In an article recently published in Blood, Elliott et al1 reported that commercially available anti–erythropoietin receptor (anti-EpoR) antibodies are hardly usable to predict EpoR expression because of their low specificity and affinity. Although we agree with the authors that many of these antibodies work poorly, we strongly disagree with them concerning 2 main points: (1) the ability of Santa Cruz Biotechnology C-20 antibody (Santa Cruz, CA) to efficiently detect the EpoR in Western blot (WB) analysis, and (2) the apparent molecular mass of the EpoR.
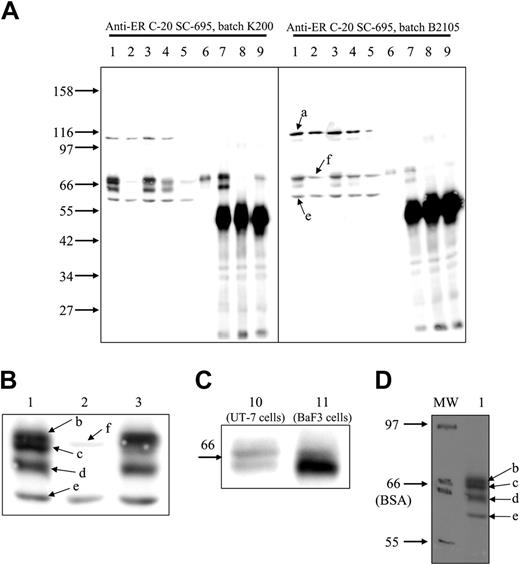
For a long time, we have been using C-20 anti-EpoR antibodies from Santa Cruz Biotechnology (catalog no. SC-695) to reveal EpoR in WB (see, for example, Verdier et al2 and Walrafen et al3 ), and we always detect EpoR with a high sensitivity and a good specificity. To demonstrate the efficiency of this antibody, we used UT-7 cells, since they express endogenous EpoR and they were used by Elliott et al.1 The Mo7E cell line was used as a negative control. In UT-7 cells, C-20 antibodies revealed 5 bands with molecular masses of 110 (a), 69.5 (b), 67.6 (c), 64 (d), and 58 kDa (e). In Mo7E cells, only bands a and e were clearly visible, with a faint band at 68.4 kDa (f) that was masked by bands b and c in UT-7 cells (Figure 1). We identified bands b, c, and d as the EpoR because they are not present in Mo7E extracts (Figure 1A-B, lane 2); because as the EpoR2 they are short-lived proteins (Figure 1A, lane 5); and because they are immunoprecipitated by another anti-EpoR antibody (C-2363 ; Figure 1A, lane 7). Bands b and c represent the cell surface form of the EpoR, since they are shifted after Epo stimulation (Figure 1A-B lanes 1 and 3), they are downregulated by sustained Epo stimulation (Figure 1A, lane 4), and they are precipitated by biotinylated Epo/streptavidin (Figure 1A, lane 6) and by Epo/anti-Epo antibodies (Figure 1A, lane 9). Band d, which is strongly increased in EpoR-transfected BaF3 cells, corresponds to the maturing EpoR present in the endoplasmic reticulum3 (Figure 1C). To explain the difference between the results published by Elliott et al1 and ours, we suspected that the antibody batches were different. We tested the same extracts using another batch (B2105). This latter batch revealed mainly bands a, e, and f, which are contaminating proteins also present in Mo7E extracts (Figure 1A). Thus, we conclude that the ability of Santa Cruz Biotechnology C-20 antibody to detect EpoR is strongly variable depending on the batch of the antibody used, some batches presenting very high sensitivity and specificity, whereas others are poorly usable. We propose use of UT-7 cell extracts as standard to verify antibody specificity.
Elliott et al1 found that the apparent molecular mass of EpoR is significantly lower than previously reported. We verified this point and observed that EpoR comigrated with bovine serum albumin, demonstrating that the molecular masses reported by Elliott et al1 are underestimated (Figure 1D).
Western blot detection of EpoR with C-20 anti-EpoR antibodies. Whole-cell extracts were prepared from unstimulated UT-7 cells (lane 1), Mo7E cells (lane 2), UT-7 cells stimulated for 10 minutes (lanes 3 and 10) or for 90 minutes (lane 4) with Epo, UT-7 cells incubated for 6 hours with cycloheximide (lane 5), and EpoR-transfected BaF3 cells stimulated for 10 minutes with Epo (lane 11). UT-7 cells stimulated for 10 minutes with Epo or biotinylated Epo (lane 6) were solubilized as previously described,4 and cell extracts were precipitated with streptavidin (lane 6), a laboratory-made anti-GST–EpoR antibody (lane 7), anti-GST antibodies (lane 8), and anti-Epo antibodies (lane 9). Portions corresponding to 250 × 103 cells (whole-cell extracts) or to 106 cells (precipitations) were separated on 8.5% (A-B,D) or 10% polyacrylamide gels (C) and transferred to nitrocellulose and analyzed by WB using Santa Cruz Biotechnology C-20 antibodies (catalog no. SC-695) batch K200 (A left, B-D) or batch B2105 (A right). Images were recorded on a LAS 3000 FujiFilm camera. (A-C) Chemiluminescence detection of the proteins recognized by C-20 antibody. Panel B is an enlarged view of panel A, lanes 1-3. (D) Molecular weight markers from Biolabs (Saint-Quentin en Yvelines, France) (catalog no. P7702S) were stained with Ponceau Red after transfer on nitrocellulose and pencil labeled (MW). The upper and lower limits of the BSA band are indicated. After processing for WB analysis, 2 images of the nitrocellulose sheet were successively recorded to detect chemiluminescence and pencil labels, respectively. The images were overlaid and analyzed using MultiGauge software (Fujifilm), giving the molecular masses indicated in the text (molecular mass of the EpoR mature band, 67.6 to 70.4 kDa; molecular mass of the EpoR maturing band, 64 kDa).
Western blot detection of EpoR with C-20 anti-EpoR antibodies. Whole-cell extracts were prepared from unstimulated UT-7 cells (lane 1), Mo7E cells (lane 2), UT-7 cells stimulated for 10 minutes (lanes 3 and 10) or for 90 minutes (lane 4) with Epo, UT-7 cells incubated for 6 hours with cycloheximide (lane 5), and EpoR-transfected BaF3 cells stimulated for 10 minutes with Epo (lane 11). UT-7 cells stimulated for 10 minutes with Epo or biotinylated Epo (lane 6) were solubilized as previously described,4 and cell extracts were precipitated with streptavidin (lane 6), a laboratory-made anti-GST–EpoR antibody (lane 7), anti-GST antibodies (lane 8), and anti-Epo antibodies (lane 9). Portions corresponding to 250 × 103 cells (whole-cell extracts) or to 106 cells (precipitations) were separated on 8.5% (A-B,D) or 10% polyacrylamide gels (C) and transferred to nitrocellulose and analyzed by WB using Santa Cruz Biotechnology C-20 antibodies (catalog no. SC-695) batch K200 (A left, B-D) or batch B2105 (A right). Images were recorded on a LAS 3000 FujiFilm camera. (A-C) Chemiluminescence detection of the proteins recognized by C-20 antibody. Panel B is an enlarged view of panel A, lanes 1-3. (D) Molecular weight markers from Biolabs (Saint-Quentin en Yvelines, France) (catalog no. P7702S) were stained with Ponceau Red after transfer on nitrocellulose and pencil labeled (MW). The upper and lower limits of the BSA band are indicated. After processing for WB analysis, 2 images of the nitrocellulose sheet were successively recorded to detect chemiluminescence and pencil labels, respectively. The images were overlaid and analyzed using MultiGauge software (Fujifilm), giving the molecular masses indicated in the text (molecular mass of the EpoR mature band, 67.6 to 70.4 kDa; molecular mass of the EpoR maturing band, 64 kDa).
Anti-Epo receptor antibodies do not predict Epo receptor expression
In this issue, Verdier et al comment on our recent publication, concluding that some anti-EpoR antibodies have limited utility for detecting EpoR expression.1 We reported that the C-20 anti-EpoR antibody from Santa Cruz Biotechnology (Santa Cruz, CA) detected 5 protein bands in UT-7/Epo cells including 59-kDa and 66-kDa proteins. Using both direct and indirect methods, we showed that the 59-kDa protein, not the 66-kDa protein, is EpoR: (1) the 59-kDa protein migrated similarly to recombinant, FLAG-tagged, full-length EpoR; (2) the 59-kDa protein levels decreased following EPOR shRNA treatment of cells, whereas 66-kDa protein levels remained unchanged; (3) the 59-kDa protein was absent in EPOR-knockout fetal liver but present in wild-type fetal liver; (4) C-20 immunoprecipitated the 59-kDa protein, not the 66-kDa protein, from UT-7/Epo cells (the immunocomplexed 59-kDa protein was detected by 2 other anti-EpoR antibodies, 07-311 and M-20); (5) the 59-kDa, not the 66-kDa, protein bands contained EpoR peptide sequences; and (6) peptides derived from heat-shock proteins specifically inhibited C-20 binding to the 66-kDa protein.
Agreeing that many antibodies are unsuitable for detecting EpoR, the authors also do not challenge our conclusion that C-20 should not be used for immunohistochemistry. However, they disagree about the utility of C-20 for detecting EpoR proteins in Western blots and that the apparent molecular mass of EpoR is approximately 59 kDa, not 66/78 kDa as described in the product information sheet provided by Santa Cruz.
Verdier et al claimed that 3 protein bands larger than 59 kDa (64, 67.6, and 69.5 kDa) detected in UT-7 cells by C-20 were different forms of EpoR, based on indirect methods. First, they showed that the 64-kDa protein was not detected by C-20 in EpoR-negative Mo7E cells, although an approximately 68-kDa protein was detected in the same cells. Second, they showed that the levels of these proteins were altered following treatment with cycloheximide or stimulation with Epo. However, neither agent necessarily selectively alters EpoR levels. Third, they used an in-house anti-EpoR antibody (C-236), biotinylated Epo/streptavidin, and an anti-Epo antibody to immunoprecipitate the putative EpoR proteins, but each yielded different protein patterns. For example, 2 proteins were immunoprecipitated by C-236 and only one by biotinylated Epo/streptavidin or the anti-Epo antibody. Furthermore, when probed with a second batch of C-20 (lot B2105), the negative control anti-GST antibody appeared to precipitate the same 2 putative EpoR proteins as C-236. Finally, they showed a predominant 64-kDa protein in BaF3 cells transfected with EPOR; expression of the larger 67.6- and 69.5-kDa proteins did not increase in this experiment. Based on their results, we do not believe that Verdier et al have adequately demonstrated that the larger proteins detected by C-20 are EpoR. A possible explanation for the discrepancies between the 2 studies is that the 64-kDa protein detected by Verdier et al is the same as the 59-kDa EpoR protein we report, whereas the larger proteins are non-EpoR cross-reacting proteins. The reported size difference (59 vs 64 kDa) may be a consequence of different conditions used to resolve the proteins or the use of different marker proteins to estimate size.
The authors are employed by Amgen Inc, a manufacturer and distributor of erythropoietic-stimulating proteins.
Correspondence: Steve Elliott, Amgen Inc, One Amgen Center Dr, Thousand Oaks, CA 91320; e-mail: selliott@amgen.com.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal