Tumors may escape from immune control by the induction of CD11b+Gr-1+ myeloid suppressor cells in the spleen. In this study, we demonstrate that this cell population can be subdivided into a CD11bhiGr-1intSSCloLy6GnegM-CSFRint immature monocytic fraction and a CD11bhi+Gr-1hiSSChiLy6GhiM-CSFRneg granulocytic fraction. Upon in vitro culture, the monocytic CD11b+Gr-1+ cell fraction is sufficient for cytotoxic T lymphocyte (CTL) suppression, which is linked to the gradual differentiation of these monocytic cells into mature F4/80+ CD68+ macrophages. These CTL-suppressive macrophages are alternatively activated (M2), as demonstrated by the expression of known and novel M2 signature genes. In search of M2-associated genes involved in the suppressive activity, it is shown that stimulation of peroxisome proliferator-activated receptor γ (PPARγ) and inhibition of phospholipase A2 (PLA2) activity cooperate to alleviate CTL suppression. Of importance, purified tumor-associated macrophages display a similar M2 phenotype and are suppressive for antitumor CTLs, via a mechanism that can be almost completely reversed by PPARγ ligands. Overall, our data identify PLA2 and especially PPARγ as new potential therapeutic targets to subvert macrophage-mediated CTL suppression in cancer.
Introduction
Tumors may escape from immune control by the induction of distinct immunosuppressive cell types, among which myeloid cells occupy a prominent position.1 In recent years, it has become clear that tumor growth, in both humans and mice, is associated with the accumulation of immature myeloid cells in the secondary lymphoid organs and the blood.2-8 In mice, these cells are characterized by the coexpression of CD11b and Gr-1 surface markers, a rather crude classification covering ill-defined populations of immature monocytic and granulocytic cell types. CD11b+Gr-1+ cells have been shown to suppress CD8+ T-cell activity in a cell-contact and antigen-specific fashion, which can mechanistically be explained by the high production of reactive oxygen species (ROSs) in response to integrin-mediated contact with the responding T cell.9-10 Other studies established an association between the suppressive activity and the arginase-1–producing capacity of the myeloid population.11-13 Arginase-1, through activating the reductase domain of inducible NO synthase (iNOS), can increase superoxide production in myeloid cells, thereby leading to T-cell suppression.11-12 Alternatively, by depleting extracellular l-arginine levels, arginase-1 may decrease CD3ζ expression and as such impair T-cell function.13-14 However, our own previous work demonstrates that arginase-1 activity is not necessarily implicated in the T-cell down-modulation by arginase-1+ myeloid suppressor cells (MSCs) from mice with progressing tumors, suggesting the existence of as-yet-unexplored suppressive mechanisms in these cells.15
Elevated arginase-1 activity is one of the hallmarks of IL-4– and/or IL-13–elicited alternatively activated myeloid cells (M2), as opposed to IFNγ- and/or TNF-induced classically activated myeloid cells (M1).16 Recently, the M2 concept has been broadened to myeloid cells developing in the presence of immune complexes and TLR ligands (M2b), or IL-10 (M2c),17 sharing overlapping functions: down-regulation of type I cytokine-driven inflammation and stimulation of angiogenesis and wound healing.18 This type of cell has been observed in different pathologies, including parasite infections19-21 and tumors.22 In a tumor context, M2 cells are, at least in part due to their immunosuppressive properties, associated with an exacerbation of the disease, whereas the induction of cytotoxic M1 contributes to tumor rejection.23-26 Therefore, a better understanding of the molecular and functional profile of cancer-associated M2 is warranted to use those cells as potential targets in cancer therapy.
In the present study, we aimed at obtaining a better insight into the origin, molecular profile, and cytotoxic T lymphocyte (CTL)–suppressive mechanism of in vitro–generated splenic MSCs from mice with progressing T-cell lymphoma tumors, and translating this information to the characteristics of in vivo–generated suppressive myeloid cells. We demonstrated that the monocytic fraction of splenic CD11b+Gr-1+ cells is sufficient for the generation of M2-oriented, CTL-suppressive macrophages. M2-associated genes, peroxisome proliferator-activated receptor γ (PPARγ) and phospholipase A2 (PLA2), were identified as targets for suppression-lifting intervention, not only on in vitro–generated MSCs but, more importantly, also on M2-oriented tumor-associated macrophages.
Materials and methods
Mice and tumor growth
Six- to 9-week-old female AKR mice (H-2k; Harlan, Horst, The Netherlands) were implanted subcutaneously in the flank with 2 × 106 BW-Sp3 cancer cells, and 5 to 7 weeks later mice were used for experiments.
Cell lines and media
The generation of BW-Sp3 and the BW-Sp3(B7-1) cells was described earlier.27-28 Cancer cells were maintained in RPMI 1640 medium, supplemented with 10% heat-inactivated FCS, 0.03% l-glutamine, 100 mg/mL streptomycin, and 100 mg/mL penicillin (Invitrogen, Frederick, MD). For splenocyte cultures, this medium was supplemented with 1 mM nonessential amino acids, 1 mM sodium pyruvate (Invitrogen), and 0.02 mM 2-ME (ME medium).
Fluorescence-activated cell sorter (FACS) staining
Cells were stained for 20 minutes at 4°C using conventional protocols: preincubation with anti-FcγR Ab (2.4G2) before adding at 1 μg/106 cells anti-CD11b/PE or anti-CD11b/FITC (M1/70), anti–Gr-1/FITC (RB6-8C5), anti-Ly6G/FITC (1A8), anti-Ly6C/FITC (AL-21), anti-F4/80/PE (CI:A3-1), anti-CD31/PE (MEC13.3), anti-CD68/PE (FA-11), anti-CD11c/PE (HL3), or PE/FITC-conjugated isotype controls. The 7/4 antibody (gift of Dr Gordon Brown, Sir William Dunn School of Pathology, Oxford, United Kingdom) and anti–M-CSFR (AFS98; gift of Dr Pieter Leenen, Erasmus Medical Center, Rotterdam, the Netherlands) were used as purified Abs and hybridoma supernatant, respectively, detected by polyclonal anti–rat Ig/PE. All antibodies were from BD Biosciences (San Jose, CA), except anti-F4/80 and anti-CD68 (Serotec, Raleigh, NC).
For intracellular cytokine staining, 5 × 105 cells were stimulated for 6 hours with anti-CD3. GolgiPlug (BD Biosciences) was added during the last 4 hours. Following CD8/FITC (53-6.7) staining, cells were stained intracellularly using Cytofix/Cytoperm (BD Biosciences), according to the manufacturer's instructions. Antibodies were anti–IFN-γ/PE (XMG1.2) and rat IgG1 isotype/PE (R3-34) (BD Biosciences).
For detection of ROSs, cells were incubated with 2 μM dihydroethytium (DHE; Molecular Probes, Eugene, OR) for 1 hour, or 2 μM dichlorodihydrofluorescein diacetate (DCFDA; Molecular Probes) for 30 minutes at 37°C. Cells were washed, incubated at 4°C with anti-CD11b/PE, and analyzed on a FACSVantage SE flow cytometer (BD Biosciences).
In vivo depletion of granulocytes
The anti–Gr-1 Ab (RB6-8C5) was purified from hybridoma supernatant, using standard techniques, and tested for LPS contamination, using the LAL assay (BioWhittaker, Walkersville, MD). LPS-free Ab (100 μg) was injected intraperitoneally in 5- to 7-week tumor-bearing mice and 24 hours later spleens were taken.
Purification and depletion of cell populations
To isolate tumor-associated macrophages, tumors were dissected and chopped into small pieces before incubation with an enzyme mixture dissolved in RPMI 1640 for 30 minutes at 37°C: 400 U/mL collagenase type IV, 0.05 mg/mL collagenase type I, 0.025 mg/mL hyaluronidase (Sigma-Aldrich, St Louis, MO), 0.01 mg/mL Dnase I, 0.2 U/mL soybean trypsin inhibitor (Boehringer Mannheim, Mannheim, Germany). Single-cell suspensions were made in magnetic-activated cell sorter (MACS) buffer (degassed PBS, 0.5% BSA, 2 mM EDTA) and incubated (15 minutes, 8°C) with anti-CD11b microbeads (5 μL beads/107 cells; Miltenyi Biotec, Auburn, CA). CD11b+ cells were isolated on LS columns (Miltenyi Biotec), using the MidiMACS system (Miltenyi Biotec) according to the manufacturer's instructions.
To isolate peritoneal macrophages, peritoneal cells were collected in 0.34 M sucrose, resuspended in MACS buffer, incubated with anti-CD19 microbeads, and passed over LD columns (both Miltenyi Biotec), using the MidiMACS system (depletion of CD11b+CD19+ cells). Flow-through was collected, and remaining CD11b+ cells were isolated on LS columns. These cells were more than 90% CD11b+F4/80+ macrophages.
Depletion of splenic CD11b+ cells was performed using anti-CD11b microbeads and LD columns. To purify CD11b+Gr-1+ cells, splenocytes were first depleted of CD19+ and CD4+ cells, using anti-CD19 microbeads, anti-CD4 microbeads, and LD columns. Flow-through was collected, and remaining CD11b+ cells were isolated on LS columns as described in the first paragraph of this section. These cells were more than 90% CD11b+Gr-1+.
Evaluation of CTL activity
Splenocytes from naive AKR or BW-Sp3 progressors were restimulated at 2 × 107 splenocytes/well with 106 irradiated (110 Gray [11 000 rad]) BW-Sp3(B7-1) cells in 6-well plates (Falcon; Becton Dickinson, Lincoln Park, NJ) for 5 days in ME-medium, without addition of cytokines. When appropriate, the following compounds were added to the cultures at day 2: 200 U/mL superoxide dismutase (SOD, O2– scavenger; Sigma-Aldrich), 200 U/mL catalase (H2O2 scavenger; Sigma), 500 ng/mL anti–PD-L2 (eBioscience, San Diego, CA), 6 μM baicalein (12/15 LOX inhibitor; Sigma), 10 μM GW9662 (PPARγ inhibitor; Sigma), 5 μM rosiglitazone (PPARγ ligand; Cayman Chemical, Ann Arbor, MI), 1 μM 15-deoxy-Δ12,14 -prostaglandin J2 (15d-PGJ2, PPARγ ligand; Cayman Chemical), 3 μM aristolochic acid (PLA2 inhibitor; Sigma), 2.5 μM AACOCF3 (cPLA2 inhibitor; Biomol International, Plymouth Meeting, PA), 2.5 μM PACOCF3 (iPLA2 inhibitor; Calbiochem, San Diego, CA), and 0.5 μM ONO-RS-082 (sPLA2 inhibitor; Biomol International). Stock solutions of the latter 8 compounds were made in DMSO. Therefore, a corresponding volume of DMSO was added to cultures without these compounds.
Alternatively, nonadherent cells were removed after 2 days of culture and restimulated for another 3 days in a new plate. To test cytotoxicity to CTLs, the compounds were added to these nonadherent cell cultures.
For each of these cultures, the nonadherent cells were recuperated after 5 days and loaded on Ficoll-Paque (Pharmacia Biotech, Piscataway, NJ) for gradient centrifugation. Viable cells were tested for their cytotoxicity toward 111In-labeled BW-Sp3(B7-1), as described earlier.15 Where appropriate, percent CTL recovery was calculated as follows: ([% specific lysis Total + inhibitor] – [% specific lysis Total])/([% specific lysis Nad] – [% specific lysis Total]) × 100.
For flow cytometry and functional assays, adherent cells were recovered at different time points, using Cell Dissociation Buffer (Gibco, Carlsbad, CA) and gentle scraping.
RNA extraction and quantitative reverse-transcriptase–polymerase chain reaction (RT-PCR) analysis
Total RNA (1 μg), prepared using TRIzol reagent (Invitrogen), was reverse-transcribed using oligo(dT) and SuperScript II RT (Invitrogen). Quantitative real-time PCR was performed in a Bio-Rad iCycler, with Bio-Rad iQ SYBR Green Supermix (Bio-Rad, Hercules, CA), using primers listed in Table 1. For all primers, each PCR cycle consisted of 1 minute at 94°C, 45 seconds at 55°C, and 1 minute at 72°C. Gene expression was normalized using ribosomal protein S12 as housekeeping gene.
List of gene-specific primer pairs used for quantitative real-time PCR
Gene product . | Forward primer . | Reverse primer . |
|---|---|---|
| FIZZ1 | 5′-CCCAGGATGCCAACTTTGAA-3′ | 5′-GGCCCATCTGTTCATAGTCT-3′ |
| Ym | 5′-GGGCATACCTTTATCCTGAG-3′ | 5′-CCACTGAAGTCATCCATGTC-3′ |
| Fibronectin-1 | 5′-CAGCGGATGAGCTGCACATG-3′ | 5′-GTTCTCTGATTGTATCTCTGTG-3′ |
| Arginase-1 | 5′-GCTGTCTTCCCAAGAGTTGGG-3′ | 5′-ATGGAAGAGACCTTCAGCTAC-3′ |
| CCL8 | 5′-TCTACGCAGTGCTTCTTTGC-3′ | 5′-CCACTTCTGTGTGGGGTCTA-3′ |
| CCL24 | 5′-GCTGCACGTCCTTTATTTCC-3′ | 5′-GGCTAAACCTCGGTGCTATT-3′ |
| E-cadherin | 5′-ACTTGGGGACAGCAACATCA-3′ | 5′-GGGTTTAAATCGGCCAGCAT-3′ |
| MGL-1 | 5′-ATGATGTCTGCCAGAGAACC-3′ | 5′-ATCACAGATTTCAGCAACCTTA-3′ |
| MGL-2 | 5′-GATAACTGGCATGGACATATG-3′ | 5′-TTTCTAATCACCATAACACATTC-3′ |
| Selenoprotein P | 5′-TTCTGCAGGCATCCAGATTG-3′ | 5′-CACAAGACGGCCACATCTGT-3′ |
| Coagulation factor XIIIa | 5′-CCAGGAATTAAGCAAGACATC-3′ | 5′-TGCCCTTACTTTCTAGTTCTC-3′ |
| Retinoic acid Rγ | 5′-CTAGTGCCTTCTACCCTTGA-3′ | 5′-AACAGCGGGGTAGGAGATAT-3′ |
| PD-L2 | 5′-GCTTGAGGCGCAGAACACTG-3′ | 5′-TGACCGCAGAGAATGCACTG-3′ |
| TREM-2b | 5′-CTCCGAGGCCGAGAGGC-3′ | 5′-GGGCACCCTCGAAACTCGA-3′ |
| PLA2GIVa | 5′-GAGGGGCTTTATTCCACACG-3′ | 5′-GTCCAAAATCCCCGACTCAT-3′ |
| Cathepsin L | 5′-TGGACTCGGAGGAGTCTTAC-3′ | 5′-TCTGTTCCTTCATAGCCATAG-3′ |
| PLA2GVII/PAF-AH | 5′-TGCACCAGAACTTTGACGAC-3′ | 5′-CAACTGTGTAACAAGCAAGTTC-3′ |
| Macrophage mannose receptor | 5′-CTCGTGGATCTCCGTGACAC-3′ | 5′-GCAAATGGAGCCGTCTGTGC-3′ |
| Prosaposin | 5′-GCCATTGTCATAGCACAGAG-3′ | 5′-CTTCACTCATGAGCTGAACAG-3′ |
| Cysteinyl leukotriene R1 | 5′-GAGCCTCCACAGAACAATCA-3′ | 5′-TAGCAGAGGATCAAAGCAACA-3′ |
| PPARγ | 5′-AGAGTCTGCTGATCTGCGAG-3′ | 5′-GGCATACTCTGTGATCTCTTG-3′ |
| iNOS | 5′-TGGAGCCAAGGCCAAACACAG-3′ | 5′-TCCACCAGGAGATGTTGAAC-3′ |
| 12/15 lipoxygenase | 5′-CAGCTGGATTGGTTCTACTG-3′ | 5′-CTCTGAACTTCTTCAGCACAG-3′ |
| PLA2GIId | 5′-ATTGTTGCTATGCCCACCTG-3′ | 5′-GCACACAGTTGCCTTTCACA-3′ |
| PLA2GV | 5′-GACCGTTGTTATGGGCAACT-3′ | 5′-GAGCCTCATTGGACAGAAGG-3′ |
| PLA2GXIIa | 5′-CCACTGTTTGGCGTTCATCT-3′ | 5′-GGAACTCCTCGTCACAGTCG-3′ |
| PLA2GVIa | 5′-ATGGGAAAGCAGGAGATGGT-3′ | 5′-TCATTTCAGCACACCCCTTC-3′ |
Gene product . | Forward primer . | Reverse primer . |
|---|---|---|
| FIZZ1 | 5′-CCCAGGATGCCAACTTTGAA-3′ | 5′-GGCCCATCTGTTCATAGTCT-3′ |
| Ym | 5′-GGGCATACCTTTATCCTGAG-3′ | 5′-CCACTGAAGTCATCCATGTC-3′ |
| Fibronectin-1 | 5′-CAGCGGATGAGCTGCACATG-3′ | 5′-GTTCTCTGATTGTATCTCTGTG-3′ |
| Arginase-1 | 5′-GCTGTCTTCCCAAGAGTTGGG-3′ | 5′-ATGGAAGAGACCTTCAGCTAC-3′ |
| CCL8 | 5′-TCTACGCAGTGCTTCTTTGC-3′ | 5′-CCACTTCTGTGTGGGGTCTA-3′ |
| CCL24 | 5′-GCTGCACGTCCTTTATTTCC-3′ | 5′-GGCTAAACCTCGGTGCTATT-3′ |
| E-cadherin | 5′-ACTTGGGGACAGCAACATCA-3′ | 5′-GGGTTTAAATCGGCCAGCAT-3′ |
| MGL-1 | 5′-ATGATGTCTGCCAGAGAACC-3′ | 5′-ATCACAGATTTCAGCAACCTTA-3′ |
| MGL-2 | 5′-GATAACTGGCATGGACATATG-3′ | 5′-TTTCTAATCACCATAACACATTC-3′ |
| Selenoprotein P | 5′-TTCTGCAGGCATCCAGATTG-3′ | 5′-CACAAGACGGCCACATCTGT-3′ |
| Coagulation factor XIIIa | 5′-CCAGGAATTAAGCAAGACATC-3′ | 5′-TGCCCTTACTTTCTAGTTCTC-3′ |
| Retinoic acid Rγ | 5′-CTAGTGCCTTCTACCCTTGA-3′ | 5′-AACAGCGGGGTAGGAGATAT-3′ |
| PD-L2 | 5′-GCTTGAGGCGCAGAACACTG-3′ | 5′-TGACCGCAGAGAATGCACTG-3′ |
| TREM-2b | 5′-CTCCGAGGCCGAGAGGC-3′ | 5′-GGGCACCCTCGAAACTCGA-3′ |
| PLA2GIVa | 5′-GAGGGGCTTTATTCCACACG-3′ | 5′-GTCCAAAATCCCCGACTCAT-3′ |
| Cathepsin L | 5′-TGGACTCGGAGGAGTCTTAC-3′ | 5′-TCTGTTCCTTCATAGCCATAG-3′ |
| PLA2GVII/PAF-AH | 5′-TGCACCAGAACTTTGACGAC-3′ | 5′-CAACTGTGTAACAAGCAAGTTC-3′ |
| Macrophage mannose receptor | 5′-CTCGTGGATCTCCGTGACAC-3′ | 5′-GCAAATGGAGCCGTCTGTGC-3′ |
| Prosaposin | 5′-GCCATTGTCATAGCACAGAG-3′ | 5′-CTTCACTCATGAGCTGAACAG-3′ |
| Cysteinyl leukotriene R1 | 5′-GAGCCTCCACAGAACAATCA-3′ | 5′-TAGCAGAGGATCAAAGCAACA-3′ |
| PPARγ | 5′-AGAGTCTGCTGATCTGCGAG-3′ | 5′-GGCATACTCTGTGATCTCTTG-3′ |
| iNOS | 5′-TGGAGCCAAGGCCAAACACAG-3′ | 5′-TCCACCAGGAGATGTTGAAC-3′ |
| 12/15 lipoxygenase | 5′-CAGCTGGATTGGTTCTACTG-3′ | 5′-CTCTGAACTTCTTCAGCACAG-3′ |
| PLA2GIId | 5′-ATTGTTGCTATGCCCACCTG-3′ | 5′-GCACACAGTTGCCTTTCACA-3′ |
| PLA2GV | 5′-GACCGTTGTTATGGGCAACT-3′ | 5′-GAGCCTCATTGGACAGAAGG-3′ |
| PLA2GXIIa | 5′-CCACTGTTTGGCGTTCATCT-3′ | 5′-GGAACTCCTCGTCACAGTCG-3′ |
| PLA2GVIa | 5′-ATGGGAAAGCAGGAGATGGT-3′ | 5′-TCATTTCAGCACACCCCTTC-3′ |
Statistical analysis
All comparisons were tested for statistical significance (P < .05) via the unpaired t test, using GraphPad Prism 3.0 software (GraphPad Software, San Diego, CA).
Results
Identification of markers discriminating between monocyte- and granulocyte-oriented splenic CD11b+Gr-1+ MSCs
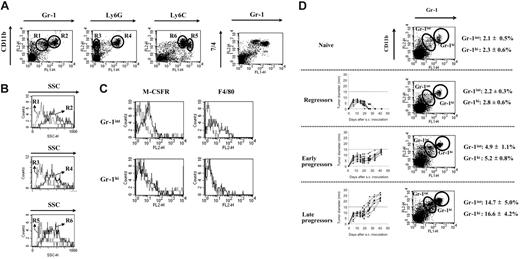
Although the heterogeneity of splenic immature CD11b+Gr-1+ MSCs in mice with progressing tumors has been recognized,8 this population has up to now been studied as one entity. In order to refine the composition of MSCs, we used the BW-Sp3 T-cell lymphoma model for which the expansion of CD11b+Gr-1+ cells in the spleens of AKR mice with progressing tumors (progressors) has been reported.15 FACS analysis on freshly isolated splenocytes from 5- to 7-week tumor-bearing mice revealed that, within the CD11b+Gr-1+ fraction, 2 populations exist, differing mainly in the Gr-1 and to a lesser extent also the CD11b expression level (Figure 1A). Of interest, cells staining intermediately for Gr-1 and high for CD11b (CD11bhiGr-1int; gate R1) had a low SSC profile (SSClo), suggestive of monocyte-oriented myeloid cells, while the Gr-1 high expressers that were also the highest CD11b expressers (CD11bhi+Gr-1hi; gate R2) had a higher SSC (SSChi), suggestive of a granulocytic cell type (Figure 1B). To further establish the nature of both MSC subpopulations, additional markers with a monocyte/granulocyte discriminative potential were evaluated. In the first instance, Ly6G and Ly6C seemed interesting candidates, since the anti–Gr-1 antibody (RB6-8C5) recognizes a shared epitope on these molecules.29 Ly6G is specifically expressed on granulocytic cells,29 while Ly6C expression is higher on immature monocytic than on granulocytic cells.30 It was demonstrated that the CD11bhi SSClo cells did not express the Ly6G marker (gate R3), but were the highest expressers of Ly6C (gate R5) (Figure 1A-B), corroborating their monocytic lineage. In contrast, the CD11bhi+ SSChi population was uniformly positive for Ly6G (gate R4) and Ly6C (gate R6), the latter being expressed at a somewhat lower level compared with the CD11bhi SSClo cells. Hence, the CD11bhi+ SSChi cells displayed typical characteristics of the granulocyte lineage.
Taylor et al31 used the 7/4 antigen to define myeloid cell heterogeneity in the spleen, with high expression levels on Gr-1hi neutrophils, but even higher levels on Gr-1int monocytes. Accordingly, splenic Gr-1int MSCs of BW-Sp3 progressors expressed higher 7/4 levels than the Gr-1hi MSCs, strengthening their identification as monocytic and granulocytic cells, respectively (Figure 1A).
Final evidence for the distinction between both MSC populations came from the finding that the M-CSF receptor (M-CSFR), a prototypic monocyte/macrophage-lineage marker, was expressed only on the Gr-1int population (Figure 1C). In addition, F4/80 expression—another monocyte/macrophage lineage marker—was detected only on the Gr-1int cells, although not all of these cells scored positive (Figure 1C), a finding that was also reported for peripheral blood monocytes.32 Altogether, our data subdivide the CD11b+Gr-1+ splenic MSCs into CD11bhi Gr-1int SSClo Ly6Gneg Ly6Chi+ 7/4hi+ M-CSFRint monocytic cells and CD11bhi+Gr-1hiSSChiLy6GhiLy6Chi7/4hiM-CSFRneg granulocytic cells.
Finally, it should be remarked that these subpopulations already existed at low percentages in naive spleens (Figure 1D) and that both were equally expanded in function of tumor load. Subcutaneous BW-Sp3 inoculation can result in a dramatically different tumor outcome in different recipients, going from complete tumor rejection in some mice to tumor progression in others.15,27-28,33 At 6 weeks after tumor inoculation, no expansion of MSCs was seen in mice that had rejected their tumor (regressors); an intermediate expansion (2- to 3-fold) in mice whose tumors only recently started to progress after a stabilization phase (early progressors); and a strong expansion (6- to 8-fold) in mice with a longer history of progression and larger tumors (late progressors) (Figure 1D).
Splenic CD11b+Gr-1+ cells from mice with progressing tumors consist of a monocytic and a granulocytic fraction. Splenocytes from BW-Sp3 tumor-bearing mice with progressing tumors (average tumor diameter ± 20 mm), 5 to 7 weeks after inoculation, were labeled with PE- or FITC-conjugated mAbs recognizing surface antigens for FACS analysis. (A) Dot plots representing costainings of PE-labeled anti-CD11b with FITC-labeled anti–Gr-1, anti-Ly6G, or anti-Ly6C. To detect 7/4 expression, cells were incubated with purified anti-7/4 mAb, followed by PE-labeled anti–rat IgG. Isotype-matched control mAbs did not significantly stain the cells (data not shown). (B) SSC profiles of cells gated in regions R1 (dotted line) versus R2 (full line); R3 (dotted line) versus R4 (full line); and R5 (dotted line) versus R6 (full line). (C) Splenocytes were coincubated with FITC-labeled anti–Gr-1 and PE-labeled anti-F4/80. To detect M-CSFR expression, cells were incubated with purified anti–M-CSFR mAb, followed by PE-labeled anti–rat IgG. Histograms represent isotype (dotted line) and antigen-specific (full line) staining on the gated Gr-1int and Gr-1hi populations. Profiles are representative of 1 progressor spleen of at least 5 investigated. (D) Six weeks after tumor inoculation, a subdivision was made between mice that rejected their tumor (regressors), mice with a short history of tumor progression and an average tumor diameter 15 mm or smaller (early progressors), or mice with a longer history of tumor progression and average tumor diameter of more than 15 mm (late progressors) before death. Tumor growth curves of individual mice in each group are shown. Splenocytes from a minimal of 5 mice in each group were stained with PE-labeled anti-CD11b and FITC-labeled anti–Gr-1. A dot plot from one mouse, representative for each group, is given. Average percentages ± SD of Gr-1int and Gr-1hi cells within the total splenocyte population are shown for each group.
Splenic CD11b+Gr-1+ cells from mice with progressing tumors consist of a monocytic and a granulocytic fraction. Splenocytes from BW-Sp3 tumor-bearing mice with progressing tumors (average tumor diameter ± 20 mm), 5 to 7 weeks after inoculation, were labeled with PE- or FITC-conjugated mAbs recognizing surface antigens for FACS analysis. (A) Dot plots representing costainings of PE-labeled anti-CD11b with FITC-labeled anti–Gr-1, anti-Ly6G, or anti-Ly6C. To detect 7/4 expression, cells were incubated with purified anti-7/4 mAb, followed by PE-labeled anti–rat IgG. Isotype-matched control mAbs did not significantly stain the cells (data not shown). (B) SSC profiles of cells gated in regions R1 (dotted line) versus R2 (full line); R3 (dotted line) versus R4 (full line); and R5 (dotted line) versus R6 (full line). (C) Splenocytes were coincubated with FITC-labeled anti–Gr-1 and PE-labeled anti-F4/80. To detect M-CSFR expression, cells were incubated with purified anti–M-CSFR mAb, followed by PE-labeled anti–rat IgG. Histograms represent isotype (dotted line) and antigen-specific (full line) staining on the gated Gr-1int and Gr-1hi populations. Profiles are representative of 1 progressor spleen of at least 5 investigated. (D) Six weeks after tumor inoculation, a subdivision was made between mice that rejected their tumor (regressors), mice with a short history of tumor progression and an average tumor diameter 15 mm or smaller (early progressors), or mice with a longer history of tumor progression and average tumor diameter of more than 15 mm (late progressors) before death. Tumor growth curves of individual mice in each group are shown. Splenocytes from a minimal of 5 mice in each group were stained with PE-labeled anti-CD11b and FITC-labeled anti–Gr-1. A dot plot from one mouse, representative for each group, is given. Average percentages ± SD of Gr-1int and Gr-1hi cells within the total splenocyte population are shown for each group.
The monocytic fraction of splenic CD11b+Gr-1+ MSCs is sufficient for the generation of CTL-suppressive macrophages in vitro
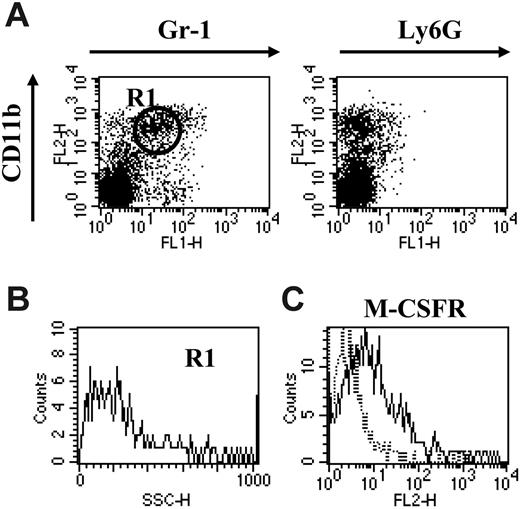
CD11b+Gr-1+ MSCs have been shown before to be responsible for the suppression of anti–BW-Sp3 CTLs upon in vitro culture.15 Having established the heterogeneity of splenic MSCs, we next wondered whether both subpopulations were strictly needed for this suppressive activity. Upon administration of anti–Gr-1 mAb to BW-Sp3 late progressors, the majority of the CD11b+Gr-1+ cells remaining in the spleen 24 hours after treatment were of the CD11bhiGr-1int phenotype (gate R1), lacking Ly6G (Figure 2A), with low SSC (Figure 2B) but expressing M-CSFR (Figure 2C), reminiscent of the monocytic MSC fraction. In quantitative terms, the percentage of splenic monocytic CD11b+Gr-1+ cells in the anti–Gr-1–treated mice (13.2% ± 3.1%) was on average in the same range as in untreated progressor mice (14.7% ± 5.0%).
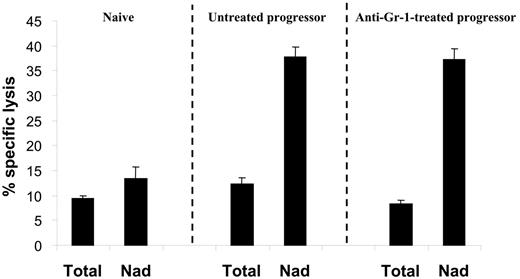
In order to assess the impact of granulocyte depletion on the suppression of tumor-specific CTL activity, splenocytes from either naive, untreated progressor, or anti–Gr-1–treated progressor mice were restimulated in vitro with irradiated BW-Sp3(B7-1) cells for 5 days before testing the cytotoxic activity. In both anti–Gr-1–treated and untreated progressor spleens, CTL activity was as low as in naive spleens (Figure 3, Total). However, as opposed to naive spleens, strong anti–BW-Sp3 CTL activity was recovered by dissociating the plastic-nonadherent cells from the plastic-adherent splenocyte fraction in both types of progressors (Figure 3, Nad), reflecting the presence of CTL-suppressive cells in the progressor adherent population. Hence, the monocyte fraction of the CD11b+Gr-1+ splenocytes seemed to be sufficient for the generation of suppressive adherent cells in vitro.
Anti–Gr-1 treatment specifically depletes the granulocytic fraction of splenic CD11b+Gr-1+cells. (A) BW-Sp3 late progressors (tumor diameter ± 20 mm), 5 to 7 weeks after inoculation, were injected intraperitoneally with 100 μg purified and LPS-free anti–Gr-1 antibodies, and 24 hours later splenocytes were stained with PE-labeled anti-CD11b and FITC-labeled anti–Gr-1 or anti-Ly6G. Surface staining profiles are represented in dot plots. (B) SSC profile of cells gated in R1. (C) Cells prepared in panel A were incubated with purified anti-MCSFR, followed by PE-labeled anti–rat IgG and FITC-labeled anti–Gr-1. Histograms represent isotype (dotted line) and M-CSFR–specific (full line) staining on the Gr-1int–gated population. Profiles shown in panels A-C are representative of one anti–Gr-1–treated spleen of 4 investigated.
Anti–Gr-1 treatment specifically depletes the granulocytic fraction of splenic CD11b+Gr-1+cells. (A) BW-Sp3 late progressors (tumor diameter ± 20 mm), 5 to 7 weeks after inoculation, were injected intraperitoneally with 100 μg purified and LPS-free anti–Gr-1 antibodies, and 24 hours later splenocytes were stained with PE-labeled anti-CD11b and FITC-labeled anti–Gr-1 or anti-Ly6G. Surface staining profiles are represented in dot plots. (B) SSC profile of cells gated in R1. (C) Cells prepared in panel A were incubated with purified anti-MCSFR, followed by PE-labeled anti–rat IgG and FITC-labeled anti–Gr-1. Histograms represent isotype (dotted line) and M-CSFR–specific (full line) staining on the Gr-1int–gated population. Profiles shown in panels A-C are representative of one anti–Gr-1–treated spleen of 4 investigated.
Monocytic CD11b+Gr-1+ cells are sufficient for the generation of CTL-suppressive cells in vitro. Splenocytes from either naive AKR or tumor size–matched BW-Sp3 progressors, with or without anti–Gr-1 treatment, were restimulated in vitro with irradiated BW-Sp3(B7-1) cells for 5 days. To test the function of adherent cells, a comparison was made between untouched cultures (Total) or cultures where the nonadherent cells were removed from the adherent population after 2 days, followed by another 3-day restimulation of the nonadherent cells (Nad). The cytotoxic activity in all cultures was tested using 111In-labeled BW-Sp3(B7-1) cells as targets at a 100:1 effector-target ratio. Results show the average 111In-release ± SD of triplicates. Similar results were obtained from 3 independent experiments.
Monocytic CD11b+Gr-1+ cells are sufficient for the generation of CTL-suppressive cells in vitro. Splenocytes from either naive AKR or tumor size–matched BW-Sp3 progressors, with or without anti–Gr-1 treatment, were restimulated in vitro with irradiated BW-Sp3(B7-1) cells for 5 days. To test the function of adherent cells, a comparison was made between untouched cultures (Total) or cultures where the nonadherent cells were removed from the adherent population after 2 days, followed by another 3-day restimulation of the nonadherent cells (Nad). The cytotoxic activity in all cultures was tested using 111In-labeled BW-Sp3(B7-1) cells as targets at a 100:1 effector-target ratio. Results show the average 111In-release ± SD of triplicates. Similar results were obtained from 3 independent experiments.
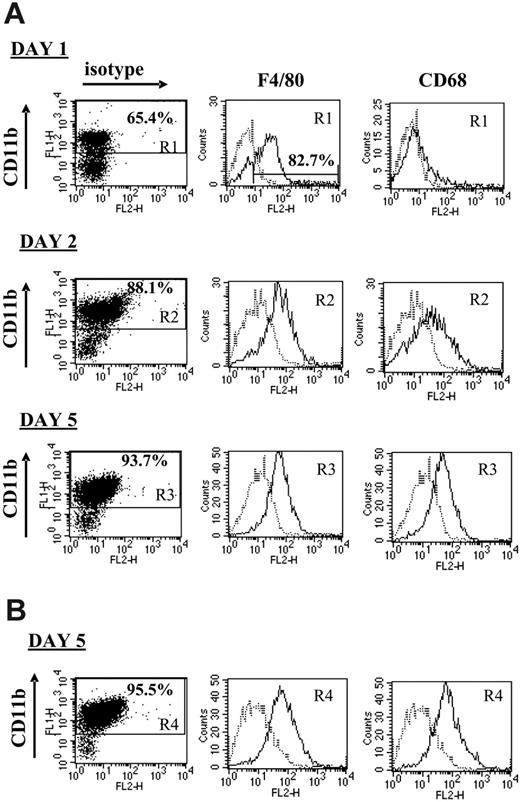
Monocytes are the precursors of both dendritic cells (DCs) and macrophages, for each of which suppressive/tolerogenic subpopulations have been described.19,34 It was therefore of interest to evaluate the phenotype of the plastic-adherent suppressor cells, using the prototypical macrophage markers F4/80 and CD68, as an indicator of mature elicited macrophages, and the DC marker CD11c. After 1 day of culture of untreated progressor splenocytes, about 65% of the adherent cells were CD11bhi, the majority of which (82.68% in the experiment shown in Figure 4A) were also positive for F4/80. However, hardly any CD68 expression could be detected on these cells at this stage, suggesting that no fully matured macrophages had been formed yet (Figure 4A). By the next day, the adherent splenocytes consisted of more than 85% CD11bhi cells, almost all of which were mature macrophages uniformly coexpressing F4/80 and CD68, a phenotype that remained unchanged until day 5 (Figure 4A). No CD11c+ DCs could be detected at any time point, suggesting that the splenic CD11b+Gr-1+ cells exclusively differentiated into macrophages under these culture conditions (data not shown). Culturing the granulocyte-depleted progressor splenocytes for 5 days resulted in a similar formation of CD11bhiF4/80hiCD68hiCD11cneg mature macrophages, demonstrating that the monocytic CD11b+Gr-1+ fraction was sufficient as precursors for these macrophages (Figure 4B).
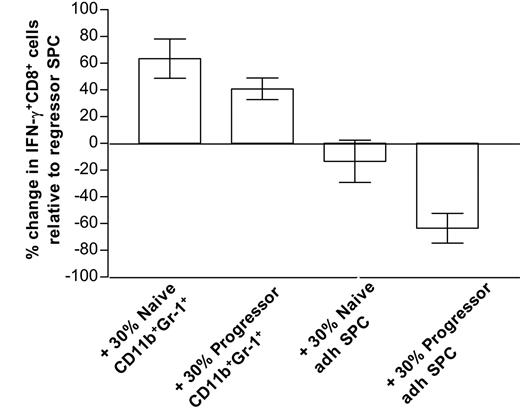
Overall, our data firmly establish an association between suppression and the in vitro generation of mature macrophages but do not exclude a role for the CD11b+Gr-1+ precursors in CTL dysfunction. To directly compare the suppressive activity of both types of cells, a short-term assay was mandatory to preclude the possibility that CD11b+Gr-1+ cells would differentiate during the time course of the experiment. Therefore, purified CD11b+Gr-1+ cells or 5-day adherent splenocytes from either naive mice or late progressors were added at a 30% ratio to anti-CD3–stimulated regressor splenocytes (which contain activated antitumor CTLs, but only low amounts of CD11b+Gr-1+ cells, Figure 1D), and 6 hours later IFN-γ production by CD8+ cells was tested via intracellular FACS staining. Of interest, only late progressor—but not naive—adherent splenocytes significantly suppress the amount of IFN-γ–producing cells, while both type CD11b+Gr-1+ cells rather stimulate CD8+ IFN-γ production (Figure 5). Hence, under the given conditions, in vitro–differentiated macrophages from progressor spleens seem to be superior in suppressing T-cell functions compared with their naive counterparts or CD11b+Gr-1+ precursors.
CTL-suppressive macrophages are M2 oriented
Depending on the environment they encounter, macrophages can evolve into a continuum of distinct subsets, of which M1 and M2 are the 2 extremes. Of note, we previously reported a high arginase activity in the suppressor cells of the BW-Sp3 tumor model,15 suggesting an M2 orientation. To further expand this finding, we quantified—using real-time PCR—the expression of selected M2-associated genes, comparing the 5-day plastic-adherent cells from restimulated late progressor and naive spleens. The latter population also contained about 85% F4/80hiCD68hi macrophages (data not shown), making its composition comparable with the progressor cells. The tested genes were selected from a collection of M2-associated transcripts, obtained in our lab from a subtracted cDNA library between M1 and M2 elicited in a Trypanosoma brucei brucei infection model.35
Table 2 (“P/N at least 2”) demonstrates that many of these M2-associated genes were at least 2-fold up-regulated in the progressor adherent splenocytes compared with the naive. Namely, the high induction of well-documented murine M2 markers, such as arginase-1,16 fibronectin-1,36 and especially Ym35 and FIZZ1,35 strengthens the notion that these cells were strongly M2 polarized. The presence of other IL-4– and/or IL-13–inducible genes (CCL24,37 E-cadherin,38 MGL-1,39 MGL-2,39 coagulation factor XIIIa,40 PD-L2,41 macrophage mannose receptor,42 cysteinyl leukotriene R1,43 and PPARγ40,44 ) further contributed to the M2 nature of the MSC-derived macrophages and probably reflected the enhanced production of these cytokines in the progressor versus the naive splenocyte cultures (data not shown). Other genes listed in Table 2 (“P/N ≥ 2”) had not been linked to M2 cells before, although functionally most of them fit within some recurrent themes, which might partially overlap and represent known functions of M2: genes involved in anti-inflammation (selenoprotein P,45 coagulation factor XIIIa,46 PLA2GVII/PAF-AH,47 PPARγ48 ), wound healing (fibronectin-1,36 arginase-1,16 coagulation factor XIIIa49 ), lipid metabolism (FIZZ1,50 prosaposin,51 PPARγ52 ) and the related arachidonic acid metabolism (PLA2GIVa,53 PLA2GVII/PAF-AH,47 cysteinyl leukotriene R1,43 PPARγ48 ), Th2-associated chemotaxis (CCL8,54 CCL2437 ), and pattern recognition receptors of the C-type lectin family (MGL-1,55 MGL-2,55 macrophage mannose receptor42 ).
CTL-suppressive macrophages are M2 oriented
Gene product . | Accession no. . | P/N . | Function . |
|---|---|---|---|
| P/N at least 2 | |||
| FIZZ1 | NM_020509 | 438.3 ± 245.9 | Lipid and sugar metabolism |
| Ym | NM_009892 (Ym1); NM_145126 (Ym2) | 357.3 ± 163.3 | Lectin |
| Fibronectin-1 | NM_010233 | 101.3 ± 6.9 | Wound healing |
| Arginase-1 | NM_007482 | 95.6 ± 39.4 | Wound healing, immunomodulation/ROS production |
| CCL8 (MCP-2) | NM_021443 | 17.7 ± 5.2 | Th2-type chemotaxis |
| CCL24 (eotaxin-2) | NM_019577 | 12.0 ± 5.2 | Th2-type chemotaxis |
| E-cadherin | NM_009864 | 11.9 ± 6.6 | Cell-cell adhesion |
| MGL-1 | NM_010796 | 10.3 ± 2.0 | C-type lectin (pattern recognition) |
| MGL-2 | NM_145137 | 9.5 ± 0.8 | C-type lectin (pattern recognition) |
| Selenoprotein P | NM_009155 | 8.5 ± 2.5 | Anti-inflammation |
| Coagulation factor XIIIa | NM_028784 | 5.0 ± 1.2 | Wound healing |
| Retinoic acid Rγ | NM_011244 | 4.7 ± 2.1 | Transcription factor |
| PD-L2 | NM_021396 | 4.6 ± 2.0 | Immunomodulation |
| TREM-2b | AY024349 | 4.6 ± 2.5 | Pattern recognition |
| PLA2GIVa | NM_008869 | 4.1 ± 1.4 | cPLA2; arachidonic acid metabolism |
| Cathepsin L | NM_009984 | 3.7 ± 0.9 | Antigen processing |
| PLA2GVII/PAF-AH | NM_013737 | 3.5 ± 0.8 | Anti-inflammation (arachidonic acid metabolism) |
| Macrophage mannose receptor | NM_008625 | 3.3 ± 1.0 | C-type lectin (pattern recognition) |
| Prosaposin | NM_011179 | 2.9 ± 0.6 | Sphingolipid metabolism |
| Cysteinyl leukotriene R1 | NM_021476 | 2.7 ± 0.6 | Immunomodulation (arachidonic acid metabolism) |
| PPARγ | NM_011146 | 2.4 ± 0.4 | Lipid metabolism, anti-inflammation (arachidonic acid metabolism) |
| P/N less than 2 | |||
| iNOS | NM_010927 | 0.6 ± 0.2 | NO/ROS production |
| 12/15-lipoxygenase | NM_007440 | 0.8 ± 0.3 | Anti-inflammation (arachidonic acid metabolism) |
| PLA2GIId | NM_011109 | 0.9 ± 0.2 | sPLA2, arachidonic acid metabolism |
| PLA2GV | NM_011110 | 1.5 ± 0.5 | sPLA2, arachidonic acid metabolism |
| PLA2GXIIa | NM_023196 | 1.6 ± 0.4 | sPLA2, arachidonic acid metabolism |
| PLA2GVIa | NM_016915 | 2.0 ± 0.5 | iPLA2, arachidonic acid metabolism |
Gene product . | Accession no. . | P/N . | Function . |
|---|---|---|---|
| P/N at least 2 | |||
| FIZZ1 | NM_020509 | 438.3 ± 245.9 | Lipid and sugar metabolism |
| Ym | NM_009892 (Ym1); NM_145126 (Ym2) | 357.3 ± 163.3 | Lectin |
| Fibronectin-1 | NM_010233 | 101.3 ± 6.9 | Wound healing |
| Arginase-1 | NM_007482 | 95.6 ± 39.4 | Wound healing, immunomodulation/ROS production |
| CCL8 (MCP-2) | NM_021443 | 17.7 ± 5.2 | Th2-type chemotaxis |
| CCL24 (eotaxin-2) | NM_019577 | 12.0 ± 5.2 | Th2-type chemotaxis |
| E-cadherin | NM_009864 | 11.9 ± 6.6 | Cell-cell adhesion |
| MGL-1 | NM_010796 | 10.3 ± 2.0 | C-type lectin (pattern recognition) |
| MGL-2 | NM_145137 | 9.5 ± 0.8 | C-type lectin (pattern recognition) |
| Selenoprotein P | NM_009155 | 8.5 ± 2.5 | Anti-inflammation |
| Coagulation factor XIIIa | NM_028784 | 5.0 ± 1.2 | Wound healing |
| Retinoic acid Rγ | NM_011244 | 4.7 ± 2.1 | Transcription factor |
| PD-L2 | NM_021396 | 4.6 ± 2.0 | Immunomodulation |
| TREM-2b | AY024349 | 4.6 ± 2.5 | Pattern recognition |
| PLA2GIVa | NM_008869 | 4.1 ± 1.4 | cPLA2; arachidonic acid metabolism |
| Cathepsin L | NM_009984 | 3.7 ± 0.9 | Antigen processing |
| PLA2GVII/PAF-AH | NM_013737 | 3.5 ± 0.8 | Anti-inflammation (arachidonic acid metabolism) |
| Macrophage mannose receptor | NM_008625 | 3.3 ± 1.0 | C-type lectin (pattern recognition) |
| Prosaposin | NM_011179 | 2.9 ± 0.6 | Sphingolipid metabolism |
| Cysteinyl leukotriene R1 | NM_021476 | 2.7 ± 0.6 | Immunomodulation (arachidonic acid metabolism) |
| PPARγ | NM_011146 | 2.4 ± 0.4 | Lipid metabolism, anti-inflammation (arachidonic acid metabolism) |
| P/N less than 2 | |||
| iNOS | NM_010927 | 0.6 ± 0.2 | NO/ROS production |
| 12/15-lipoxygenase | NM_007440 | 0.8 ± 0.3 | Anti-inflammation (arachidonic acid metabolism) |
| PLA2GIId | NM_011109 | 0.9 ± 0.2 | sPLA2, arachidonic acid metabolism |
| PLA2GV | NM_011110 | 1.5 ± 0.5 | sPLA2, arachidonic acid metabolism |
| PLA2GXIIa | NM_023196 | 1.6 ± 0.4 | sPLA2, arachidonic acid metabolism |
| PLA2GVIa | NM_016915 | 2.0 ± 0.5 | iPLA2, arachidonic acid metabolism |
Splenocytes from 3 individual BW-Sp3 progressors and 2 individual naive mice were restimulated in vitro with irradiated BW-Sp3(B7-1) for 5 days, after which RNA was extracted and cDNA prepared from the adherent splenocytes. Expression of each of the listed genes was determined via quantitative real-time PCR and normalized for the housekeeping gene ribosomal protein S12. The fold induction of gene expression (mean ± SD of 3 individual mice) in progressor compared with naive adherent splenocytes (mean ± SD of 2 individual mice) is shown (P/N).
P/N at least 2 indicates all tested genes with a minimal 2-fold induction in progressor adherent splenocytes, a criterion that is not met by the genes listed under P/N less than 2.
The CTL-suppressive adherent splenocyte population largely consists of mature macrophages. Splenocytes from either untreated (A) or anti–Gr-1–treated (B) 5- to 7-week BW-Sp3 progressors were restimulated in vitro with irradiated BW-Sp3(B7-1). After 1, 2, or 5 days of culture, nonadherent splenocytes were washed away and adherent cells were recovered. Each adherent cell population was incubated with FITC-labeled anti-CD11b, in combination with PE-labeled isotype control, anti-F4/80, anti-CD68, or anti-CD11c. Histograms represent isotype (dotted line) and surface Ag–specific (full line) staining within the gated CD11b+ population (gate R1, R2, R3 or R4, depending on the condition tested). One of 3 representative experiments is shown.
The CTL-suppressive adherent splenocyte population largely consists of mature macrophages. Splenocytes from either untreated (A) or anti–Gr-1–treated (B) 5- to 7-week BW-Sp3 progressors were restimulated in vitro with irradiated BW-Sp3(B7-1). After 1, 2, or 5 days of culture, nonadherent splenocytes were washed away and adherent cells were recovered. Each adherent cell population was incubated with FITC-labeled anti-CD11b, in combination with PE-labeled isotype control, anti-F4/80, anti-CD68, or anti-CD11c. Histograms represent isotype (dotted line) and surface Ag–specific (full line) staining within the gated CD11b+ population (gate R1, R2, R3 or R4, depending on the condition tested). One of 3 representative experiments is shown.
Only progressor-adherent splenocytes suppress anti-CD3–stimulated IFN-γ production by CD8+cells. Splenocytes (SPCs) from mice that almost fully rejected their tumor (regressors) were stimulated for 6 hours with anti-CD3 (with Golgi blockade during the last 4 hours), either alone (control), in the presence of 30% purified splenic CD11b+Gr-1+ cells from naive AKR or late progressors, or in the presence of 30% 5-day adherent splenocytes (adh SPC) from naive AKR or late progressors. Subsequently, cells were stained with FITC-labeled anti-CD8, fixed, permeabilized, and stained intracellularly with PE-labeled anti–IFN-γ or isotype control. For each condition, the percentage of IFN-γ+ cells within the gated CD8+ population was calculated. Bars represent the percentage increase or decrease in the number of IFN-γ+CD8+ cells relative to control regressor splenocytes. Bars are averages ± SD from 2 experiments. Note that hardly any IFN-γ production could be seen in any condition without anti-CD3 stimulation.
Only progressor-adherent splenocytes suppress anti-CD3–stimulated IFN-γ production by CD8+cells. Splenocytes (SPCs) from mice that almost fully rejected their tumor (regressors) were stimulated for 6 hours with anti-CD3 (with Golgi blockade during the last 4 hours), either alone (control), in the presence of 30% purified splenic CD11b+Gr-1+ cells from naive AKR or late progressors, or in the presence of 30% 5-day adherent splenocytes (adh SPC) from naive AKR or late progressors. Subsequently, cells were stained with FITC-labeled anti-CD8, fixed, permeabilized, and stained intracellularly with PE-labeled anti–IFN-γ or isotype control. For each condition, the percentage of IFN-γ+ cells within the gated CD8+ population was calculated. Bars represent the percentage increase or decrease in the number of IFN-γ+CD8+ cells relative to control regressor splenocytes. Bars are averages ± SD from 2 experiments. Note that hardly any IFN-γ production could be seen in any condition without anti-CD3 stimulation.
PPARγ ligands and PLA2 inhibitors cooperate to diminish M2-associated CTL suppression
Next we wondered whether any of the M2-associated gene products, listed in Table 2 (“P/N at least 2”), had an impact on the CTL-suppressive mechanism of these macrophages. Hereto, splenocytes from 5- to 7-week tumor-bearing mice were restimulated in vitro for 5 days, with or without specific inhibitors to block M2 gene function added at day 2.
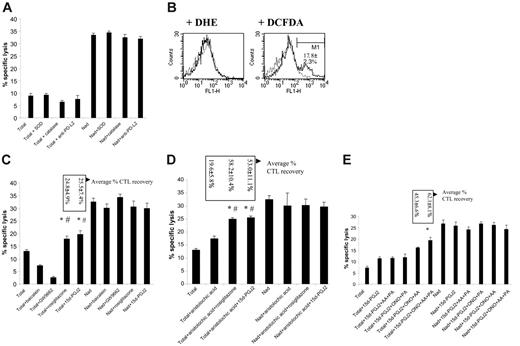
PPARγ ligands and PLA2 inhibitors cooperate to reverse M2-mediated CTL suppression. (A) Splenocytes from 5- to 7-week BW-Sp3 progressors were restimulated in vitro with irradiated BW-Sp3(B7-1), and a distinction was made between (1) CTL activity in a 5-day culture without inhibitors (Total). (2) CTL activity in 5-day cultures with inhibitors added at day 2 (Total + inhibitor). Inhibitors used were as follows: SOD (O2–), catalase (H2O2), and neutralizing anti–PD-L2 mAb. (3) CTL activity of nonadherent splenocytes, removed from the adherent fraction after 2 days of culture and restimulated in a fresh plate for another 3 days (Nad). (4) Nad cultures supplemented with inhibitors (Nad + inhibitor). The cytotoxic activity in all cultures was tested using 111In-labeled BW-Sp3(B7-1) cells as targets at a 100:1 effector-target ratio. Results show the average 111In-release ± SD of triplicates. One exemplary experiment is shown, but all inhibitors have been tested in at least 3 independent experiments with comparable results. (B) After 3 days of restimulation, without inhibitors, plastic-adherent cells were recovered from the plate and incubated with 2 μM DHE for 1 hour or 2 μM DCFDA for 30 minutes at 37°C. After washing, cells were labeled with anti-CD11b/PE and analyzed. Histograms represent gated CD11b+ cells without dye (dotted line), or with either DHE or DCFDA (full line) from one experiment. The average percentage of DCFDA-positive (gate M1) ± SD from 3 independent experiments is given. (C) Similar experiment to that in panel A. Inhibitors used were as follows: baicalein (12/15 LOX), GW9662 (PPARγ). Agonists used were as follows: rosiglitazone (PPARγ), 15d-PGJ2 (PPARγ). One exemplary experiment of 4 is shown. Boxes represent average percentage of CTL recovery ± SD of 4 experiments. *P < .05, percentage of CTL recovery higher comparing Total + PPARγ ligands versus Total + baicalein; #P < .05, percentage of CTL recovery higher comparing Total + PPARγ ligands versus Total + GW9662. (D) Similar experiment as panel A. Inhibitor used was as follows: aristolochic acid (PLA2). Agonists used were as follows: rosiglitazone (PPARγ), 15d-PGJ2 (PPARγ). Again, one exemplary experiment of 4 is shown. Boxes represent average percentage of CTL recovery ± SD of 4 experiments.*P < .05, percentage CTL recovery higher comparing Total + aristolochic acid + PPARγ ligands versus Total + PPARγ ligands; #P < .05, percentage CTL recovery higher comparing Total + aristolochic acid + PPARγ ligands versus Total + aristolochic acid. (E) Similar experiment to that in panel A. Inhibitors used were as follows: AACOCF3 (= AA; cPLA2), PACOCF3 (= PA; iPLA2), ONO-RS-082 (= ONO; sPLA2). Agonist used was as follows: 15d-PGJ2 (PPARγ). One exemplary experiment of 2 is shown. Boxes represent average percentage CTL recovery ± SD of 2 experiments. *P < .05, percentage CTL recovery higher comparing Total + 15d-PGJ2 + ONO + AA + PA versus Total + 15d-PGJ2.
PPARγ ligands and PLA2 inhibitors cooperate to reverse M2-mediated CTL suppression. (A) Splenocytes from 5- to 7-week BW-Sp3 progressors were restimulated in vitro with irradiated BW-Sp3(B7-1), and a distinction was made between (1) CTL activity in a 5-day culture without inhibitors (Total). (2) CTL activity in 5-day cultures with inhibitors added at day 2 (Total + inhibitor). Inhibitors used were as follows: SOD (O2–), catalase (H2O2), and neutralizing anti–PD-L2 mAb. (3) CTL activity of nonadherent splenocytes, removed from the adherent fraction after 2 days of culture and restimulated in a fresh plate for another 3 days (Nad). (4) Nad cultures supplemented with inhibitors (Nad + inhibitor). The cytotoxic activity in all cultures was tested using 111In-labeled BW-Sp3(B7-1) cells as targets at a 100:1 effector-target ratio. Results show the average 111In-release ± SD of triplicates. One exemplary experiment is shown, but all inhibitors have been tested in at least 3 independent experiments with comparable results. (B) After 3 days of restimulation, without inhibitors, plastic-adherent cells were recovered from the plate and incubated with 2 μM DHE for 1 hour or 2 μM DCFDA for 30 minutes at 37°C. After washing, cells were labeled with anti-CD11b/PE and analyzed. Histograms represent gated CD11b+ cells without dye (dotted line), or with either DHE or DCFDA (full line) from one experiment. The average percentage of DCFDA-positive (gate M1) ± SD from 3 independent experiments is given. (C) Similar experiment to that in panel A. Inhibitors used were as follows: baicalein (12/15 LOX), GW9662 (PPARγ). Agonists used were as follows: rosiglitazone (PPARγ), 15d-PGJ2 (PPARγ). One exemplary experiment of 4 is shown. Boxes represent average percentage of CTL recovery ± SD of 4 experiments. *P < .05, percentage of CTL recovery higher comparing Total + PPARγ ligands versus Total + baicalein; #P < .05, percentage of CTL recovery higher comparing Total + PPARγ ligands versus Total + GW9662. (D) Similar experiment as panel A. Inhibitor used was as follows: aristolochic acid (PLA2). Agonists used were as follows: rosiglitazone (PPARγ), 15d-PGJ2 (PPARγ). Again, one exemplary experiment of 4 is shown. Boxes represent average percentage of CTL recovery ± SD of 4 experiments.*P < .05, percentage CTL recovery higher comparing Total + aristolochic acid + PPARγ ligands versus Total + PPARγ ligands; #P < .05, percentage CTL recovery higher comparing Total + aristolochic acid + PPARγ ligands versus Total + aristolochic acid. (E) Similar experiment to that in panel A. Inhibitors used were as follows: AACOCF3 (= AA; cPLA2), PACOCF3 (= PA; iPLA2), ONO-RS-082 (= ONO; sPLA2). Agonist used was as follows: 15d-PGJ2 (PPARγ). One exemplary experiment of 2 is shown. Boxes represent average percentage CTL recovery ± SD of 2 experiments. *P < .05, percentage CTL recovery higher comparing Total + 15d-PGJ2 + ONO + AA + PA versus Total + 15d-PGJ2.
Arginase-1, in concert with iNOS, has been reported to be a major contributor to the production of T-cell suppressive ROSs by M2.11-14,20 However, arginase/iNOS-dependent ROS production was not involved in suppression mediated by splenic M2 in this tumor model.15 Yet, we could not a priori exclude a role for ROSs produced by other enzymes, such as NADPH oxidase. Therefore, we tested ROS production by progressor adherent splenocytes after 3 days of culture—at which time point these cells consisted mainly of mature macrophages—using the oxidation-sensitive dyes DHE (specific interaction with superoxide) and DCFDA (interaction with hydrogen peroxide, hydroxyl radicals, peroxynitrite, and to a lesser extent superoxide). DHE did not color these macrophages at all, while only a minority (on average 17.81% ± 2.34%) was stained by DCFDA, demonstrating the predominantly nonoxidative nature of these M2-oriented cells (Figure 6B). Accordingly, the addition of ROS-scavenging enzymes, such as SOD and catalase, to the CTL cultures was unable to reverse CTL suppression (Figure 6A).
PD-L2 is another M2-associated molecule with a known function in inhibiting T-cell activation, through direct interaction with the inhibitory receptor PD-1.56 PD-L2 mRNA is up-regulated in the splenic suppressor macrophages (Table 2 “P/N at least 2”), resulting in a readily detectable surface expression of the protein (data not shown). However, addition of neutralizing anti–PD-L2 antibodies to the cultures did not recover CTL activity, excluding PD-L2 as a candidate suppressor molecule in this model (Figure 6A).
Another possible pathway of T-cell inactivation is initiated by the IL-4–induced macrophage 12/15-lipoxygenase (LOX) activity, thereby generating potential ligands for PPARγ, a member of the ligand-dependent nuclear receptor family.57 The mRNA for both 12/15-LOX and PPARγ was detected in the progressor adherent splenocytes, though only PPARγ expression was up-regulated compared with naive adherent splenocytes (Table 2 “P/N at least 2”). To assess this putative suppressive mechanism, CTL activity was investigated in the presence of baicalein, a selective 12/15-LOX inhibitor, or of the PPARγ antagonist GW9662. Of interest, as shown in Figure 6C, antitumor CTL activity was actually further decreased by both compounds. This effect could not be attributed to a direct toxicity of the compounds toward CTLs, since addition of baicalein or GW9662 to nonadherent splenocyte cultures did not alter the observed cytotoxicity (Figure 6C). This finding suggested that PPARγ stimulation in macrophages could be beneficial for CTL function. Therefore, potent synthetic and endogenous PPARγ-specific ligands—rosiglitazone and 15d-PGJ2, respectively—were tested for their impact on CTL activity. Of importance, the selected doses of the ligands did not affect the viability of the suppressor population (data not shown), nor did they have an impact on the CTL activity of nonadherent splenocyte cultures (Figure 6C). In the total splenocyte cultures, both ligands could partially restore CTL activity, but the effect remained limited to about 25% recovery (Figure 6C).
Endogenous PPARγ ligands are lipid mediators, generated through the arachidonic acid metabolism. The initiating regulatory enzymes of this pathway are the phospholipases A2, which are broadly classified into secretory (sPLA2), cytosolic Ca2+-dependent (cPLA2), and cytosolic Ca2+-independent (iPLA2) PLA2s.58 Of interest, though only the cPLA2 group IVa mRNA was up-regulated more than 2-fold in the suppressor macrophages (Table 2 “P/N at least 2”), other sPLA2 or iPLA2 members also tended to have a slightly higher expression in these cells (Table 2 “P/N less than 2”). Therefore, we wondered whether modulating PLA2 activity, either alone or in combination with PPARγ stimulation, influenced CTL activity. Supplementing the cultures with a nontoxic dose of a general PLA2 inhibitor, aristolochic acid, recovered CTL activity on average by 19.53% ± 5.79% (Figure 6D). However, in combination with rosiglitazone or 15d-PGJ2, the increase in cytotoxicity reached 58.19% ± 10.38% and 53.04% ± 11.10%, respectively (Figure 6D), demonstrating that PPARγ stimulation and PLA2 inhibition can cooperate in diminishing suppression by M2 cells.
Subsequently, we investigated whether the suppression-reversing effects of aristolochic acid could be attributed to the inhibition of a particular subfamily of PLA2. To this end, combinations of sPLA2-specific (ONO-RS-082), cPLA2-specific (AA-COCF3), and iPLA2-specific (PACOCF3) inhibitors were added to the CTL cultures together with 15d-PGJ2, and antitumor cytotoxicity was monitored. As shown in Figure 6E, a combination of ONO-RS-082, AACOCF3, and 15d-PGJ2 was minimally needed for the CTL activity to rise above the level seen with 15d-PGJ2 alone. Yet, the best recovery of CTLs was repeatedly noticed when combining all inhibitors with 15d-PGJ2 (Figure 6E), suggestingthat sPLA2, cPLA2, and iPLA2 activities contributed to PLA2-mediated suppression.
PPARγ ligands subvert CTL suppression by M2-oriented tumor-associated macrophages
We next asked whether similar CTL-suppressive mechanisms would apply to freshly isolated tumor-associated macrophages (TAMs), since at least part of these cells was reported to be derived from the in vivo differentiation of CD11b+Gr-1+ splenocytes.12 To isolate TAMs, tumor single-cell suspensions were made and the CD11b+ fraction was collected via magnetic sorting. About 95% of the viable CD11b+ tumor-associated cells were F4/80hiCD68hi, identifying them as mature macrophages (data not shown). In order to discern specific characteristics of TAMs, these cells were compared with tumor-distal F4/80hi peritoneal macrophages from the same tumor-bearing mice.
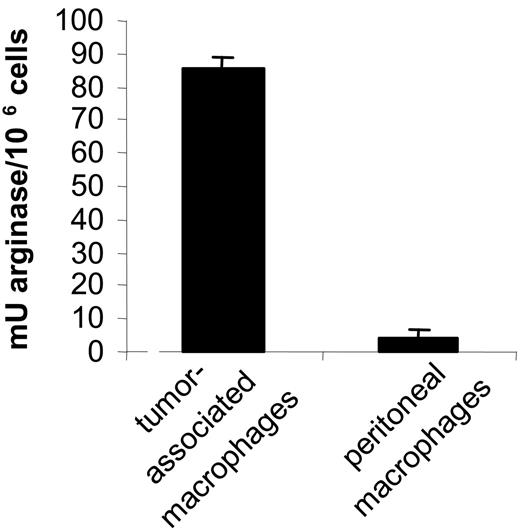
In the first instance, we determined the activation state of TAMs. Therefore, the expression of a selected group of genes was assessed in both types of macrophages via real-time PCR. Clearly, the most prominent M2 markers—FIZZ1, Ym, fibronectin-1, and arginase-1—were all several fold higher in tumor-associated macrophages (Table 3). This finding was corroborated by an elevated arginase enzymatic activity in TAM versus peritoneal macrophages (Figure 7), altogether suggesting that the tumor-infiltrating macrophages had a more pronounced M2 phenotype than their tumordistal counterparts.
Tumor-associated macrophages are M2 oriented
M2 marker genes . | Tumor/peritoneal macrophages . |
|---|---|
| FIZZ1 | 16.3 ± 6.1 |
| Ym | 9.9 ± 3.6 |
| Fibronectin-1 | 4.8 ± 1.5 |
| Arginase-1 | 21.3 ± 9.7 |
| PPARγ | 8.9 ± 4.6 |
M2 marker genes . | Tumor/peritoneal macrophages . |
|---|---|
| FIZZ1 | 16.3 ± 6.1 |
| Ym | 9.9 ± 3.6 |
| Fibronectin-1 | 4.8 ± 1.5 |
| Arginase-1 | 21.3 ± 9.7 |
| PPARγ | 8.9 ± 4.6 |
Tumor-associated and peritoneal macrophages from 2 tumor-bearers were obtained via magnetic sorting using anti-CD11b microbeads, after which RNA was extracted and cDNA prepared from these samples. Expression of each of the listed genes was determined via quantitative real-time PCR and normalized for the housekeeping gene ribosomal protein S12. The fold induction of gene expression (mean ± SD of 2 individual mice) in tumor-associated macrophages compared with peritoneal macrophages (mean ± SD of 2 individual mice) is shown.
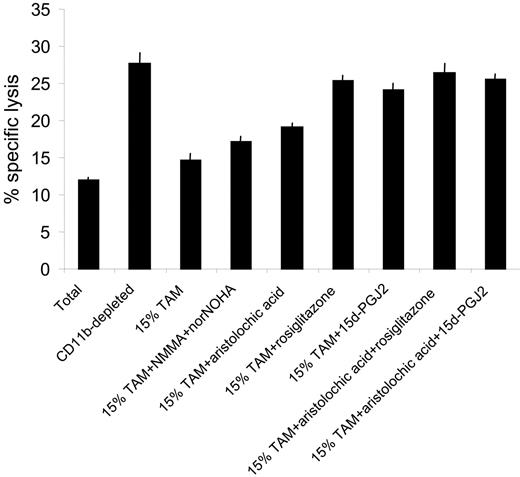
Next we asked whether the M2-oriented TAMs were suppressive for antitumor CTLs and whether suppression could be modulated by PPARγ ligands and PLA2 inhibitors. In this context, it is important to note that the PPARγ gene is more prominently expressed in TAMs compared with tumor-distal macrophages (Table 3). Hence, splenocytes from progressors were first depleted of CD11b+ cells via MACS, replenished with 15% freshly purified TAMs, and restimulated in vitro for 5 days, with or without PPARγ ligands and/or PLA2 inhibitors. Figure 8 demonstrates that depletion of CD11b+ cells resulted in a greatly enhanced CTL activity in progressor spleens, but addition of 15% TAMs reduced the cytotoxicity almost back to the level seen in the total splenocyte culture. Reminiscent of the suppressive mechanism by in vitro–generated M2 macrophages, L-NMMA/norNOHA supplementation had only a modest effect on CTL activity and aristolochic acid only partially reversed TAM-mediated suppression (Figure 8). However, addition of rosiglitazone or 15d-PGJ2, whether or not combined with aristolochic acid, almost completely restored CTL activity, suggesting that PPARγ stimulation could be an efficient tool to subvert TAM-mediated T-cell suppression.
Discussion
The accumulation of immature myeloid cells in the lymphoid organs and the blood is a widespread phenomenon in tumor-bearing mice and patients with advanced cancer.2-8 Functionally, these cells are associated with impaired T-cell responses and as such present a major hurdle for current immunotherapy trials.9-15 In mice, these myeloid cells are characterized as CD11b+Gr-1+ cells. Although morphologic analysis suggested that these 2 markers actually cover a mixture of granulocytes and monocytes-macrophages at different stages of differentiation,8 this knowledge was not translated into a subdivision of CD11b+Gr-1+ cells based on differential surface marker expression. In this study, we detected 2 main subpopulations within the splenic CD11b+Gr-1+ cells from mice with progressing tumors, both of which express high levels of CD11b but differ in SSC profile and expression of Gr-1, Ly6G, Ly6C, 7/4, and M-CSFR. One of the subpopulations is characterized by high expression levels of Gr-1 and Ly6G, which in combination with a high SSC are unique features of granulocytes/neutrophils.29,31,59 The other population, characterized as CD11bhiGr-1intSSCloLy6GnegLy6Chi+7/4hi+M-CSFRint, can be considered as cells belonging to the monocyte/macrophage lineage. Remarkably, the majority of these monocytic cells express high levels of Ly6C, indicative of immature monocytes.32 Also the other criteria for immature monocytes, as put forward by Sunderkötter et al,32 are met by these splenic Gr-1int cells: absence of CD16/32, MHC II, and CD11c; low levels of CD43; and high levels of CD62L (J.A.V.G., unpublished data, February 2005). Thus, the CD11b+Gr-1+ population, expanded in the spleen of mice with large tumors, consists of immature monocytic and granulocytic fractions.
Tumor-associated macrophages have a higher arginase activity than tumor-distal macrophages. Arginase activity was evaluated in 106 tumor-associated macrophages and peritoneal macrophages as described earlier.15 The mean of 2 individual mice ± SD is shown.
Tumor-associated macrophages have a higher arginase activity than tumor-distal macrophages. Arginase activity was evaluated in 106 tumor-associated macrophages and peritoneal macrophages as described earlier.15 The mean of 2 individual mice ± SD is shown.
PPARγ ligands subvert CTL suppression by tumor-associated macrophages. Splenocytes from 5- to 7-week BW-Sp3 progressors were restimulated in vitro with irradiated BW-Sp3(B7-1) for 5 days, and a distinction was made between (1) CTL activity in total splenocyte cultures (Total), (2) CTL activity in splenocytes magnetically depleted in CD11b+ cells (CD11b-depleted), (3) CTL activity in CD11b-depleted splenocytes, supplemented with 15% freshly prepared tumor-associated macrophages at the initiation of the culture (15% TAMs), and (4) CTL activity in CD11b-depleted splenocytes, supplemented with 15% freshly prepared tumor-associated macrophages and the appropriate inhibitors at the initiation of the culture (15% TAM + inhibitors). The cytotoxic activity in all cultures was tested using 111In-labeled BW-Sp3(B7-1) cells as targets at a 100:1 effector-target ratio. Results show the average 111In-release ± SD of triplicates.
PPARγ ligands subvert CTL suppression by tumor-associated macrophages. Splenocytes from 5- to 7-week BW-Sp3 progressors were restimulated in vitro with irradiated BW-Sp3(B7-1) for 5 days, and a distinction was made between (1) CTL activity in total splenocyte cultures (Total), (2) CTL activity in splenocytes magnetically depleted in CD11b+ cells (CD11b-depleted), (3) CTL activity in CD11b-depleted splenocytes, supplemented with 15% freshly prepared tumor-associated macrophages at the initiation of the culture (15% TAMs), and (4) CTL activity in CD11b-depleted splenocytes, supplemented with 15% freshly prepared tumor-associated macrophages and the appropriate inhibitors at the initiation of the culture (15% TAM + inhibitors). The cytotoxic activity in all cultures was tested using 111In-labeled BW-Sp3(B7-1) cells as targets at a 100:1 effector-target ratio. Results show the average 111In-release ± SD of triplicates.
CD11b+Gr-1+ cells can intrinsically differentiate, both in vitro and in vivo, to macrophages, DCs, and even endothelial cells.4,60 This notion becomes important when assessing the T-cell suppressive activity of CD11b+Gr-1+ cells via assays requiring in vitro culture, thereby possibly initiating the differentiation of these cells. For example, upon 24 hours of in vitro culture, CD11b+Gr-1+ cells suppress only CD8+ T-cell activity,9 but they acquire the ability to block CD4-mediated responses after several days in vitro.11 In our experimental set-up, F4/80+CD68+ macrophages make up the large majority of plastic-adherent cells as early as 2 days after initiating the culture. Of interest, CD68 surface expression, which has previously been associated with fully matured, elicited, or activated macrophages,61 seems to mark the transition from culture day 1 to day 2. The presence of low levels of GM-CSF and high levels of IL-6 and IL-10 in our cultures, skewing monocyte differentiation in the direction of macrophages,62-63 could be one explanation for the efficient macrophage differentiation (J.A.V.G., unpublished data, July 2001); the presence of irradiated cancer cells, which often block DC differentiation,64 could be another. In any case, the precursors of these macrophages reside within the monocytic fraction of the CD11b+Gr-1+ cells, since specific depletion of the granulocytic CD11b+Gr-1+ cells does not preclude the formation of these macrophages, while a depletion of all CD11b+Gr-1+ cells does.15 It should be remarked, however, that although the granulocytic CD11b+Gr-1+ cells are not strictly necessary for macrophage formation, our data do not exclude that some of these immature and relatively plastic cells could be turned into macrophages under the given culture conditions.
Antitumor CTL activity in these cultures is generally low but can be rescued when separating the CTLs from the plastic-adherent cells after 2 days of culture, suggesting an association between the presence of macrophages from day 2 onward and CTL suppression. Indeed, in granulocyte-depleted cultures, both macrophage formation and CTL suppression are intact. Direct proof of late progressor adherent splenocyte-mediated T-cell suppression came from the observation that these cells restrict the number of IFN-γ–producing CD8+ T cells upon short-term (6 hour) anti-CD3 stimulation. Remarkably, CD11b+Gr-1+ cells rather stimulate IFN-γ production in this setup, suggesting that freshly isolated, nondifferentiated CD11b+Gr-1+ cells are either not suppressive at all or at least unable to suppress anti-CD3–mediated T-cell activation.
We identified these CTL-suppressive macrophages as M2-oriented cells, based on the up-regulated expression (compared with naive adherent splenocytes) of well-established M2 markers, such as FIZZ1,35 Ym,35 fibronectin-1,36 arginase-1,16 and the MGLs,39 or other IL-4/IL-13–regulated genes (“Results”). This M2 skewing correlates with the detection of IL-4, IL-13, and IL-10 in the culture supernatant, while the M1-driving cytokine IFNγ is absent (J.A.V.G., unpublished data, July 2001). In addition, a number of novel M2 markers, not regulated by Th2 cytokines, but consistently found in in vivo–elicited M2 in different pathologies (PLA2 GVII/PAF-AH, prosaposin, TREM-2b)72 were also present. Of note, real-time PCR was performed on plastic-adherent populations enriched in macrophages (85%) but not on entirely pure macrophages. Therefore, we cannot a priori exclude that some of the genes listed in Table 2 (“P/N at least 2”) are also expressed in other cells besides myeloid cells or macrophages. However, we favor the notion that the observed differences in gene expression reflect mainly the differences between macrophage populations, since (1) all the listed genes are known to be expressed in myeloid cells, and, even more importantly, many of them (including Ym, PD-L2, TREM-2b, Cathepsin L, Macrophage mannose receptor, MGL-1, MGL-2) are predominantly expressed in myeloid cells such as macrophages and DCs, the latter of which are not present in our cultures;, and (2) all genes listed in Table 2 (“P/N at least 2”) have been, based on literature data and our own studies on peritoneal macrophages from a Trypanosoma brucei brucei infection model (G.H.G. and G.R., unpublished data), associated with M2-oriented macrophages. In addition, the most prominent genes were also up-regulated in purified TAMs.
Kusmartsev and Gabrilovich12 reported that splenic Gr-1+ cells are precursors of F4/80+ TAMs in vivo. Therefore, in order to correlate the characteristics of in vitro–generated CD11b+Gr-1+–derived M2 macrophages with the in vivo situation, we purified TAMs from mice with progressing BW-Sp3 tumors. In the first instance, it was important to find that TAMs were also M2 oriented, making a comparison between the in vitro and in vivo situation not far fetched. However, it should be noted that in a side-by-side comparison, most M2 markers have a significantly higher expression in the in vitro–generated macrophages compared with TAMs, suggesting that the 5-day in vitro culture strongly polarizes these cells.
Another point of comparison between both types of macrophages is their suppressive activity toward antitumor CTLs. Since macrophages make up a significant portion of BW-Sp3 tumors (J.A.V.G., unpublished data, June 2005), these cells could therefore have an important impact on intratumoral T-cell activity. Recently, the combined activity of arginase-1 and iNOS was shown, in a number of murine tumor models, to be important for the suppressive activity of tumor-infiltrating CD11b+ myeloid cells12 and splenic CD11b+Gr-1+ cells.11 By lowering l-arginine availability, arginase-1 can switch on the reductase domain of iNOS, thereby generating ROSs.11 Remarkably, neither the in vitro–generated splenic macrophages15 nor the TAMs from the BW-Sp3 model use this suppressive mechanism, despite high arginase enzyme activity in these cells. The reason for the discrepancy with other models is not clear, though it could be linked to the fact that the mouse AKR strain used in this study is strongly type-2 oriented and, hence, biased to the development of M2 cells with possibly different characteristics compared with C57Bl/6 or Balb/c M2.64 For example, enhanced arginase-1 activity and lowered l-arginine concentrations were shown to impede iNOS mRNA translation and protein stability in some macrophages.65
We describe the stimulation of PPARγ by synthetic (rosiglitazone) and endogenous (15d-PGJ2) ligands as a novel strategy to diminish CTL suppression by both in vitro–generated splenic M2 and TAMs. Of interest, the efficiency of this treatment on TAMs, representing true in vivo–differentiated macrophages, is much higher. In macrophages, stimulation of this nuclear receptor has profound anti-inflammatory effects,66-67 suggesting that inflammation-linked molecules could be responsible for TAM-mediated suppression. Several of such molecules, including STAT-1,12 IFNγ,12 TNF,12,68 and prostaglandins,68 have been implicated in TAM suppressive activity before, so further research will be needed to clarify this issue in this model.
In addition, when PPARγ stimulation alone has a limited effect—as is the case for the in vitro–generated M2—results could be improved by a combination treatment with PLA2 inhibitors. Note that the best results were obtained with a combination of cPLA2, sPLA2, and iPLA2 inhibitors. All 3 PLA2 types collaborate to maximize the mobilization of arachidonic acid,58 which is the precursor of the eicosanoids, in cells. Inhibition of PLA2 could therefore diminish the production of potentially T-cell–suppressive eicosanoids,69 but could also lead to an up-regulation of PPARγ transcripts,70 possibly explaining the cooperative action of both treatments.
Current data provide a rationale for the use of PPARγ ligands and/or PLA2 inhibitors to improve antitumor immunity, for example in immunotherapeutic settings. Of note, PPARγ ligands such as the synthetic thiazolidinediones (eg, rosiglitazone) are currently used to treat diabetes but have also demonstrated antitumor effects in preclinical models.71
Prepublished online as Blood First Edition Paper, March 9, 2006; DOI 10.1182/blood-2005-09-3777.
Supported by a “Prospective Research for Brussels” postdoctoral grant from the Brussels government (J.A.V.G.); by a doctoral grant from the Vrije Universiteit Brussel (S.M.); by a postdoctoral fellowship from the “Institute for Promotion and Innovation by Science and Technology in Flanders” (IWT-Vlaanderen) (G.R.); by a grant from IWT-Vlaanderen for “Generisch Basisonderzoek aan de Universiteiten” (IWT-GBOU); and by the “Fund for Scientific Research Flanders” (FWO-Vlaanderen).
J.A.V.G. designed and performed research, analyzed data, and wrote the paper; S.M., Y.L., L.B., and K.D.G. performed research; G.H.G. and G.R. designed and performed research; and P.D.B. designed research.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank Ella Omasta, Marie-Thérèse Detobel, and Eddy Vercauteren for excellent technical assistance; Alain Beschin for critically reviewing the paper; Dr Pieter Leenen for providing anti-MCSFR mAb; and Dr Gordon Brown for providing the 7/4 mAb.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal