Abstract
Recently, a gain-of-function MPL mutation, MPLW515L, was described in patients with JAK2V617F-negative myelofibrosis with myeloid metaplasia (MMM). To gain more information on mutational frequency, disease specificity, and clinical correlates, genomic DNA from 1182 patients with myeloproliferative and other myeloid disorders and 64 healthy controls was screened for MPL515 mutations, regardless of JAK2V617F mutational status: 290 with MMM, 242 with polycythemia vera, 318 with essential thrombocythemia (ET), 88 with myelodysplastic syndrome, 118 with chronic myelomonocytic leukemia, and 126 with acute myeloid leukemia (AML). MPL515 mutations, either MPLW515L (n = 17) or a previously undescribed MPLW515K (n = 5), were detected in 20 patients. The diagnosis of patients with mutant MPL alleles at the time of molecular testing was de novo MMM in 12 patients, ET in 4, post-ET MMM in 1, and MMM in blast crisis in 3. Six patients carried the MPLW515L and JAK2V617F alleles concurrently. We conclude that MPLW515L or MPLW515K mutations are present in patients with MMM or ET at a frequency of approximately 5% and 1%, respectively, but are not observed in patients with polycythemia vera (PV) or other myeloid disorders. Furthermore, MPL mutations may occur concurrently with the JAK2V617F mutation, suggesting that these alleles may have functional complementation in myeloproliferative disease.
Introduction
The identification of JAK2V617F in virtually all patients with polycythemia vera (PV) and in 50% to 75% of those with either essential thrombocythemia (ET) or de novo myelofibrosis with myeloid metaplasia (MMM) has shed new light on the molecular pathogenesis of these disorders but has also raised important questions in this regard. JAK2V617F is a somatic mutation that constitutively activates the JAK2 tyrosine kinase,1-3 which normally plays a critical, nonredundant role in mediating signal transduction downstream of several cytokine receptors and is indispensable for definitive erythropoiesis and normal myeloid lineage differentiation.4,5 JAK2V617F confers cytokine hypersensitivity1-3 and has been demonstrated in erythropoietin-independent erythroid colonies (EECs) derived from PV and ET patients.6 Furthermore, JAK2V617F expression in murine bone marrow transplant models results in a type of myeloproliferative disease (MPD) that resembles PV, albeit with important strain-related phenotypic differences.1,7
Several lines of evidence, however, have supported the existence of mutations other than JAK2V617F in MPD. For example, studies of familial MPD, including familial PV, indicate that JAK2V617F may be acquired secondarily, possibly as a “patterning” mutation, in a manner that may be independent of disease duration.8-11 Furthermore, the phenotypic pleiotropy associated with the acquisition of JAK2V617F and the variable burden of the mutant allele in MPD raise the question of whether JAK2V617F is sufficient for MPD pathogenesis. In addition, recent studies using X-chromosome inactivation pattern analysis demonstrated that most patients with JAK2V617F-negative ET and MMM exhibit clonal hematopoiesis, thus pointing to the presence of an alternative disease-promoting mutation.12-14 Furthermore, quantitation of the JAK2V617F allele in one of these studies indicated that in some MPD patients, only a fraction of clonally derived granulocytes harbored the mutant JAK2 allele, suggesting not only that JAK2V617F was acquired secondarily but that some MPD patients acquire more than one mutation during disease progression.
This possibility has been borne out by the recent identification of the MPLW515L mutation in 4 of 45 JAK2V617F-negative MMM patients.15 As is JAK2V617F, MPLW515L is an acquired mutation that induces constitutive, cytokine-independent activation of the JAK-STAT pathway. Furthermore, the expression of MPLW515L in murine bone marrow resulted in an MPD phenotype that recapitulated certain clinical and histopathologic features of MMM and ET. Several questions, however, remain unanswered: Are MPLW515L and JAK2V617F mutually exclusive mutations? What is the prevalence and disease distribution (phenotype) of MPLW515 mutations? Are MPL515 mutations clinically important? These questions are addressed in the current study through a bi-institutional study of a large cohort of patients with a spectrum of myeloid disorders. In future studies, identification of affected patients in a prospective fashion should enable additional studies that examine clonal distribution, gene expression profiling, clonal evolution over time, and drug sensitivity of primary cells to JAK2 inhibitors.
Patients, materials, and methods
Sample collection and processing
The current study was approved by the institutional review boards of both the Mayo Clinic and the Dana Farber Cancer Institute. All patients provided informed written consent for study sample collection and permission for research use. Peripheral blood (PB) and bone marrow (BM) study samples were accrued from patients seen in the Mayo Clinic MPD practice or from participants in the Harvard Myeloproliferative Disorders Study.2 DNA was prepared from granulocyte PB mononuclear cells (PBMNCs) or archived BM cell pellets, as previously described.16 In general, neutrophil DNA was used in all patients with MMM and PV and in approximately half of those with ET. BM cells were used in the remainder of the ET patients and for those with AML, MDS, and CMML. Diagnoses for all specific disease categories were determined according to criteria set by the World Health Organization-sponsored committee for the classification of hematologic malignancies,17 and molecular testing was performed on tissue samples collected from patients at variable times after the initial diagnosis.
Genotyping of MPL515 allele
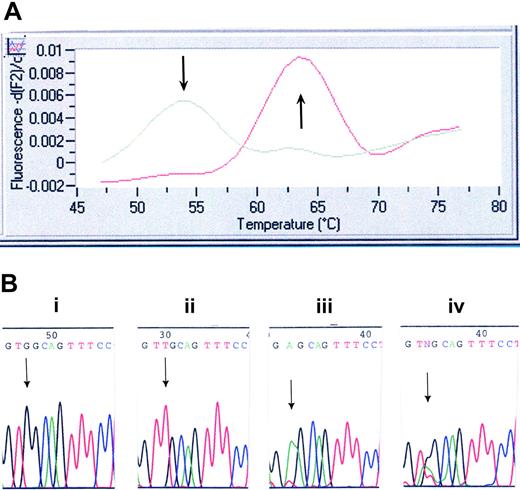
Genotyping of the initial 403 Mayo specimens was performed with the use of a LightCycler (Roche Applied Bioscience, Indianapolis, IN) assay we developed and by direct DNA sequencing. Once the LightCycler (Roche Applied Bioscience) assay was validated, only specimens (of the subsequent 396 Mayo Clinic specimens, including those of 64 healthy controls) flagged as positive were sequenced to confirm the presence of the mutation. All 447 Harvard samples were screened by DNA sequencing alone. Primers and probes were designed and selected with the use of LightCycler Probe Design software 2.0 (Roche Applied Bioscience). Each 20-μL reaction mix contained 4 μL LC FastStart DNA MasterPLUS HybProbe master mix, 150 ng DNA template, 0.2 μM sensor probe (5′-3′) (CTgCCACCTCAgCAgCALCRed640) and 0.2 μM anchor probe (5′-3′) (fluorescein-AggCCCAggACggCg-P), 1 μM forward primer (5′-TgggCCgAAgTCTgACCCTTT-3′), and 0.5 μM reverse primer (5′-ACAgAgCgAACCAAgAATgCCTgT-3′). The following PCR protocol was used: denaturation at 95°C for 10 minutes and amplification (95°C for 10 seconds, 57°C for 10 seconds, and 72°C for 15 seconds) for 40 cycles. Melting-curve analysis was performed as follows: melt product at 95°C, hold at 45°C for 30 seconds, then ramp to 85°C at a slope of 0.2°/sec. The sensor probe was designed to be a perfect match for the wild-type MPL sequence. This probe dissociates at 54°C when bound to mismatched sequence (W515L) and at 64°C when bound to the perfectly matched wild-type sequence (Figure 1A). PCR products were purified (QIAGEN PCR Purification Kit, Valencia, CA) and subjected to bidirectional sequence analysis on the ABI PRISM 3730 DNA Analyzer (Applied Biosystems) using the following primers: forward, 5′-ggTgACCgCTCTgCA TCTAgTgCT-3′; reverse, 5′-CACCTggTCCACCgCCAgTCT-3′.
Sensitivity of MPL515 LightCycler assay
DNA from a healthy control (homozygous for MPL515-WT) and from a patient homozygous for MPLW515L was mixed in various proportions to estimate assay sensitivity for mutant allele detection. Serial dilutions showed the assay sensitivity to be 3% to 5% (ie, mutant allele burden greater than 3%-5% was reproducibly detected; data not shown).
Genotyping of JAK2V617F allele
Genotyping was performed by direct DNA sequencing (Mayo Clinic specimens), as previously described.18 JAK2V617F mutational status was confirmed in all MPL515 mutation-positive samples with a previously described allele-specific PCR sensitive to 0.01% to 0.1%.18,19 Genotyping of the Harvard specimens for JAK2V67F was performed by mass spectrometry and quantitative real-time PCR assays, as previously described.2,12
Mutation screening for MPLW515L/K. (A) LightCycler assay for MPLW515L. Melting curve analysis displays that probe bound to a mismatch dissociates at 54°C, whereas probe bound across the wild-type sequence dissociates at 64°C. (B) DNA chromatogram illustrating wild-type MPL515 sequence (i), the TGG→TTG conversion in MPLW515L (ii), the TGG→AAG conversion in MPLW515K (iii), and the presence of all 3 alleles (ie, wild-type MPL, MPLW515L, and MPLW515K) in 2 patients (iv).
Mutation screening for MPLW515L/K. (A) LightCycler assay for MPLW515L. Melting curve analysis displays that probe bound to a mismatch dissociates at 54°C, whereas probe bound across the wild-type sequence dissociates at 64°C. (B) DNA chromatogram illustrating wild-type MPL515 sequence (i), the TGG→TTG conversion in MPLW515L (ii), the TGG→AAG conversion in MPLW515K (iii), and the presence of all 3 alleles (ie, wild-type MPL, MPLW515L, and MPLW515K) in 2 patients (iv).
Results
Samples from 1182 patients and 64 healthy controls were screened for the presence of MPL515 mutations; 735 samples were screened at the Mayo Clinic, and 447 samples were screened at Harvard (Table 1). Overall, 20 patients were found to carry MPL515 mutations (Table 2). Of these, 4 MMM patients, all from the Harvard cohort carrying MPLW515L, have been previously reported.15 Seventeen patients carried the previously reported MPLW515L mutation (Figure 1A-B), and 5 patients exhibited a previously undescribed mutation involving the same codon (MPLW515K) (Figure 1B; Table 2). Surprisingly, DNA sequencing showed all 3 alleles (wild-type MPL, MPLW515L, MPLW515K) present in 2 patients (patients 8 and 9 in Table 2; both de novo MMM), pointing to a mixed clonal state (Figure 1B). Study of 2 consecutively obtained archived BM specimens (patient 8) showed the presence of both MPL mutations at low levels (5%-10% of wild-type allele) in the initial specimen. The burden of both MPL mutant alleles was increased (40% of wild-type allele) in the subsequent specimen (collected 6 months later; data not shown). Although these data support the coexistence of MPL515L and MPL515K alleles, the possibility of sample contamination cannot be definitively excluded; hence, we consider this observation to be preliminary and subject to confirmation in future studies. Six of the 20 (30%) patients carried both MPLW515L and JAK2V617F mutations—2 ET, 3 de novo MMM, and 1 post-ET MMM. MPL515 mutations were restricted to patients with a history of either MMM or ET; specific diagnosis at the time of molecular testing was de novo MMM in 12 patients, ET in 4 patients, post-ET MMM in 1 patient, and MMM in blast crisis in 3 patients (Table 2). Clinical and laboratory features of the 20 patients affected by MPL mutations (11 males; median age, 58.5 years; median follow-up, 67 months) are presented in Table 2.
Diagnosis and JAK2V617F mutational status of 1182 patients with a spectrum of myeloid disorders
. | . | Mayo patients . | . | . | Harvard patients . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
. | N . | No. of patients . | JAK2V617F positive, no. (%) . | MPL515L or -515K positive, no. (%) . | No. of patients . | JAK2V617F positive, no. (%) . | MPL515L or -515K positive, no. (%) . | ||||
| All patients | 1182 | 735 | 252 | 13 | 447 | 335 | 7 | ||||
| MMM | 290 | 198 | 106 (54) | 8 (4) | 92 | 46 (50) | 5 (5) | ||||
| De novo | NA | 159 | 75 | 8 | NA | NA | 4 | ||||
| After ET | NA | 10 | 8 | 0 | NA | NA | 1 | ||||
| After PV | NA | 29 | 23 | 0 | NA | NA | n/a | ||||
| ET | 318 | 167 | 76 (46) | 2 (1) | 151 | 106 (70) | 2 (1) | ||||
| PV | 242 | 38 | 38 (100) | 0 | 204 | 183 (90) | 0 | ||||
| AML | 126 | 126 | 29 (23) | 3 (2) | 0 | n/a | n/a | ||||
| With antecedent MPD | NA | 37 | 23 | 3 | 0 | n/a | n/a | ||||
| MDS | 88 | 88 | 0 | 0 | 0 | n/a | n/a | ||||
| CMML | 118 | 118 | 3 (3) | 0 | 0 | n/a | n/a | ||||
. | . | Mayo patients . | . | . | Harvard patients . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
. | N . | No. of patients . | JAK2V617F positive, no. (%) . | MPL515L or -515K positive, no. (%) . | No. of patients . | JAK2V617F positive, no. (%) . | MPL515L or -515K positive, no. (%) . | ||||
| All patients | 1182 | 735 | 252 | 13 | 447 | 335 | 7 | ||||
| MMM | 290 | 198 | 106 (54) | 8 (4) | 92 | 46 (50) | 5 (5) | ||||
| De novo | NA | 159 | 75 | 8 | NA | NA | 4 | ||||
| After ET | NA | 10 | 8 | 0 | NA | NA | 1 | ||||
| After PV | NA | 29 | 23 | 0 | NA | NA | n/a | ||||
| ET | 318 | 167 | 76 (46) | 2 (1) | 151 | 106 (70) | 2 (1) | ||||
| PV | 242 | 38 | 38 (100) | 0 | 204 | 183 (90) | 0 | ||||
| AML | 126 | 126 | 29 (23) | 3 (2) | 0 | n/a | n/a | ||||
| With antecedent MPD | NA | 37 | 23 | 3 | 0 | n/a | n/a | ||||
| MDS | 88 | 88 | 0 | 0 | 0 | n/a | n/a | ||||
| CMML | 118 | 118 | 3 (3) | 0 | 0 | n/a | n/a | ||||
MMM indicates myelofibrosis with myeloid metaplasia; ET, essential thrombocythemia; PV, polycythemia vera; AML, acute myeloid leukemia; MPD, myeloproliferative disorder; MDS, myelodysplastic syndrome; CMML, chronic myelomonocytic leukemia; NA, accurate subclassification not available because of study design; and n/a, not applicable
Clinical, laboratory and molecular characteristics of patients with MPL515 mutations (at the time of MPL515 testing)
Patient no. . | Sex/age, y, at diagnosis . | Diagnosis . | JAK2V617F status . | MPL515 status . | MPL515 mutant peak/wild-type peak ratio, % . | Cytogenetics . | Hgb, g/dL . | WBC count, × 109/L . | Platelet count, × 109/L . | AML . | Status at last follow-up . | Duration of follow-up, mo . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mayo study | ||||||||||||
| 1 | M/32 | De novo MMM | WT | W515L | < 50 | Normal | 11.4 | 5.5 | 436 | No | Alive | 40 |
| 2 | M/46 | De novo MMM | Heterozygous | W515L | < 50 | del(13q) | 9.5 | 5.3 | 379 | No | Deceased | 91 |
| 3 | M/64 | De novo MMM | WT | W515L | < 50 | del(20q) | 9.9 | 5.7 | 182 | No | Alive | 94 |
| 4 | M/60 | De novo MMM | WT | W515K | < 50 | Normal | 8.3 | 4.5 | 215 | No | Alive | 30 |
| 5 | M/61 | De novo MMM | WT | W515L | < 50 | ND | 11.5 | 36.6 | 139 | No | Alive | 22 |
| 6 | M/75 | Post-de novo MMM AML (M0) | WT | W515L | > 50 | t(1;7) t(1;21)* | 9.0 | 72.1 | 161 | n/a | Deceased | 34 |
| 7 | F/52 | Post-de novo MMM AML (M2) | WT | W515K | 100 | Normal | 9.0 | 10.6 | 430 | n/a | Alive | 67 |
| 8 | M/49 | De novo MMM | Heterozygous | W515L + W515K | < 50 | Normal | 8.2 | 4.6 | 40 | No | Alive | 23 |
| 9 | F/53 | De novo MMM | Heterozygous | W515L + W515K | < 50 | Complex abnormalities | 6.3 | 8.5 | 124 | Yes | Deceased | 20 |
| 10 | M/58 | De novo MMM | WT | W515K | 100 | del(11q)* | 11.5 | 10.3 | 146 | No | Alive | 108 |
| 11 | M/71 | ET | WT | W515L | < 50 | Normal | 14.0 | 6.9 | 959 | No | Alive | 168 |
| 12 | F/67 | ET | WT | W515L | > 50 | t(1;21) | 9.1 | 4.1 | 1539 | Yes | Deceased | 172 |
| 13 | M/81 | Post-ET MMM AML (M2) | WT | W515L | 100 | t(8;21) | 9.6 | 47.4 | 161 | n/a | Deceased | 72 |
| Harvard study | ||||||||||||
| 14 | F/59 | De novo MMM | WT | W515L | NA | Normal | NA | NA | NA | No | Alive | NA |
| 15 | F/5 | De novo MMM | WT | W515L | NA | Normal | 9.4 | 7.5 | 261 | No | Alive | NA |
| 16 | M/58 | De novo MMM | WT | W515L | NA | del(16q)* | 10.8 | 7.2 | 120 | Yes | Deceased | 36 |
| 17 | F/68 | De novo MMM | WT | W515L | NA | Normal | 10.1 | 17.7 | 559 | No | Alive | 96 |
| 18 | F/45 | ET | Heterozygous | W515L | NA | Normal | NA | NA | NA | No | Alive | 24 |
| 19 | F/56 | ET | Heterozygous | W515L | NA | Normal | NA | NA | 658 | No | Alive | 168 |
| 20 | F/67 | Post-ET MMM | Heterozygous | W515L | NA | Normal | NA | NA | 650 | No | Alive | NA |
Patient no. . | Sex/age, y, at diagnosis . | Diagnosis . | JAK2V617F status . | MPL515 status . | MPL515 mutant peak/wild-type peak ratio, % . | Cytogenetics . | Hgb, g/dL . | WBC count, × 109/L . | Platelet count, × 109/L . | AML . | Status at last follow-up . | Duration of follow-up, mo . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mayo study | ||||||||||||
| 1 | M/32 | De novo MMM | WT | W515L | < 50 | Normal | 11.4 | 5.5 | 436 | No | Alive | 40 |
| 2 | M/46 | De novo MMM | Heterozygous | W515L | < 50 | del(13q) | 9.5 | 5.3 | 379 | No | Deceased | 91 |
| 3 | M/64 | De novo MMM | WT | W515L | < 50 | del(20q) | 9.9 | 5.7 | 182 | No | Alive | 94 |
| 4 | M/60 | De novo MMM | WT | W515K | < 50 | Normal | 8.3 | 4.5 | 215 | No | Alive | 30 |
| 5 | M/61 | De novo MMM | WT | W515L | < 50 | ND | 11.5 | 36.6 | 139 | No | Alive | 22 |
| 6 | M/75 | Post-de novo MMM AML (M0) | WT | W515L | > 50 | t(1;7) t(1;21)* | 9.0 | 72.1 | 161 | n/a | Deceased | 34 |
| 7 | F/52 | Post-de novo MMM AML (M2) | WT | W515K | 100 | Normal | 9.0 | 10.6 | 430 | n/a | Alive | 67 |
| 8 | M/49 | De novo MMM | Heterozygous | W515L + W515K | < 50 | Normal | 8.2 | 4.6 | 40 | No | Alive | 23 |
| 9 | F/53 | De novo MMM | Heterozygous | W515L + W515K | < 50 | Complex abnormalities | 6.3 | 8.5 | 124 | Yes | Deceased | 20 |
| 10 | M/58 | De novo MMM | WT | W515K | 100 | del(11q)* | 11.5 | 10.3 | 146 | No | Alive | 108 |
| 11 | M/71 | ET | WT | W515L | < 50 | Normal | 14.0 | 6.9 | 959 | No | Alive | 168 |
| 12 | F/67 | ET | WT | W515L | > 50 | t(1;21) | 9.1 | 4.1 | 1539 | Yes | Deceased | 172 |
| 13 | M/81 | Post-ET MMM AML (M2) | WT | W515L | 100 | t(8;21) | 9.6 | 47.4 | 161 | n/a | Deceased | 72 |
| Harvard study | ||||||||||||
| 14 | F/59 | De novo MMM | WT | W515L | NA | Normal | NA | NA | NA | No | Alive | NA |
| 15 | F/5 | De novo MMM | WT | W515L | NA | Normal | 9.4 | 7.5 | 261 | No | Alive | NA |
| 16 | M/58 | De novo MMM | WT | W515L | NA | del(16q)* | 10.8 | 7.2 | 120 | Yes | Deceased | 36 |
| 17 | F/68 | De novo MMM | WT | W515L | NA | Normal | 10.1 | 17.7 | 559 | No | Alive | 96 |
| 18 | F/45 | ET | Heterozygous | W515L | NA | Normal | NA | NA | NA | No | Alive | 24 |
| 19 | F/56 | ET | Heterozygous | W515L | NA | Normal | NA | NA | 658 | No | Alive | 168 |
| 20 | F/67 | Post-ET MMM | Heterozygous | W515L | NA | Normal | NA | NA | 650 | No | Alive | NA |
MPL515 mutant peak/wild-type peak ratio was the ratio on sequencing chromatogram × 100. MMM indicates myelofibrosis with myeloid metaplasia; ET, essential thrombocythemia; AML, acute myeloid leukemia; WT, wild type; Hgb, hemoglobin; n/a, not applicable; NA, not available; and ND, not done
Cytogenetic abnormalities developed after identification of MPL515 mutations
Among the 13 Mayo Clinic patients with MPL mutations, 11 had a history of MMM, 2 had a history of ET, and none had a history of familial MPD (Table 2). Among the former, 3 were in blast crisis at the time of molecular testing (2 AML-M2, 1 AML-M0) and one transformed to unclassified AML within 1 year of testing. Another patient had advanced, treatment-refractory disease with severe bone pain, progressed to accelerated-phase disease with 15% circulating blasts, and died within 2 years of the study time point. Two patients were known to have had chronic thrombocytosis before the diagnosis of de novo MMM, and each has had a stable postdiagnosis clinical course for 3 and 5 years, respectively. Similarly, 2 other patients with intermediate risk disease20 showed slow progression after 2 and 4 years from the time of molecular testing. The last 2 patients with MMM presented with high-risk disease and have been followed up for less than 2 years from the time of testing. Of the 2 Mayo Clinic patients with MPL mutations and ET, one has had a 14-year disease duration with stable clinical course, while the other patient died of AML after 14 years of disease and after having received treatment with hydroxyurea, busulfan, radiophosphorus, and anagrelide. Review of bone marrow histology and cytogenetic data did not disclose unexpected findings in patients with MPL mutations. Because of the method of patient sample accrual (see “Patients, materials, and methods”), clinical details for the 7 patients from the Harvard study were limited; 5 carried the diagnosis of MMM (4 de novo MMM, 1 post-ET MMM) and 2 had ET (Table 2). Thrombohemorrhagic complications were not a prominent feature of the disease in any of the aforementioned patients from the Mayo Clinic or Harvard.
Discussion
The initial identification of a gain-of-function MPL mutation (MPLW515L) provided proof-of-principle for a novel mechanism of constitutive JAK-STAT signaling in acquired MPD lacking the JAK2V617F mutation.15 In a substantially expanded analysis of 1182 patients with a spectrum of acute and chronic myeloid disorders, we found 20 patients carrying MPL515 mutations, indicating that this is a relatively infrequent event. A significant proportion (25%) of the 20 patients carried a previously undescribed mutation, MPLW515K. Although further analysis of the MPLW515K allele is necessary, it seems likely that it is a gain-of-function mutation and points to the MPL codon 515 as a hot spot for activating mutations in acquired MPD. These findings are also consistent with the observation that the deletion of the amphipathic KWQFP motif in the transmembrane-cytoplasmic hinge region of murine or human MPL (underlined residue corresponds to W515) constitutively activates downstream signaling in a ligand-independent fashion.21 The previous identification of a highly penetrant gain-of-function MPL mutation in the transmembrane (TM) domain (MPLS505N) in a Japanese pedigree with familial ET22 and the spontaneously occurring MPLW508S mutation in murine MPL (corresponds to W515S in human MPL)23 further support the critical role of the TM and membrane-distal domains in regulating MPL function.
Although MPLW515 mutations are infrequent in MPD, valuable insights can be gleaned about the relationship between mutant alleles in the JAK-STAT pathway and phenotypic manifestations of disease. First, we observed MPLW515 mutations only in patients with ET or MMM, never in patients with PV. This suggests that MPLW515 mutations may favor megakaryocytic lineage fate determination as opposed to erythroid fates. In contrast, the almost invariant presence of the JAK2V617F allele in patients with PV and the high frequency of PV patients with duplication of the JAK2V617F allele compared with patients with ET or MMM suggest that the JAK2V617F allele favors erythroid fate determination in hematopoietic progenitors. The hypothesis that MPL mutation favors megakaryocytic fate but that JAK2V617F favors erythroid fate is supported by observations in murine models of disease, in which overexpression of JAK2V617F results in a PV-like phenotype without thrombocytosis,1,15 whereas the expression of MPLW515L in a murine model results in a phenotype characterized by marked thrombocytosis.15 The JAK2V617F allele is present in the hematopoietic stem cell compartment in humans,24 and this observation supports the notion that the mutant allele must have some instructive role in the phenotypic manifestation of the diseases. Perhaps of most interest, 6 patients had both the MPLW515L and the JAK2V617F mutations. In all cases, the MPL515 mutant allele was present in excess of the JAK2V617F allele. Although the former was detectable by a low-sensitivity screening method (DNA sequencing) (MPL 515L or 515K/MPL 515W peak ratio is greater than 50% in 5 of 13 Mayo Clinic patients) (Table 2), the JAK2V617F allele was detectable only by the highly sensitive allele-specific PCR assay. From a pathogenetic standpoint, our current experimental approach does not distinguish between the 2 possibilities regarding whether MPL and JAK2 mutations arise in independent clones or whether the JAK2V617F-bearing cell emerges as a subclone of the MPL mutation-carrying progenitor cell. Patients with both MPLW515L and JAK2V617F had a diagnosis of ET or MMM, suggesting a potent effect of MPLW515L in supporting megakaryocytic hyperplasia and thrombocytosis, but it will be of interest to determine whether this subset of ET patients has a higher average hemoglobin level that those with MPLW515L alone. Further studies to characterize the mutations in prospectively purified hematopoietic progenitors and murine modeling will be necessary to further clarify this issue.
Prepublished online as Blood First Edition Paper, July 25, 2006; DOI 10.1182/blood-2006-04-018879.
Supported in part by grants from the Myeloproliferative Disorders Foundation (A.D.P. and A.T.). Y.P. is a Research Training Fellow of the Howard Hughes Medical Institute. D.G.G. is an Investigator of the Howard Hughes Medical Institute.
The authors declare no competing financial interests.
A.D.P., D.G.G., and A.T. wrote the paper; A.D.P., R.L.L., T.L., Y.P., D.G.G., and A.T. conceived and designed the study, analyzed the data, and performed the research; A.D.P., R.L.L., R.A.M., M.W., D.P.S., M.A.E., A.P.W., W.J.H., M.R.L., D.G.G., and A.T. contributed vital reagents and collected data; and R.F.M. participated in molecular studies and reviewed pathology data.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal