DOI 10.1182/blood-2005-11-4354.
A complete list of Cancer and Leukemia Group B institutions and principal investigators who participated in this study appears in the “Appendix.”
Supported, in part, by grants from the National Cancer Institute to the Cancer and Leukemia Group B (CA101140, CA31946, CA77658, CA33601, CA41287, CA47545, CA03927, CA47577, CA35279, and CA32291), CA16058, and the Coleman Leukemia Research Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal