Abstract
HIV-1 Nef and HIV-1–specific cytotoxic T lymphocytes (CTLs) have important and opposing roles in the immunopathogenesis of HIV-1 infection. Nef-mediated down-modulation of HLA class I on infected cells can confer resistance to CTL clearance, but the factors determining the efficiency of this process are unknown. This study examines the impact of Nef on the antiviral activity of several CTL clones recognizing epitopes from early and late HIV-1 proteins, restricted by HLA-A, -B, and -C molecules. CTL-targeting epitopes in early proteins remained susceptible to the effects of Nef, although possibly to a lesser degree than CTL-targeting late protein epitopes, indicating that significant Nef-mediated HLA down-regulation can precede even the presentation of early protein-derived epitopes. However, HLA-C–restricted CTLs were unaffected by Nef, consistent with down-regulation of cell-surface HLA-A and -B but not HLA-C molecules. Thus, CTLs vary dramatically in their susceptibility to Nef interference, suggesting differences in the relative importance of HLA-A– and HLA-B– versus HLA-C–restricted CTLs in vivo. The data thus indicate that HLA-C–restricted CTLs may have an under-appreciated antiviral role in the setting of Nef in vivo and suggest a benefit of promoting HLA-C–restricted CTLs for immunotherapy or vaccine development.
Introduction
Nef is a highly expressed 27-kDa myristoylated protein that is specific to HIV-1, HIV-2, and SIV, but not found in other retroviruses. Thus, as an “accessory protein” without a known direct role in viral replication, the observed maintenance of its reading frame in vivo indicates important roles in viral persistence. Many candidate functions have been attributed to Nef through in vitro studies, including CD4 down-regulation, enhanced viral infectivity, cellular activation, HLA class I and II down-regulation, and many others (for reviews, see Geyer et al1 and Piguet and Trono2 ). Although the precise immunopathogenic roles of such effects remain unclear, the attenuation of disease in HIV-1–infected humans3 and SIV-infected macaques4 after infection with Nef-defective viruses indicates a central role for this protein in HIV-1 infection in vivo.
Multiple lines of evidence demonstrate that Nef-mediated HLA class I down-regulation on infected cells is likely an important mechanism contributing to HIV-1 persistence through evasion of CD8+ cytotoxic T lymphocytes (CTLs). HIV-1–specific CTLs, which exert a key protective antiviral effect (for reviews, see Yang and Walker5 and Yang6 ), clear infected cells by recognition of viral epitopes presented by HLA class I molecules. Several studies have documented the ability of Nef to reduce cell-surface HLA expression, either through increased endocytosis7 or impaired trafficking from the Golgi apparatus.8,9 This effect is sufficient to interfere with the abilities of HIV-1–specific CTL clones to clear infected cells10,11 and suppress viral replication in vitro.11,12 Moreover, in macaques infected with difficult-to-revert Nef mutations specifically ablating HLA class I down-regulatory function, SIV has been observed to undergo rapid nef sequence evolution to reconstitute this function through an alternative motif,13 underscoring the relevance of this Nef activity in vivo.
The factors that affect the ability of Nef to interfere with CTL antiviral activity are poorly understood. It is unclear whether HIV-1–specific CTLs vary in their susceptibility to this effect, because they differ in specificity and other functional properties that could affect the impact of Nef. HIV-1 proteins are expressed from early (tat, rev, and nef, which are spliced) versus late (gag, pol, env, vpr, vpu, and vif, which are partially spliced or unspliced) RNA transcripts.14-16 Prior studies showing a Nef effect on CTL antiviral activity have used exclusively late protein-targeted CTLs.10-12 Thus, whereas expression of late protein epitopes can be impaired by Nef-mediated HLA down-regulation before CTL recognition, it is not known whether the more rapid presentation of early protein epitopes17,18 could allow CTL recognition before Nef mediates HLA down-regulation. Moreover, HIV-1–specific CTLs can recognize epitopes presented by HLA-A, -B, or -C molecules, although prior studies have only examined susceptibility of HLA-A– and -B–restricted CTLs to Nef.10-12 Although expression of HLA-C molecules has been shown to be unaffected by Nef,19 it is thus unknown whether C-restricted CTLs are resistant to Nef effects. Given the likely contribution of Nef to HIV-1 persistence in the face of the CTL response in vivo, defining the influence of CTL specificity on the impact of Nef will be important to understand the determinants of antiviral efficacy of CTLs in vivo.
Materials and methods
HIV-1–permissive cells
Jurkat (expressing HLA B*07 and C*07) and T1 cells (expressing HLA A*02 and C*03), as well as a T1/primary CD4+ T lymphocyte hybridoma (expressing HLA B*57) were maintained as previously described.20-22 Primary CD4+ T lymphoblasts (HLA B*07, C*03, C*15) were generated from HLA-typed healthy HIV-1–seronegative donors using CD3/CD8 bispecific antibodies as previously described.20,22 Cells in log phase growth were acutely infected for the HIV-1 inhibition experiments using CTL clones that were HLA matched with the target cells for the restricting alleles.
HIV-1 stocks
Production of infectious virions from plasmid NL4-3–based HIV-1 DNA was performed as previously described.23 Briefly, plasmid p83-2.1 (containing the NL4-3 genome including gag-pol, up to a unique EcoRI site), and the appropriate mutant of p83-10 (containing the NL4-3 genome including env-nef, beyond the EcoRI site) were linearized with EcoRI and coelectroporated into T1 cells to produce replication-competent virions. Low-passage virus stocks were frozen in aliquots at –80°C, and viral titer (50% tissue-culture infectious dose, TCID50/mL) was determined by end-point dilution as previously described.24 Wild-type Nef viruses were produced using p83-10 plasmids containing no mutations in nef; defective Nef viruses were produced using p83-10 containing a methionine to alanine mutation at amino acid 20 (M20A)11,25 or a large deletion resulting in truncation after the first 12 amino acids.26,27 The former allowed testing of Nef-specific CTL clones in this system because Nef-derived CTL epitopes were still expressed.
The specific constructs used for the inhibition assays also were engineered to contain the recognized epitope for the tested CTL clones as appropriate in cases where NL4-3 sequences contained nonrecognized epitope polymorphisms. For example, p83-2.1 (containing the A*02-restricted Gag p17 epitope SLYNTVATL, amino acids 77-85) was derived from p83-2 (containing the variant SLYNTIAVL) by polymerase chain reaction (PCR) point mutagenesis and confirmatory sequencing, as previously described.28
CTL clones
HIV-1–specific CTL clones were obtained by limiting dilution cloning from peripheral blood mononuclear cells (PBMCs) of HIV-1–infected individuals, characterized for epitope specificity and HLA restriction, and maintained by periodic restimulation with anti-CD3 antibody and irradiated feeder mononuclear cells as previously described.20,21,29 A full listing of these clones is given in Table 1.
Nef effects on inhibition of HIV-1 replication by CTL clones
Clone . | Epitope (location) . | HLA . | Nef ratio (SD) . | No. of repeats . |
|---|---|---|---|---|
| Late protein/A or B restricted | ||||
| S1-SL9-3.23T | SLYNTVATL (Gag p17 77-85) | A*02 | 0.21 (0.15) | 5 |
| S36-SL9-10.18T | SLYNTVATL (Gag p17 77-85) | A*02 | 0.47 (0.38) | 2 |
| S36-SL9-10.26T | SLYNTVATL (Gag p17 77-85) | A*02 | 0.30 | 1 |
| 18030D23 | SLYNTVATL (Gag p17 77-85) | A*02 | 0.12 (0.14) | 4 |
| S42953-SL9-10.18 | SLYNTVATL (Gag p17 77-85) | A*02 | -0.08 | 1 |
| 161J×A12 | SLYNTVATL (Gag p17 77-85) | A*02 | -0.08 | 1 |
| 115D3 | SLYNTVATL (Gag p17 77-85) | A*02 | 0.06 | 1 |
| S14-KF11-1.3 | KAFSPEVIPMF (Gag p24 30-40) | B*57 | 0.46 | 1 |
| S14-KF11-10.6 | KAFSPEVIPMF (Gag p24 30-40) | B*57 | 0.62 | 1 |
| 68A62 | ILKEPVHGV (RT 309-317) | A*02 | 0.24 (0.24) | 8 |
| S34-KW10-10.38T | KIATESIVIW (RT 374-383) | B*57 | 0.03 | 1 |
| S34-KW10-10.55T | KIATESIVIW (RT 374-383) | B*57 | 0.26 | 1 |
| KS7 | IPRRIRQGL (Env gp41 332-340) | B*07 | 0.29 | 1 |
| Early protein/A or B restricted | ||||
| S42758-RL10-3.22T | RPAEPVPLQL (Rev 66-75) | B*07 | 0.23 (0.20) | 4 |
| S14-HQ10-1.1 | HTQGYFPDWQ (Nef 116-125) | B*57 | 0.46 (0.11) | 2 |
| S14-HQ10-1.3 | HTQGYFPDWQ (Nef 116-125) | B*57 | 0.37 | 1 |
| S14-YT9-10.4 | YFPDWQNYT (Nef 120-128) | B*57 | 0.50 (0.01) | 2 |
| S14-YT9-10.8 | YFPDWQNYT (Nef 120-128) | B*57 | 0.47 | 1 |
| S36-YT9-3.36 | YFPDWQNYT (Nef 120-128) | B*57 | 0.75 | 1 |
| C restricted | ||||
| 161J×B28 | RAIEAQQHL (Env gp41 46-54) | C*03 | 0.95 | 1 |
| 161J-JD19 | RAIEAQQHL (Env gp41 46-54) | C*03 | 0.76 | 1 |
| SE7/126E | RAIEAQQHL (Env gp41 46-54) | C*15 | 1.10 | 1 |
| S22-AL9-3.7 | AAVDLSHFL (Nef 83-91) | C*03 | 0.94 (0.11) | 2 |
| S16-RY11-10.41 | RRQDILDLWVY (Nef 105-115) | C*07 | 1.05 (0.10) | 3 |
Clone . | Epitope (location) . | HLA . | Nef ratio (SD) . | No. of repeats . |
|---|---|---|---|---|
| Late protein/A or B restricted | ||||
| S1-SL9-3.23T | SLYNTVATL (Gag p17 77-85) | A*02 | 0.21 (0.15) | 5 |
| S36-SL9-10.18T | SLYNTVATL (Gag p17 77-85) | A*02 | 0.47 (0.38) | 2 |
| S36-SL9-10.26T | SLYNTVATL (Gag p17 77-85) | A*02 | 0.30 | 1 |
| 18030D23 | SLYNTVATL (Gag p17 77-85) | A*02 | 0.12 (0.14) | 4 |
| S42953-SL9-10.18 | SLYNTVATL (Gag p17 77-85) | A*02 | -0.08 | 1 |
| 161J×A12 | SLYNTVATL (Gag p17 77-85) | A*02 | -0.08 | 1 |
| 115D3 | SLYNTVATL (Gag p17 77-85) | A*02 | 0.06 | 1 |
| S14-KF11-1.3 | KAFSPEVIPMF (Gag p24 30-40) | B*57 | 0.46 | 1 |
| S14-KF11-10.6 | KAFSPEVIPMF (Gag p24 30-40) | B*57 | 0.62 | 1 |
| 68A62 | ILKEPVHGV (RT 309-317) | A*02 | 0.24 (0.24) | 8 |
| S34-KW10-10.38T | KIATESIVIW (RT 374-383) | B*57 | 0.03 | 1 |
| S34-KW10-10.55T | KIATESIVIW (RT 374-383) | B*57 | 0.26 | 1 |
| KS7 | IPRRIRQGL (Env gp41 332-340) | B*07 | 0.29 | 1 |
| Early protein/A or B restricted | ||||
| S42758-RL10-3.22T | RPAEPVPLQL (Rev 66-75) | B*07 | 0.23 (0.20) | 4 |
| S14-HQ10-1.1 | HTQGYFPDWQ (Nef 116-125) | B*57 | 0.46 (0.11) | 2 |
| S14-HQ10-1.3 | HTQGYFPDWQ (Nef 116-125) | B*57 | 0.37 | 1 |
| S14-YT9-10.4 | YFPDWQNYT (Nef 120-128) | B*57 | 0.50 (0.01) | 2 |
| S14-YT9-10.8 | YFPDWQNYT (Nef 120-128) | B*57 | 0.47 | 1 |
| S36-YT9-3.36 | YFPDWQNYT (Nef 120-128) | B*57 | 0.75 | 1 |
| C restricted | ||||
| 161J×B28 | RAIEAQQHL (Env gp41 46-54) | C*03 | 0.95 | 1 |
| 161J-JD19 | RAIEAQQHL (Env gp41 46-54) | C*03 | 0.76 | 1 |
| SE7/126E | RAIEAQQHL (Env gp41 46-54) | C*15 | 1.10 | 1 |
| S22-AL9-3.7 | AAVDLSHFL (Nef 83-91) | C*03 | 0.94 (0.11) | 2 |
| S16-RY11-10.41 | RRQDILDLWVY (Nef 105-115) | C*07 | 1.05 (0.10) | 3 |
CTL clones were tested as described in Figure 1, comparing suppression of Nef-competent (NL4-3 wild-type Nef) to Nef-defective (the same virus with either a large deletion in nef or a mutation resulting in a methionine to alanine mutation at amino acid 20, M20A) HIV-1 for approximately 7 days. The effect of Nef was calculated as an inhibition ratio, as described in Figure 1 (mean, SD, and number of repeats given when multiple independent experiments were performed with the same clone).
Assay for inhibition of HIV-1 by CTL clones
The suppression of HIV-1 in acutely infected cells by HIV-1–specific clones was assayed as previously described.20,22 Briefly, target cells were infected at a multiplicity of infection of approximately 0.01 TCID50 /cell and cocultured with CTLs at an effector-target ratio of 0.25:1, either in 96-well plates (1.25 × 104 CTLs with 5 × 104 target cells in 200 μL medium) or 24-well plates (1.25 × 105 CTLs with 5 × 105 target cells in 2 mL medium). Viral replication was measured by quantitative p24 enzyme-linked immunosorbent assay (ELISA; Dupont, Boston, MA), and inhibition was calculated by comparing replication in cells with or without CTLs.
Statistics
The 2-tailed Student t test was performed for comparisons of Nef effects on viral inhibition between different groups of CTL clones, using Microsoft Excel 2004 (Microsoft, Redmond, WA) on a G4 Power Macintosh Apple Computer (Apple, Cupertino, CA).
Results
Individual CTL clones are reproducibly susceptible to the effects of Nef
Multiple studies have demonstrated that HIV-1–specific CTL antiviral activity can be antagonized by Nef-mediated HLA class I down-modulation,10-12 although the reproducibility of this effect for individual CTL clones has never been evaluated in detail.
A system was established to allow standardized comparisons of Nef effects on CTL suppression of HIV-1 replication across repeat experiments comparing wild-type and defective Nef viruses (either Nef deletion or mutation with a methionine to alanine at amino acid 20, M20A, both of which fail to down-regulate HLA,26 and demonstrate similar effects on the antiviral activity of CTL clones, data not shown). For each experiment, the log unit efficiency of inhibition was determined for each virus at approximately day 7 (log10 units of p24 reduction by the CTL clone divided by total log units of p24 produced in the absence of CTLs), and the ratio of wild-type Nef to defective Nef log10 unit efficiency inhibition then was calculated to yield a “Nef effect ratio” (Figure 1). Because individual CTL clones appeared to vary in their susceptibility to Nef (compare Figure 1A and B), a panel of clones was tested for the reproducibility of Nef effects in this manner. A clone recognizing an A*02-restricted epitope in RT (ILKEPVHGV), 2 clones recognizing an HLA A*02-restricted epitope in Gag p17 (SLYNTVATL), a clone recognizing a B*07-restricted epitope in Rev (RPAEPVPLQL), and a clone recognizing a C*07-restricted epitope in Nef (RRQDILDLWVY) were evaluated in multiple independent experiments (Figure 2). The Nef effect ratios for each clone clustered over multiple independent measurements, demonstrating the reproducibility of Nef interference with the antiviral effects of CTLs. Interestingly, 4 of the CTL clones were similarly affected (Nef effect ratio, ∼0.2), whereas one appeared entirely unaffected by Nef (ratio, ∼1.0). This clone was Nef specific and C*07 restricted, raising the question of whether its specificity or HLA restriction conferred resistance to Nef-mediated HLA down-regulation.
Measurement of Nef effects on the antiviral activities of CTL clones. CTL clones 42758-RL10-3.22 (Rev specific, B*07 restricted) and S16-RY11-10.41 (Nef specific, C*07 restricted) were tested for inhibition of NL4-3–based viruses containing wild-type Nef or Nef with the M20A mutation (rendering it unable to down-regulate HLA class I) in parallel. The target cells were acutely infected Jurkat cells (expressing both B*07 and C*07). Replication as assessed by measuring supernatant p24 antigen (log units picograms/milliliter) is plotted over time. (A) For clone 42758-RL10-3.22, inhibition of the wild-type virus at day 6 was 0.8 log10 units (6.3-5.5) and inhibition efficiency was 0.13 (0.8 ÷ 6.3). Inhibition of the M20A virus at day 6 was 5.4 log10 units (6.3-0.9) and inhibition efficiency was 0.86 (5.4 ÷ 6.3). Thus, the Nef effect ratio of wild-type to defective Nef virus inhibition was 0.15 (0.13 ÷ 0.86). (B) Similarly for clone S16-RY11-10.41, inhibition efficiency of the wild-type and M20A viruses were 0.49 and 0.59, respectively, yielding a Nef effect ratio of 0.83.
Measurement of Nef effects on the antiviral activities of CTL clones. CTL clones 42758-RL10-3.22 (Rev specific, B*07 restricted) and S16-RY11-10.41 (Nef specific, C*07 restricted) were tested for inhibition of NL4-3–based viruses containing wild-type Nef or Nef with the M20A mutation (rendering it unable to down-regulate HLA class I) in parallel. The target cells were acutely infected Jurkat cells (expressing both B*07 and C*07). Replication as assessed by measuring supernatant p24 antigen (log units picograms/milliliter) is plotted over time. (A) For clone 42758-RL10-3.22, inhibition of the wild-type virus at day 6 was 0.8 log10 units (6.3-5.5) and inhibition efficiency was 0.13 (0.8 ÷ 6.3). Inhibition of the M20A virus at day 6 was 5.4 log10 units (6.3-0.9) and inhibition efficiency was 0.86 (5.4 ÷ 6.3). Thus, the Nef effect ratio of wild-type to defective Nef virus inhibition was 0.15 (0.13 ÷ 0.86). (B) Similarly for clone S16-RY11-10.41, inhibition efficiency of the wild-type and M20A viruses were 0.49 and 0.59, respectively, yielding a Nef effect ratio of 0.83.
Nef can mediate HIV-1 resistance against CTL-recognizing epitopes in early proteins
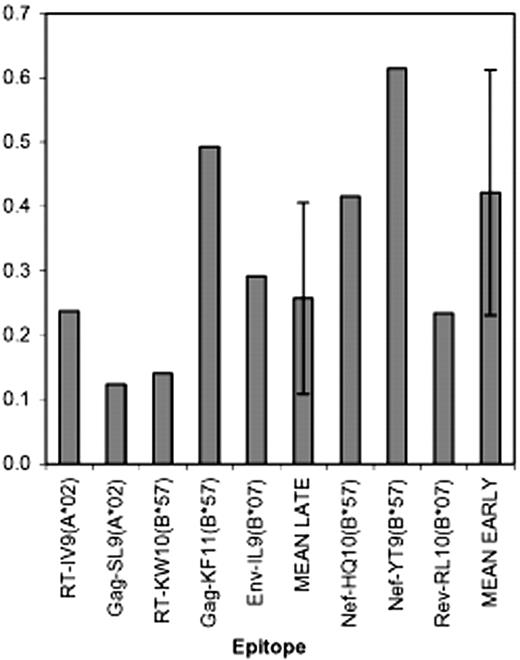
All prior reports of Nef effects on CTL anti-HIV-1 activity used clones recognizing epitopes in Gag, Pol, or Env, which are all late HIV-1 proteins compared with Tat, Rev, and Nef, whose expression is several hours earlier in the viral life cycle.14-16 It is thus unknown whether CTL-targeting early protein epitopes, particularly Nef (Figure 2), might have a kinetic advantage in relation to HLA down-regulation by Nef. To explore this, CTL clones (restricted by HLA-A and -B) targeting epitopes from early proteins (Rev and Nef) versus late proteins (Gag, Pol, and Env) were tested for ability to suppress replication of Nef-competent versus Nef-defective HIV-1 strains. Despite targeting of early protein epitopes, the Nef- and Rev-specific CTLs remained susceptible to functional interference by Nef (Figure 3; Table 1). There was a trend (not reaching statistical significance, P = .279) suggesting that CTLs recognizing early proteins might be less affected than those recognizing late proteins (mean Nef effect ratio of 0.26 ± 0.15 for late protein CTLs versus 0.42 ± 0.19 for early protein CTLs), particularly those targeting Nef. Nevertheless, all clones were clearly less antiviral in the presence of wild-type Nef. These results indicate that down-modulation of HLA by Nef can affect CTLs recognizing early proteins and that the kinetic advantage of early protein targeting does not necessarily overcome this Nef-mediated mechanism of immune evasion.
Reproducibility of Nef effects on the antiviral activities of HIV-1–specific CTL clones on repeat testing. Five CTL clones were tested as described in Figure 1. The means (± SEM) are plotted for multiple independent experiments with each clone. Comparisons revealed that 68A62, S1-SL9-3.23T, 18030D23, and 42758-RL10-3.22 were statistically indistinguishable from each other (2-tailed t test P ≥ .26 for all comparisons between these clones), but clone S16-RY11-10.41 was statistically distinct from the others (P ≤ .01 for all comparisons to the other clones).
Reproducibility of Nef effects on the antiviral activities of HIV-1–specific CTL clones on repeat testing. Five CTL clones were tested as described in Figure 1. The means (± SEM) are plotted for multiple independent experiments with each clone. Comparisons revealed that 68A62, S1-SL9-3.23T, 18030D23, and 42758-RL10-3.22 were statistically indistinguishable from each other (2-tailed t test P ≥ .26 for all comparisons between these clones), but clone S16-RY11-10.41 was statistically distinct from the others (P ≤ .01 for all comparisons to the other clones).
Comparisons of Nef effects on CTL-recognizing late protein versus early protein epitopes. For the 8 epitopes shown (5 from the late proteins RT, Gag, and Env, and 3 from the early proteins Nef and Rev), CTL clones were tested for Nef effects as described in Figure 1. The bar plots represent average results for each epitope. In the case that there were multiple repeats of individual clones (as described in Figure 2 and Table 1), these were averaged; if there were multiple clones from the same HIV-1–infected person, these were then again averaged (to apply equal weight to CTL clones from different persons). Finally, results across all persons were averaged (if clones recognizing the same epitope from multiple persons were tested) to generate the single values plotted for each epitope. MEAN LATE indicates the average for the 5 late protein-derived epitopes (± 1 SD) and MEAN EARLY indicates the average for the 3 early protein-derived epitopes.
Comparisons of Nef effects on CTL-recognizing late protein versus early protein epitopes. For the 8 epitopes shown (5 from the late proteins RT, Gag, and Env, and 3 from the early proteins Nef and Rev), CTL clones were tested for Nef effects as described in Figure 1. The bar plots represent average results for each epitope. In the case that there were multiple repeats of individual clones (as described in Figure 2 and Table 1), these were averaged; if there were multiple clones from the same HIV-1–infected person, these were then again averaged (to apply equal weight to CTL clones from different persons). Finally, results across all persons were averaged (if clones recognizing the same epitope from multiple persons were tested) to generate the single values plotted for each epitope. MEAN LATE indicates the average for the 5 late protein-derived epitopes (± 1 SD) and MEAN EARLY indicates the average for the 3 early protein-derived epitopes.
HLA-C–restricted, HIV-1–specific CTLs are functionally unaffected by Nef-mediated HLA down-regulation
Although Nef has been shown to reduce total cell-surface HLA class I in several reports, a more detailed study subsequently has demonstrated that Nef selectively down-regulates HLA-A and -B but not -C molecules.19 The lack of Nef impact on the antiviral function of an HLA-C–restricted clone (Figures 1, 2) was consistent with this finding. To determine whether this is a generalized phenomenon for all HLA-C–restricted CTLs, multiple C-restricted CTL clones were tested for their ability to suppress replication of wild-type versus Nef-defective viruses (Figure 4 and Table 1). In sharp contrast to the HLA-A– and -B–restricted CTLs, 5 different C-restricted CTL clones were essentially unaffected by Nef (mean Nef effect ratio 0.32 ± 0.17 for all A- and B-restricted epitopes versus 1.00 ± 0.10 for C-restricted epitopes). Three of these clones recognized the same Env epitope (RAIEAQQHL), but 2 were restricted by C*03 and one by C*15. The other 2 recognized Nef epitopes restricted by C*03 (AAVDLSHFL) and C*07 (RRQDILDLWVY). Thus agreeing with the prior report that Nef does not down-regulate HLA-C molecules, Nef generally reduces the antiviral activity of HLA-A– and -B– but not -C–restricted CTLs. Finally, these data (and Figure 1) indicate that HLA-C–restricted CTL antiviral activity is similarly potent to that of HLA-A– and -B–restricted CTLs against Nef-defective viruses and superior against Nef-competent viruses, and confirm that these effects are observed in primary CD4+ T lymphocytes.
Discussion
The interaction of CTLs with HIV-1 in vivo remains incompletely understood. Convincing qualitative evidence for an important antiviral role includes observations that the development of the CTL response correlates with the drop in viremia at the end of acute infection,30,31 that the waning of the CTL response correlates with rising viremia in late infection,32,33 and that transient CD8 depletion in SIV-infected macaques results in transient marked rises in viremia.34-36 Despite these findings, defining a quantitative measurement of HIV-1–specific CTLs that correlates to immune control of viral replication in vivo remains elusive.6 All other factors being equal, more CTLs should equate to better immune control, but comprehensive studies have shown that the magnitude of the CTL response bears little or no apparent relationship to the level of viremia.37,38
Although bias in the detection of CTL responses due to differences between screening sequences versus autologous HIV-1 sequences could be a confounder,39,40 it is likely that other factors have an impact on the antiviral effects of CTLs in vivo. Candidate explanations include viral factors such as cell tropism, replicative capacity, Nef function, and variation in epitope sequences. CTL factors could include their differentiation, activation, functional status, functional avidity, and epitope targeting. Indirect evidence for the importance of CTL targeting includes associations of certain HLA haplotypes with rapid or slow disease progression, and further support is provided by in vitro experiments demonstrating that epitope targeting determines the ability of CTLs to suppress HIV-1 replication41 due to epitope differences in expression42 or kinetics.17,43 The current study raises another possible mechanism whereby epitope targeting could affect the antiviral efficiency of CTLs, namely, differential susceptibility to the down-regulation of HLA by Nef.
Prior studies on the impact of Nef on CTL antiviral activity10-12 have used clones that target epitopes in structural proteins, which are expressed later in the viral life cycle than the accessory proteins Tat, Rev, and Nef. HIV-1 proteins are expressed early or late, depending on whether their transcripts contain splice sites from overlapping reading frames. Transcripts that are fully spliced include those coding for Tat, Rev, and Nef; like mature cellular mRNAs whose introns have been spliced, these transcripts readily exit the nucleus for translation in the cytoplasm. However, the other transcripts (coding for Gag/Pol, Env, Vpr, Vpu, and Vif) contain splice sites from overlapping reading frames and require the Rev protein for efficient export from the nucleus, delaying their transport and translation by several hours by comparison.14-16 Thus epitopes from different proteins also vary in their kinetics of presentation,17,18 but the prior studies have not addressed whether this phenomenon affects the ability of Nef to mediate viral resistance to CTLs.
The kinetic relationships of Nef-mediated HLA down-regulation, early protein epitope presentation, and late protein epitope presentation are poorly understood. Prior demonstrations that late protein-specific CTL inhibition of HIV-1 replication is reduced by Nef 10-12 indicate that Nef-mediated down-regulation of HLA must occur before late epitope presentation, but conceivably epitopes from early proteins could be processed and presented at levels adequate for CTL recognition before Nef is expressed at levels sufficient to down-regulate HLA. However, the current study demonstrates that CTL-targeting epitopes in Nef and Rev remain susceptible to Nef-mediated HLA down-regulation, indicating that the down-regulation can occur early enough to interfere with presentation of early protein epitopes. Although the tested Nef-specific CTLs appeared to be affected less by Nef-mediated HLA downregulation than the late protein epitope-specific CTLs, testing of more clones will be needed to determine whether this is generally true.
It is somewhat surprising that CTL-recognizing early protein epitopes remain susceptible to down-regulation of HLA by Nef. Processing and presentation of newly translated proteins is rapid, and recognition of a target cell by a T-cell receptor may require very few peptide-HLA complexes (< 10)44 ; in contrast, it has been shown that HLA down-regulation requires high levels of Nef expression.45 The functional avidities of the Nef- and Rev-specific CTL clones tested were within the same range as the late protein-specific CTL clones (data not shown), suggesting that lower T-cell receptor sensitivity of the early epitope-specific CTLs is unlikely to be an explanation for their susceptibility to impact of Nef.
C-restricted CTLs have potent antiviral activity against HIV-1 and are unaffected by Nef-mediated HLA down-regulation. (A) CTL clones KS7 (Env specific, B*07 restricted) and SE7/126E (Env specific, C*15 restricted) were compared for their abilities to inhibit HIV-1 with wild-type or deleted Nef. The target cells were primary CD4+ T lymphocytes from a seronegative B*07+/C*15+ donor, and the assay was otherwise performed as in Figure 1. The potent level of inhibition of replication seen here for clone SE7/126E (and clone S16-RY11-10.41 in Figure 1) was typical of multiple experiments with HLA-C–restricted CTL clones. (B) Nef effects on the A- and B-restricted CTL epitopes shown in Figure 3 were compared to several C-restricted clones (listed in Table 1) that were tested and summarized in the same manner. The values for the A- and B-restricted CTLs versus the C-restricted CTLs were significantly different (2-tailed t test, P < .001). MEAN A/B indicates the average of results from HLA-A– and -B–restricted epitopes, and MEAN C indicates the average of results from HLA-C–restricted epitopes. Error bars represent the standard deviation for all A/B- or C-restricted CTLs.
C-restricted CTLs have potent antiviral activity against HIV-1 and are unaffected by Nef-mediated HLA down-regulation. (A) CTL clones KS7 (Env specific, B*07 restricted) and SE7/126E (Env specific, C*15 restricted) were compared for their abilities to inhibit HIV-1 with wild-type or deleted Nef. The target cells were primary CD4+ T lymphocytes from a seronegative B*07+/C*15+ donor, and the assay was otherwise performed as in Figure 1. The potent level of inhibition of replication seen here for clone SE7/126E (and clone S16-RY11-10.41 in Figure 1) was typical of multiple experiments with HLA-C–restricted CTL clones. (B) Nef effects on the A- and B-restricted CTL epitopes shown in Figure 3 were compared to several C-restricted clones (listed in Table 1) that were tested and summarized in the same manner. The values for the A- and B-restricted CTLs versus the C-restricted CTLs were significantly different (2-tailed t test, P < .001). MEAN A/B indicates the average of results from HLA-A– and -B–restricted epitopes, and MEAN C indicates the average of results from HLA-C–restricted epitopes. Error bars represent the standard deviation for all A/B- or C-restricted CTLs.
A previously described property of Nef is the selective downregulation of HLA-A and -B, but not -C molecules, which has been suggested as a mechanism to avoid HIV-1–infected cell clearance by HLA-C–inhibited natural killer (NK) cells.19 The implications of this finding for CTL antiviral activity against HIV-1 are poorly understood. Although several HLA-C–restricted HIV-1 CTL epitopes have been identified,46 the role of these CTLs is understudied, including their ability to suppress viral replication. The present study addresses this limitation and demonstrates that HLA-C–restricted CTLs can exert vigorous antiviral activity comparable to HLA-A– and -B–restricted CTLs when challenged with infected cells in vitro. Interestingly, in contrast to HLA-A– and -B–restricted CTLs, HLA-C–restricted CTLs are unaffected by Nef. Given the substantial contribution of Nef to HIV-1 pathogenicity and the likely role of HLA down-regulation in this process, this observation suggests a potentially more prominent role for HLA-C–restricted CTLs in controlling viral replication in vivo.
Thus it would appear that the trade-off of inability to affect HLA-C–restricted CTLs for evasion of HLA-C–inhibited NK cells is favorable for viral replication in vivo. The relative importance of these 2 antiviral mechanisms is unclear. It could be argued that HLA-C–restricted CTLs are ineffective, and thus Nef “sacrifices” down-regulation of C molecules for an advantage against NK cells, for which NK inhibitory receptors commonly recognize HLA-C. However, the observed potency of HLA-C–restricted HIV-1–specific CTLs suggests that they have antiviral capabilities comparable to HLA-A– and -B–restricted CTLs. It may be that the relatively lower occurrence of HLA-C–restricted epitope targeting quantitatively reduces the role of these CTL responses and thus contributes to making this functional trade-off favorable.
This raises the question of why relatively fewer HLA-C–restricted HIV-1 epitope responses are observed compared to HLA-A– and -B–restricted epitopes,47 when Nef reduces expression of epitopes presented by A and B molecules but does not affect those presented by C molecules. At least 2 factors may contribute. Cell-surface HLA-C expression appears to be about 10-fold lower than HLA-A and -B despite equivalent transcription, possibly due to more restricted epitope selectivity or higher binding affinity to TAP and thus requirement for greater amounts of epitope to allow delivery to the cell surface.48,49 Additionally, it has been suggested that up-regulation of HLA-C transcription by cytokines can be less than that for HLA-A and -B.50 Thus the pathway for HLA-C–bound epitope presentation at the cell surface appears to be less efficient. Another reason that HLA-A– and -B–restricted epitope immunogenicity may remain dominant is that Nef appears not to downregulate HLA molecules on dendritic cells,51 which drive the proliferation of CTLs.
Another recent study also suggests epitope dependence in the ability of Nef to interfere with CTL antiviral activity.52 Interestingly, Pol-specific CTL clones restricted by B*51 were relatively resistant to the effects of Nef. The authors hypothesized that this might be due to different levels of epitope presentation because the down-regulation of HLA by Nef is not absolute. These data suggest that the ability of Nef to allow viral evasion of CTLs restricted by HLA-A and -B molecules may vary due to epitope-specific factors such as expression, processing, and transport, which ultimately determine the levels of epitopes reaching the infected cell surface.
Finally, our findings are interesting in light of a recent study from Goulder and colleagues suggesting that HLA-B molecules appear to dominantly determine the efficacy of CTL responses against HIV-1 infection.53 On the surface, it would appear that these data are contradictory. However, Goulder and colleagues did find significant trends for some HLA-C molecules that were attributed to linkage disequilibrium between B and C genetic loci, but which could, in fact, reflect a true impact of HLA-C molecules. Furthermore, the roles of HLA-A, -B, and -C molecules were inferred by statistical associations to viremia levels and HIV-1 sequence polymorphisms; the much lower frequency of HAL-C–restricted CTLs in vivo would reduce the impact of C molecules and therefore reduce the statistical power to detect their effects compared to B molecules.
In summary, CTLs targeting different epitopes vary in their susceptibility to the down-regulation of HLA by Nef. This activity of Nef affects even CTL-recognizing early protein epitopes (including those derived from Nef itself), although it remains unclear whether these may be less affected than CTL-recognizing late protein epitopes. Finally, the inability of Nef to down-regulate HLA-C molecules results in a lack of impact on C-restricted CTLs. This observation raises questions regarding a differential role for HLA-C– versus HAL-A– and -B–restricted CTLs in HIV-1 pathogenesis and vaccine development, suggesting that C-restricted CTL responses may be substantially more difficult to generate but have greater antiviral efficacy in vivo in the setting of functional Nef.
Prepublished online as Blood First Edition Paper, August 1, 2006; DOI 10.1182/blood-2006-06-030668.
Supported by Public Health Service grants AI051970 (O.O.Y.) and AI067077 (C.B.).
The authors declare no competing financial interests.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal