Abstract
Platelets play critical roles in diverse hemostatic and pathologic disorders and are broadly implicated in various biological processes that include inflammation, wound healing, and thrombosis. Recent progress in high-throughput mRNA and protein profiling techniques has advanced our understanding of the biological functions of platelets. Platelet proteomics has been adopted to decode the complex processes that underlie platelet function by identifying novel platelet-expressed proteins, dissecting mechanisms of signal or metabolic pathways, and analyzing functional changes of the platelet proteome in normal and pathologic states. The integration of transcriptomics and proteomics, coupled with progress in bioinformatics, provides novel tools for dissecting platelet biology. In this review, we focus on current advances in platelet proteomic studies, with emphasis on the importance of parallel transcriptomic studies to optimally dissect platelet function. Applications of these global profiling approaches to investigate platelet genetic diseases and platelet-related disorders are also addressed.
Introduction
Human blood platelets play important roles in fundamental biological processes, including thrombosis, inflammation, wound repair, and stroke. Although they are anucleate and lack nuclear DNA, platelets retain small amounts of megakaryocyte-derived mRNA.1,2 Platelets also contain rough endoplasmic reticulum and polyribosomes, thus retaining the capacity for protein biosynthesis from cytoplasmic mRNA.3 Quiescent platelets display minimal translational activity, although platelet activation leads to the rapid translation of preexisting mRNA,4 with the release or derivation of platelet-secreted proteins, cytokines, exosomes, and microparticles.
The traditional paradigm that platelet mRNA content is invariant and gradually declines with cell senescence was challenged when signal-dependent pre-mRNA splicing was identified in platelets.5 Signal-dependent splicing provides a mechanism for altering the repertoire of translatable messages in response to cellular activation/stimulation. Furthermore, platelets have essential components of a functional spliceosome and selected unspliced pre-mRNAs. These spliceosomes retain a unique ability to splice pre-mRNA in the cytoplasm (as opposed to the typical nuclear location), a capability not described in any other mammalian cell. This discovery emphasizes that the molecular mechanisms of platelet function cannot be optimally dissected without accurate platelet transcript profiling.
Modern postgenomic, high-throughput approaches allow integrated studies of molecular components (at the RNA and the protein levels) involved in cell function. Platelets represent an attractive, simplified model for these studies because they lack nuclear DNA and because their genome consists of a small subset of megakaryocyte-derived mRNA transcripts. This complete pool of platelet RNAs is significantly smaller than the transcriptome of a nucleated cell.6 The entire pool of platelet proteins constitutes the platelet proteome: the initially static, but functionally dynamic, protein interactions that occur with platelet activation. In this review, we focus on recent applications of proteomic and transcriptomic technologies to dissect platelet function in normal processes and pathologic disorders.
Platelet transcript profiling: transcriptomics
Modern approaches to transcript profiling
The development of global transcript profiling technologies, such as microarray and serial analysis of gene expression (SAGE),7,8 provided novel methodologies for dissecting platelet function. Microarray analysis adapts artificially constructed grids of known DNA samples such that each element of the grid probes for a specific RNA sequence; these are then used to capture and quantify RNA transcripts.9 Microarray platforms developed to date represent closed transcript profiling systems—that is, they detect only those transcripts that correspond to specific probes imprinted on the chip. Transcripts without corresponding probes are not detected. Recent technological advancements allow accurate whole genome transcript profiling and are capable of detecting alternatively spliced transcripts.9
SAGE represents an open transcript profiling system that can detect any transcript within the SAGE library. Classical SAGE10 relies on the observation that short (less than 10 bp) sequences (tags) within 3′-mRNAs can stringently discriminate among the genes that constitute the human genome. Differentially expressed genes can be identified in a quantitative manner because the frequency of tag detection reflects the steady state mRNA level of the cellular transcriptome.10,11 Genes expressed at low levels (less than 0.01% of total mRNA) can be identified by SAGE. Modified SAGE protocols have been devised to provide more definitive identification by using longer tags, identify low-abundant transcripts efficiently through subtractive SAGE techniques, and amplify small amounts of mRNA starting material.12-14
Analyzing the platelet transcriptome
Initial characterization of platelet-derived mRNA transcripts was achieved by constructing platelet-specific cDNA libraries15 and through single-gene polymerase chain reaction (PCR)2 technology. To date, a limited number of microarray experiments using platelet-derived mRNAs have been published; these studies generally agree on platelet transcript quantitation and gene expression patterns.6,16,17 Furthermore, it is clear that this approach provides an efficient means to identify novel genes and proteins functionally expressed in human platelets.6 Not surprisingly, platelets retain fewer transcripts—ranging from approximately 1600 to 3000 mRNAs—than those found in nucleated cells6,17,18 (Table 1). This limited number of platelet-expressed transcripts presumably represents the lack of ongoing transcription in the anucleate platelet.
Platelet microarray studies
Reference . | No. genes studied . | No. present/no. marginal . | No. present . | No. arrays . | Platelet source . |
|---|---|---|---|---|---|
| Gnatenko et al6 | 12 599 | 1500/2147 | NR | 3 | Apheresis |
| Sauer et al101 | 22 200 | ∼1668/3562* | NR | 11* | Apheresis |
| Bugert et al18 | 9 850 | NR | ∼1526 | 6 | Concentrates |
| McRedmond et al17 | 12 599 | NR | 2928 | 1 (in duplicate) from 23 pooled donors | Blood (50 mL) |
Reference . | No. genes studied . | No. present/no. marginal . | No. present . | No. arrays . | Platelet source . |
|---|---|---|---|---|---|
| Gnatenko et al6 | 12 599 | 1500/2147 | NR | 3 | Apheresis |
| Sauer et al101 | 22 200 | ∼1668/3562* | NR | 11* | Apheresis |
| Bugert et al18 | 9 850 | NR | ∼1526 | 6 | Concentrates |
| McRedmond et al17 | 12 599 | NR | 2928 | 1 (in duplicate) from 23 pooled donors | Blood (50 mL) |
Numbers of genes studied were the numbers of probes (genes) represented on individual microarray slides.
NR indicates not reported.
Includes normal and essential thrombocythemic platelets.
Given that most microarray protocols are semiquantitative, microarray findings must be validated by other techniques such as quantitative polymerase chain reaction (Q-PCR). The combination of 2 complementary transcript profiling techniques, microarray and SAGE, demonstrated that 50% to 89% of platelet RNA tags are mitochondrial transcripts presumably related to persistent mitochondrial transcription in the absence of nuclear-derived transcripts.19 The overrepresentation of mitochondrial transcripts in platelets is considerably greater than that of its closest cell type (skeletal muscle), of which 20% to 25% are mitochondrial SAGE tags.20 Thus, although SAGE clearly has advantages in cellular transcript profiling, its applicability in platelet diseases appears limited. Nonetheless, the relative enrichment of mitochondria-derived transcripts does not interfere with platelet microarray studies.
The feasibility of analyzing platelet transcripts from a single platelet donor with as little as 50 ng total platelet RNA, or approximately 40 mL whole blood, has been demonstrated.16 Reliable mRNA amplification was validated by Q-PCR and by parallel hybridization of amplified and nonamplified RNA samples. In this study, gene profiling results were reproducible for 9815 of 9850 represented genes, providing initial proof that this approach can be applied in platelet-related human diagnostic studies starting from small sample volumes. Recently, microarray analysis was used to identify genes that are differentially expressed in several platelet-related diseases (see “Global profiling to study platelet-associated disorders,” below). These studies clearly establish the feasibility of platelet transcript profiling in identifying differentially expressed genes, characterizing novel platelet-expressed genes, and elucidating the molecular signature of a disease with potential application for platelet diagnostics.
MicroRNA profiling of platelets
MicroRNAs (miRNAs) are a highly conserved class of short, noncoding RNAs that regulate gene expression during cell differentiation, proliferation, and apoptosis. To dissect regulatory pathways that control megakaryocytic differentiation, miRNA expression profiling was performed on in vitro–differentiated megakaryocytes derived from CD34+ hematopoietic progenitors.21 Several miRNAs were identified, with the subsequent demonstration that miR-130a targets MAFB, a transcription factor that is up-regulated during megakaryocytic differentiation and induces the glycoprotein IIb (GPIIB) gene ITGA2B. Moreover, the up-regulation of miR-101, miR-126, miR-99a, miR-135, and miR-20 was documented in megakaryoblastic leukemic cell lines compared with in vitro–differentiated megakaryocytes and CD34+ progenitors. These data suggest an important regulatory role of miRNAs during megakaryocytopoiesis; however, a role for miRNAs in thrombopoiesis remains unestablished.
Limitations and perspectives of transcript profiling
Major limitations of modern transcript profiling approaches include reliable and reproducible detection of low-abundant transcripts, feasibility of truly quantitative transcript profiling, bulky and complex data processing, and (in the case of platelets) limited amounts of RNA. Furthermore, accurate platelet transcript profiling requires stringent attention to purification methodologies because a single nucleated cell (ie, leukocyte) contains considerably more mRNA than a platelet.22 It has become evident that the platelet transcriptome is complex and dynamically controlled at different levels, including regulation by miRNAs,21 signal-dependent pre-mRNA splicing,5 and translational control pathways such as mammalian target of rapamycin (mTOR).4 Despite significant progress in microarray chip design, accurate transcript profiling still requires validation by Q-PCR or other techniques. Efficient mRNA amplification,16 development of more sensitive whole genome microarrays (which detect alternatively spliced transcripts),9 and enhancements to bioinformatics software should obviate some of these restrictions.
Platelet protein analyses: proteomics
Modern proteomic techniques
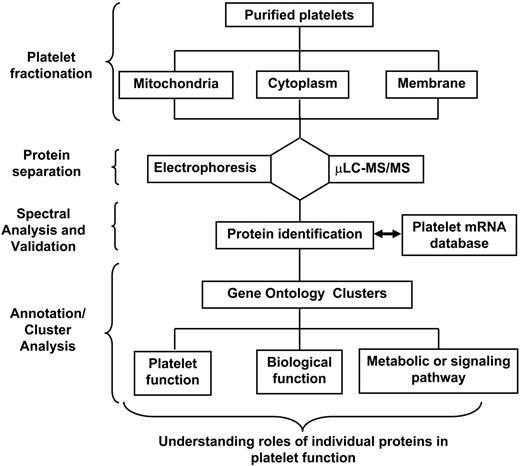
The proteome is the full set of proteins expressed by a genome under a particular set of environmental conditions.23 The strategy for platelet proteomic analysis generally incorporates platelet fractionation, protein separation, and tryptic digestion methodologies followed by protein identification (Figure 1). Improvements in each of these steps enhance the sensitivity and accuracy of protein identification. Proteomic experiments may begin with protein separation (either in-gel or non-gel); alternatively, the entire mixture of protein can be digested before protein identification (for reviews, see Perrotta and Bahou,24 de Hoog and Mann,25 Aebersold and Mann,26 and Steen and Mann27 ). The availability of the human genome sequence coupled with advances in bioinformatics, computer technology, and mass spectrometry (MS) provide for large-scale, robust, and automated analyses of the cellular proteome.
Schema outlining a general approach to platelet proteomic analysis. An ideal experiment generally incorporates platelet subfractionation methods with sophisticated mass spectrometric techniques and computational analyses to elucidate platelet biological functions.
Schema outlining a general approach to platelet proteomic analysis. An ideal experiment generally incorporates platelet subfractionation methods with sophisticated mass spectrometric techniques and computational analyses to elucidate platelet biological functions.
Several mass spectrometry technologies have been developed, including electrospray ionization (ESI-MS),28 matrix-assisted laser desorption/ionization (MALDI)29,30 with variations such as ESI-TOF (time of flight) and MALDI-TOF, liquid chromatography coupled to tandem mass spectrometry (μLC-MS/MS), and multidimensional protein identification technology (MudPIT).31,32 Self-assembled monolayers (SAMs) using trypsin attached to gold have been used to identify 107 platelet proteins, thus establishing the feasibility of proteomic biosensors.33 More recently, several techniques have been devised to directly compare cellular proteomes, including isotope-coded affinity tags (ICATs)34 and isobaric tags (iTRAQ)35-37 coupled to μLC-MS/MS.
Quantitative proteomic38 techniques are used not only to estimate protein abundance but also to compare cell proteomes. These strategies involve labeling peptides derived from individual samples with isobaric tags (ie, same mass, distinct tags) that fragment into different reporter ions on collision-induced dissociation (CID). Peak area ratios of the reporter ions are used to quantify relative peptide abundance.39 iTRAQ technology is potentially superior to ICAT-labeling strategies, which detect only cysteine-containing peptides and typically discard peptides that have been posttranslationally modified.34,39 Other quantitative techniques, such as carboxyl termini labeling of platelet-derived tryptic peptides with oxygen-18, have been used to study the differential expression of platelet proteins.40
Limitations of proteomic techniques: sensitivity and the dynamic range
The platelet proteome is subject to rapid changes in response to external signals. Accordingly, accurate and comprehensive platelet protein profiling requires strict attention to technical details to ensure reproducibility and to minimize protein losses resulting from proteolysis, sample preparation, and protein isolation.
Modern proteomic approaches are limited by large differences in the concentrations of the most and least abundant cellular proteins (approximately 5-log difference). Because data acquisition using tandem MS (MS/MS) is triggered by ion abundance levels,41 peptides derived from more abundant proteins may obscure those derived from less abundant proteins. Thus, this technique can be biased against low-abundance ion signals. In yeast, the dynamic range is insufficient to effectively sample low-abundance proteins of less than 100 copies/cell.42 Other technical limitations in proteomic techniques include detection of proteins with extremes in pI and molecular weight43 and membrane-associated or -bound proteins.44
Accurate platelet protein profiling requires that platelets be stringently purified of contaminating erythrocytes, leukocytes, and plasma proteins. Table 2 summarizes typical platelet RNA and protein yields obtained from 10 mL peripheral blood, adapting a 2-step purification procedure that incorporates centrifugation, gel filtration, and immunodepletion.6 Although the amounts of total platelet RNA (approximately 1 μg) and protein are sufficient for most experiments, detection of low-abundance proteins cannot be enhanced because methods for protein amplification (unlike gene-based PCR technologies) are nonexistent.
Total RNA and protein yields from peripheral blood
. | Platelet yields, × 109 . | . | . | . | . | ||
|---|---|---|---|---|---|---|---|
. | Blood, 10 mL . | PRP* . | Purification† . | Yield . | Sensitivity, copies/cell . | ||
| Total RNA | 3.1 | 2.1 | 1.0 | 1 μg | <0.1‡ | ||
| Protein | — | — | — | 2 mg | 12§ | ||
. | Platelet yields, × 109 . | . | . | . | . | ||
|---|---|---|---|---|---|---|---|
. | Blood, 10 mL . | PRP* . | Purification† . | Yield . | Sensitivity, copies/cell . | ||
| Total RNA | 3.1 | 2.1 | 1.0 | 1 μg | <0.1‡ | ||
| Protein | — | — | — | 2 mg | 12§ | ||
— indicates not applicable.
Centrifugation step (platelet-rich plasma isolated by centrifugation).
Gel filtration with or without CD45+ leukocyte depletion.
Calculated for microarray analysis after amplification step.
Calculated based on silver stain detection limits of 1 ng for a 50-kDa protein.
This issue of protein sensitivity is further highlighted in Table 3, which delineates platelet RNA and protein abundance distributions through composite microarray studies6 or results from a representative platelet proteomic experiment with μLC-MS/MS spectral counts as estimates of relative protein abundance.41 Although this distribution of platelet transcript abundances follows a near-normal distribution, the relative protein abundances are distributed differently. Most detected proteins are identified with 1, 2, or 3 peptide hits per protein. Indeed, these results are in agreement with another study in which 62.6% of proteins were identified by a single peptide, with most of the peptide hits corresponding to highly abundant platelet proteins.45 These data suggest that platelet protein identification using available proteomic technologies has limited ability to quantify peptides with low spectral counts17 and that protein identification is skewed toward more abundant cellular proteins.46,47 Although the number of “one-hit wonders” may potentially be reduced by the better separation of proteins before MS identification, it is unclear whether these limitations can be addressed through other quantitative proteomic approaches.34-37
Distribution of transcript and protein abundance
Relative abundance, decile . | Transcripts, % . | Proteins, % . | Average no. of peptide hits per protein . |
|---|---|---|---|
| 1 | 20.6 | 85.1 | 5.8 |
| 2 | 28.4 | 7.8 | 39.2 |
| 3 | 22.0 | 2.8 | 66.1 |
| 4 | 9.9 | 2.1 | 95.8 |
| 5 | 10.6 | 1.4 | 111.0 |
| 6 | 2.8 | 0 | — |
| 7 | 1.4 | 0 | — |
| 8 | 2.1 | 0 | — |
| 9 | 1.4 | 0 | — |
| 10 | 0.7 | 0.7 | 257 |
Relative abundance, decile . | Transcripts, % . | Proteins, % . | Average no. of peptide hits per protein . |
|---|---|---|---|
| 1 | 20.6 | 85.1 | 5.8 |
| 2 | 28.4 | 7.8 | 39.2 |
| 3 | 22.0 | 2.8 | 66.1 |
| 4 | 9.9 | 2.1 | 95.8 |
| 5 | 10.6 | 1.4 | 111.0 |
| 6 | 2.8 | 0 | — |
| 7 | 1.4 | 0 | — |
| 8 | 2.1 | 0 | — |
| 9 | 1.4 | 0 | — |
| 10 | 0.7 | 0.7 | 257 |
In this prototype experiment, solubilized platelet proteins were analyzed by μLC-MS/MS, and protein abundance was quantified from the number of peptide hits per individual protein (in this experiment, peptide hits per protein ranged from 1 to 257). Relative protein abundance was determined by the number of peptide hits per protein, and rank-ordered proteins were assigned to deciles based on abundance (1, least abundant; 10, most abundant). The percentage of proteins per decile and the average number of peptide hits per protein are also delineated. Note that most proteins (>85%) were skewed toward those of low abundance, represented by a small number of peptide hits, though transcript expression is distributed more evenly (abundance of corresponding transcripts was determined as previously reported90 ).
—indicates not applicable.
Preanalytical variability in platelet proteomics
The methods used to isolate and prepare protein samples affect the results of platelet proteomic studies. For example, findings may vary when platelet proteins are precipitated with ethanol or trichloroacetic acid.48 Delays in sample processing and type of anticoagulant used to collect blood samples may also influence the results of MS identification of platelet proteins.49 Similarly, studies of the low-molecular–weight (less than 15-kDa) peptidome demonstrate that early and gentle platelet separation is crucial for obtaining reproducible identification of platelet-released peptides and proteins.50 Other studies have emphasized the importance of protease inhibitors and uniform sample storage in improving data reproducibility.51 These studies demonstrate a clear requirement for standardization of preanalytical variables to ensure comprehensive, reproducible, and comparative (laboratory-to-laboratory) platelet proteomic analyses.
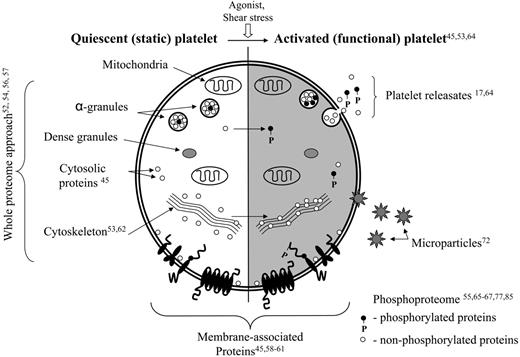
Schema of platelet ultrastructure integrated with proteomic studies. The unshaded panel delineates the proteomic studies involving quiescent platelets. The shaded panel represents those studies focusing on activation-dependent platelet end points (eg, microparticles, exosome/releasates). Superscript numbers refer to references.
Schema of platelet ultrastructure integrated with proteomic studies. The unshaded panel delineates the proteomic studies involving quiescent platelets. The shaded panel represents those studies focusing on activation-dependent platelet end points (eg, microparticles, exosome/releasates). Superscript numbers refer to references.
Platelet proteomic analyses
Platelet proteomic studies can be grouped into 2 distinct yet overlapping subcategories, proteomic analyses of quiescent platelets (the static platelet proteome) or activated platelets (the functional platelet proteome) (Figure 2). Whole proteomic strategies identified many platelet proteins, although subsequent approaches to dissect the changes that occur with platelet activation focused on platelet fractions (membrane proteins) or functional end points (eg, phosphorylation patterns, microparticles) in response to external stimuli. Such subproteomic studies use the same technologies but provide a more detailed analysis of function over time.
Quiescent platelets: the static platelet proteome. Initial studies of the platelet proteome focused on characterizing proteins in resting platelets through combinations of 2-dimensional electrophoresis (2-DE) and in-gel protein detection using monoclonal antibodies.52-54 Although successful, these approaches were limited by the dynamic range of platelet proteins, coupled with the cost and sensitivity of immunodetection. Similar techniques were subsequently applied to establish a platelet protein map and to characterize tyrosine-phosphorylated proteins in resting platelets.55 In this study, cytosolic protein fractions were separated by 2-DE, and phosphorylated proteins were immunodetected using antiphosphotyrosine antibodies and MALDI-TOF. One hundred eighty-six protein spectra corresponding to 123 unique proteins were identified through this approach. By making use of immobilized pH gradient (IPG) strips to facilitate the one-dimensional gel separation, 286 platelet proteins were identified in the acidic range (pI range 4-5).56 A more comprehensive profiling of platelet proteins (pI range 5-11) identified 760 protein features corresponding to 311 different genes, resulting in the annotation of 54% of the 2-DE (pI range 5-11) proteome map.57
Combined fractional diagonal chromatography (COFRADIC) is a non-gel–based technique in which peptide sets are sorted in a diagonal reverse-phase chromatography system through a specific modification of their side chains.58,59 This technique was used to identify 264 platelet proteins present in cytosolic and membrane fractions; the dynamic range spanned 4 to 5 orders of magnitude of protein concentration. Modifications of this technology identified a core set of 641 platelet proteins,45 the largest platelet protein set reported to date. These proteins were classified using the Gene Ontology database, revealing that 16% were membrane proteins (see “Membrane proteins,” below), and 64% were present in the cytoskeleton, endoplasmic reticulum, mitochondria, cytosol, or Golgi apparatus. Interestingly, up to 20% of proteins were classified as nucleus restricted. Given that platelets are anucleate, these proteins presumably arise from megakaryocytes during thrombopoiesis; their function in platelets is unclear.
Membrane proteins. The study of platelet integral membrane proteins and surface receptors through traditional 2-DE techniques is limited by the low solubility of these proteins, their association with the platelet membrane, their high molecular weight, and the potentially limited number of diagnostic tryptic cleavage fragments. In addition, the presence of highly abundant cytoskeleton proteins such as actin obscures less abundant platelet membrane proteins.
Two groups have adapted proteomic approaches for the study of platelet membrane proteins.45,60 Although applications using COFRADIC45 predicted the presence of 87 putative helix-spanning membrane proteins, a more focused analysis of the platelet membrane proteome was pursued by enriching membrane proteins before protein identification through μLC-MS/MS. In this study, 2 distinct solubilization methods were used to reduce the overrepresentation of cytoskeleton proteins, allowing for the aggregate detection of 233 established or putative transmembrane proteins.60 Finally, a combination of microarray and mass spectrometric techniques has been used to identify novel membrane proteins that signal during platelet aggregation and to characterize proteins that become phosphorylated on tyrosine or serine residues on platelet aggregation.61
Activated platelet proteome: activation-dependent platelet complexes. An early study of activation-dependent responses in platelets identified 27 proteins that were translocated to the actin cytoskeleton during platelet activation.62 Subsequently, a related study that focused on characterizing static protein complexes as a prelude to dissecting activation-dependent complexes was undertaken in accordance with a novel 2-DE technology that combined 1-dimensional separation of monomeric and multimeric proteins in their native state followed by a 2-dimensional denaturing step.63 In total, 63 platelet proteins were identified, among them the integral membrane proteins GPIbβ, CD9, and CD36 and the fibrinogen receptor αIIbβ3. Mitochondrial proteins were detected in the membrane fraction, several of which were organized into known complexes.
Proteins secreted during platelet activation. Proteomic strategies have been used to identify protein subsets secreted during platelet activation. Such strategies resulted in the initial identification of 82 secreted proteins17 ; this was followed by a more robust analysis of the platelet secretome.64 With the use of thrombin-stimulated platelets, a combination of MALDI-TOF and MudPIT identified more than 300 released proteins.64 Thirty-seven percent of these proteins were previously known to be released from platelets, whereas another 35% were reported to be released from other secretory cells. The remaining 28% of proteins were not known to be released by any cell type. Moreover, several of the secreted proteins have been previously identified in human atherosclerotic lesions but absent in normal vasculature. In this study, protein profiling resulted in the identification of potential targets for future drug development (ie, secretogranin III, cyclophilin A, and calumenin) and shed light on possible mechanisms of platelet adhesion that contribute to the development of atherosclerosis.
Platelet phosphoproteome. Tyrosine phosphorylation plays a key role in the activation-dependent regulation of protein function, signal transduction, and other complex cellular processes. Several studies have used phosphotyrosine-dependent antibodies and proteomic technologies to dissect intracellular signaling events that occur with thrombin activation, consistently demonstrating an ability to identify discrete proteins, subsets of which had been previously undescribed.65,66 By using the thrombin receptor activating peptide (TRAP) to specifically activate proteinase-activated receptor 1 (PAR1), 62 differentially phosphorylated proteins were detected, 41 of which were identified by μLC-MS/MS.67 Interestingly, 8 of these had never been described, and the protein repertoire was shown to originate from 31 genes, highlighting how alternative splicing expands the platelet proteome. Although these initial studies established the feasibility of using proteomic approaches to dissect ligand-dependent phosphorylation changes,68,69 it is likely that technical advances will further enhance its robustness.70
Platelet microparticles. When platelets are activated in vivo, 2 types of membrane vesicles are released, microparticles (which bud from the plasma membrane) and exosomes.71 Microparticles range in size from 0.1 to 1.0 μm, whereas exosomes are smaller and range from 40 to 100 nm. Platelet microparticles are abundant, exert procoagulant activity, and play hemostatically critical roles in several clinical disorders, including heparin-induced thrombocytopenia and immune thrombocytopenic purpura. A recent proteomic study provided the first panoramic overview of microparticles, identifying 578 proteins that constitute this subcellular proteome.72 As expected, many of these represented well-characterized platelet proteins. Surprisingly, 380 of 578 proteins had not been previously described in platelet proteomic studies, suggesting these platelet fragments have a unique protein composition.
Integrating platelet transcriptomic and proteomic studies
Recent evidence from mathematical modeling studies demonstrates the need to delineate mRNA and protein expression levels to optimally map intracellular networks. When applied to nucleated cells, these integrated platforms demonstrate some correlation between transcript and protein expression.73 In platelets, this relationship is even more complex because platelets lack a genome and ongoing RNA transcription, display minimal translational activity, have the capacity for signal-dependent translation of selected mRNAs4 and inducible (activation-dependent) transcript splicing,5 absorb select plasma proteins, and contain residual megakaryocyte-derived mRNAs and proteins.
Initial attempts to correlate platelet mRNA and protein profiles have been described.17,74 These studies demonstrate that up to 69% of secreted and cytosolic proteins were detectable at the mRNA level, suggesting relatively good correlation between proteomic and transcriptomic data in the study of end points of detection and identification.17 The authors concluded that despite the absence of gene transcription, the platelet proteome is mirrored in the transcriptome, and transcriptional analysis predicts the presence of novel proteins in the platelet. The lack of detailed quantitative correlations limits the ability to compare platelet gene and protein levels with those of nucleated cells.73
Global profiling to study platelet-associated disorders
Platelet proteomic studies
Glanzmann thrombasthenia. GT is an autosomal recessive disease characterized by the complete absence or a marked reduction in the αIIbβ3 heterodimeric complex.75,76 Compared with normal platelets, GT platelets exhibit a low tyrosine phosphorylation profile, confirming the key role of functional αIIbβ3 in the initiation of protein tyrosine phosphorylation.77 Several proteins displayed attenuated thrombin-dependent phosphorylation kinetics despite normal initial phosphorylation rates. Similar results were obtained by inhibiting thrombin aggregation of control platelets with αIIbβ3 antagonists or in the absence of stirring. These results suggest that tyrosine phosphorylation of specific proteins is dependent on thrombin activation during early and late steps of αIIbβ3 engagement in aggregation. No other clear differences were identified between normal and GT platelets.
Ischemic stroke. Genetic risk factors have been identified in patients with thrombophilia, though associations are the strongest for patients with venous thromboembolic disease.78 Thus, whereas hyperhomocysteinemia and antiphospholipid/anticardiolipin antibodies are stroke risk factors,79 congenital thrombophilic states (factor V Leiden and prothrombin gene mutations; protein C, protein S, and antithrombin III deficiencies) account for a disproportionately small subset of ischemic stroke,80 with the greatest risk in younger persons.81 In contrast, differential expression of platelet proteins may favor platelet activation and thrombus formation. The best evidence in support of this observation stems from correlative studies involving platelet cell surface glycoprotein receptors such as α2β1, in which higher expression levels are associated with increased collagen binding,82 and greater risk for ischemic heart disease in homozygotes, especially smokers.83 Similarly, platelet membrane polymorphisms have been linked to stroke in small studies, but the evidence is not strong, and it is most significant in young patients.84
A phosphoproteomic comparison of platelets from healthy controls and stroke patients was completed using antiphosphotyrosine antibodies.85 In unstimulated platelets, a discrete subset of proteins displayed significantly greater tyrosine phosphorylation in 85% of the 20 stroke patients studied than in the healthy controls. Additionally, the authors identified other tyrosinephosphorylated bands in the stroke platelets that were absent in the resting platelets of the control patients. Although these results may have some prognostic merit in monitoring patients with cerebrovascular insufficiency,85 they provide no specific protein identification and fail to distinguish cause from effect in dissecting proteomic differences that pathogenetically define platelet-associated stroke risk.
Toxicology. Benzene exposure is an established risk factor for acute myeloid leukemia and may play a role in other human blood diseases.86 Surface-enhanced laser desorption ionization-TOF MS (SELDI-TOF MS) was used to compare the blood serum proteome of 40 shoe factory workers who experienced well-characterized occupational exposure to benzene with the blood serum proteome of unexposed controls to identify potential biomarkers of benzene exposure.87 Three low-molecular–weight proteins were consistently down-regulated in exposed subjects. Two of these proteins were identified as platelet-derived CXC chemokines, platelet factor 4 (PF4), and connective tissue–activating peptide III (CTAP III). These findings suggest that lower expression of these proteins may serve as a potential biomarker of early benzene toxicity.
From the transcriptome to the proteome
With today's technology, it is feasible to adapt transcriptomic analysis to predict differences in the proteome, to validate these differences between normal and diseased platelets, and to exploit these differences to develop modern diagnostic tests.19,74,88
Direct platelet transcriptomics: essential thrombocythemia as a paradigm. The most compelling evidence that transcript profiling can distinguish diseased from normal platelet profiles is based on studies of essential thrombocythemia (ET) that were conducted with megakaryocytes and platelets.89,90 ET is a myeloproliferative disorder characterized by increased proliferation of megakaryocytes, elevated numbers of morphologically normal circulating platelets, and considerable thrombohemorrhagic events.91 Microarray profiling has been used to analyze platelet transcripts of 6 patients with ET and 5 healthy controls.90 Initial analysis demonstrated different molecular signatures capable of distinguishing normal platelets from ET platelets. Moreover, ET platelets collectively expressed higher numbers of individual transcripts than normal platelets but considerably less distinct transcripts than nucleated cells.6 Statistical analysis revealed 170 genes that were differentially expressed between ET and normal platelets, most (141 of 170 genes) of which were up-regulated in ET platelets.
Among differentially expressed genes, the transcript encoding 17β-hydroxysteroid dehydrogenase type 3 (HSD17B3) was dramatically reduced in ET platelets. This enzyme, not previously characterized in human platelets, belongs to a larger family of 17BHSDs that play a role in normal and abnormal testosterone biosynthesis.92 Transcripts for 2 other 17BHSD family members were identified in platelets; they are HSD17B11 and HSD17B12. Absence of HSD17B3 transcript expression was evident in all 6 patients with ET. These changes occurred concomitantly with elevated transcript levels of HSD17B12 in the same patient subgroup. In contrast, the expression of HSD17B3 in normal platelets was accompanied by negligible HSD17B12 expression. At the protein level, HSD17β3 enzyme activity was demonstrated in platelet lysates, confirming that human platelets retained the capacity to catalyze the final step in gonadal testosterone synthesis. These data demonstrate the potential of using transcriptomics to lend focus to proteomic studies.
Indirect transcriptomic approaches of platelet disorders. Gray platelet syndrome. Gray platelet syndrome (GPS) is a rare platelet disorder manifest by bleeding, thrombocytopenia, and a distinct lack of α-granules.93,94 Recently, microarray analysis was used to study molecular mechanisms involved in GPS, focusing on fibroblasts because these cells may be involved in the transition to myelofibrosis evident in subsets of patients with the disorder.95 By comparing microarray profiles of normal and GPS fibroblasts, the up-regulation of various proteins (fibronectin 1, thrombospondins 1 and 2, and collagen VIα) was demonstrated. Overexpression of fibronectin and thrombospondin 1 was confirmed at the RNA level by Northern blot analysis, whereas fibronectin overexpression was confirmed at the protein level by immunostaining. Although this study fails to address the role of megakaryocyte-derived proteins in the development of myelofibrosis, it does suggest that distinct protein subsets may play a role in the progression to myelofibrosis seen in some GPS patients.
Polycythemia rubra vera. Like ET, polycythemia rubra vera (PRV) is a myeloproliferative disorder associated with thrombotic complications.96 Although the molecular basis for enhanced thrombotic risk remains unknown, defects affecting the platelet thrombohemorrhagic balance are envisioned; however, ancillary leukocyte dysfunction linking the inflammatory response to risk for thrombosis cannot be excluded. To date, microarray studies have been restricted to PRV leukocytes to the exclusion of platelets.97-99 Furthermore, limited correlative proteomic studies validate the differences seen by microarray or stratify transcriptomic differences by thrombotic phenotype. Although further correlative studies are needed, PRV platelets, like ET platelets, may have a distinct proteomic profile that could help explain the pathogenesis of thrombosis in this hematopoietic disorder.
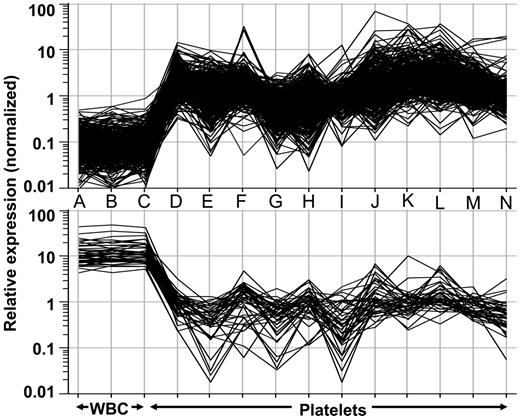
In silico gene rank–intensity plots from a first-generation platelet gene chip. Normalized data from individual microarray analyses obtained from 3 distinct leukocyte samples (A-C), 5 normal platelet samples (D-H), and 6 essential thrombocythemic platelet samples (I-N)90 were analyzed by one-way ANOVA using parametric testing to identify a 432-member gene list. (top) Relative expression of the platelet-restricted genes (n = 389). (bottom) Expression intensity of the leukocyte-restricted genes (n = 43). Note the clear difference in the expression patterns between the 2 groups (leukocyte vs platelet).
In silico gene rank–intensity plots from a first-generation platelet gene chip. Normalized data from individual microarray analyses obtained from 3 distinct leukocyte samples (A-C), 5 normal platelet samples (D-H), and 6 essential thrombocythemic platelet samples (I-N)90 were analyzed by one-way ANOVA using parametric testing to identify a 432-member gene list. (top) Relative expression of the platelet-restricted genes (n = 389). (bottom) Expression intensity of the leukocyte-restricted genes (n = 43). Note the clear difference in the expression patterns between the 2 groups (leukocyte vs platelet).
Future directions and applicability
Custom platelet chips
The expense of global transcript and platelet protein profiling precludes investigators from performing larger disease cohort studies. The development of cost-effective and reliable tools for transcript profiling would facilitate platelet-related diagnostics and experimentation. Thus, we have fabricated a custom platelet microarray that can be used in conjunction with proteomic analyses to study normal and diseased platelets. The final transcript list was established by analyzing microarray results from normal and ET platelets90 and included platelet-restricted genes without detectable expression in leukocytes, genes that appear to be associated with a thrombohemorrhagic ET phenotype, and genes whose platelet expression is more than 10-fold greater than its leukocyte expression or whose leukocyte expression is more than 10-fold greater than its platelet expression. Several Arabidopsis probe elements were included to serve as normalization controls and to minimize interslide and intraslide variability.100 After removing duplicates, the final list contains 432 genes that clearly cosegregate by cell type (Figure 3). Although initial analyses demonstrate the potential of this chip to distinguish ET platelets from normal platelets (data not shown), further studies are ongoing. Nonetheless, the development of platelet-specific gene and protein chips could lead to more widespread applicability of this technology to platelet-related and thrombotic disorders.
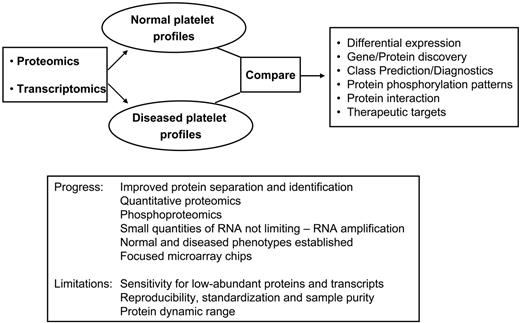
Integration of platelet transcript and protein profiling to study human diseases. The combination of proteomic and transcriptomic technologies can be applied for comparative studies between normal and diseased platelets, ultimately leading to novel diagnostic assays or to the identification of novel therapeutic targets. Potential applications include treatment of not only single-gene platelet disorders but also the broader subset of patients with platelet-related cardiovascular or cerebrovascular disease. The box summarizes current limitations and progress achieved to date.
Integration of platelet transcript and protein profiling to study human diseases. The combination of proteomic and transcriptomic technologies can be applied for comparative studies between normal and diseased platelets, ultimately leading to novel diagnostic assays or to the identification of novel therapeutic targets. Potential applications include treatment of not only single-gene platelet disorders but also the broader subset of patients with platelet-related cardiovascular or cerebrovascular disease. The box summarizes current limitations and progress achieved to date.
General profiling perspectives
Future advances in proteomic technology that incorporate miniaturization,101 coupled with an ability to integrate functional genomics and proteomics,102 will help unravel the complex biological pathways that are relevant to platelet-associated disorders. The human genome encodes between 20 000 and 40 000 genes, whereas the estimated number of functional proteins may number as much as half a million because of alternative splicing, translational regulation, and posttranslational modifications.102 Because of this large number of functional proteins, we anticipate that integrated analysis of the transcriptome and the proteome are required to optimally dissect the molecular mechanisms responsible for platelet-related diseases (Figure 4). Parallel advances in quantitative proteomic techniques will also have an impact on the field,34-37 resulting in the identification of platelet-related disease biomarkers and novel therapeutic targets.
Authorship
Contribution: D.V.G., P.L.P, and W.F.B. reviewed the literature and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Prepublished online as Blood First Edition Paper, August 22, 2006; DOI 10.1182/blood-2006-06-026518.
This work was supported by NIH grants HL49141, HL53665, HL76457; Department of Defense grant MPO48005; a Targeted Research Award from Stony Brook University; and National Institutes of Health Center grant MO1 10710-5 to the University Hospital General Clinical Research Center.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal