Abstract
In addition to transporting oxygen and carbon dioxide to and from the tissues, a range of other functions are attributed to red blood cells (RBCs) of vertebrates. Diseases compromising RBC performance in any of these functions warrant in-depth study. Furthermore, the human RBC is a vital host cell for the malaria parasite. Much has been learned from classical biochemical approaches about RBC composition and membrane organization. Here, we use mass spectrometry (MS)–based proteomics to characterize the normal RBC protein profile. The aim of this study was to obtain the most complete and informative human RBC proteome possible by combining high-accuracy, high-sensitivity protein identification technology (quadrupole time of flight and Fourier transform MS) with selected biochemical procedures for sample preparation. A total of 340 membrane proteins and 252 soluble proteins were identified, validated, and categorized in terms of subcellular localization, protein family, and function. Splice isoforms of proteins were identified, and polypeptides that migrated with anomalously high or low apparent molecular weights could be grouped into either ubiquitinylated, partially degraded, or ester-linked complexes. Our data reveal unexpected complexity of the RBC proteome, provide a wealth of data on its composition, shed light on several open issues in RBC biology, and form a departure point for comprehensive understanding of RBC functions.
Introduction
As new tools have been used, an increasing understanding of human red blood cell (RBC) protein composition and organization has emerged over the last 40 years. Many of the properties that contribute directly to RBCs' pivotal role in oxygen and carbon dioxide transport have been elucidated. Related properties, such as the cell's ability to maintain its unique discoid shape1-14 yet allow cytoskeleton rearrangements that permit it to pass through capillaries15,16 are now better understood.
In humans, the circulating mature RBC is the end stage of a development process that starts in the bone marrow as hematopoietic stem cells differentiate to nucleated RBCs. After extrusion of nuclei and degradation of endoplasmic reticulum, reticulocytes emerge in the circulation, where they rapidly develop into mature RBCs with a 4-month life span.
Since RBCs are the main gas transporter, genetic or infectious diseases compromising RBC performance can be particularly serious. Genetic diseases of the RBC are frequently linked to the lack of an RBC protein17,18 or to altered/reduced expression of normal RBC components.19,20 In addition to gas transport, evidence suggest that the RBC is involved in a range of other functions, such as transfer of GPI-linked proteins21-25 and transport of iC3b/C3b-carrying immune complexes.26 The RBC is also a critical cell in the life cycle of the malaria parasite, a protozoan parasite that is responsible for up to 2.7 million deaths each year and a massive burden of morbidity, primarily in developing countries.27 Interactions between the parasite and the RBC include a series of complex molecular events during parasite invasion of the RBC,28-30 as well as remodelling of the RBC membrane during intraerythrocytic parasite development.31,32 To improve our understanding of such diseases and to shed further light on RBC function, accurate and sensitive techniques are required to determine RBC protein profiles.
Recent technologic advances in functional genomics encompass proteomic and microarray methods, which allow global approaches for protein identification and expression analyses. However, in the absence of a nucleus, mRNA-based microarray approaches are not applicable to mature mammalian RBCs.
Three previous reports have applied different mass spectrometric techniques to analyze the RBC proteome.33-35 Those pioneering studies used technology with relatively low accuracy and low resolution. Recent technologic advances in proteomics have been dramatic36 and now allow in-depth and unambiguous characterization of the proteomes in large proteomes such as the nucleolus37 or fluorescence-activated cell sorted (FACS) malaria gametocytes.38 We have sought to extend and improve the proteomic approach by combining state-of-the-art protein identification technology, quadrupole time-of-flight mass spectrometry (MS; Q-STAR; PE Sciex, Toronto, ON, Canada), and hybrid linear ion trap, Fourier transform MS (LTQ-FT) with biochemical procedures for RBC subfractionation. In addition to sample preparative techniques to analyze membrane and cytoplasmic RBC compartments, we have focused on (1) a high analysis number, to allow statistical underpinning of protein hits even when only one peptide is identified, (2) combining the strengths of various approaches to MS to exploit the strengths and minimize the weaknesses inherent to any single approach, and (3) in-depth analysis and validation of the data sets that emerged, down to the level of individual proteins and isoforms.
In this study, 314 membrane and 252 soluble proteins were identified and validated. In addition, splice isoforms were critically analyzed. An initial categorization in terms of subcellular localization, family, and function was made through interrogation of available annotation databases and the literature.
Materials and methods
RBC preparation
Peripheral whole blood from individual human donors who provided informed consent in accordance with the Declaration of Helsinki was obtained, collected in citrate at the Leiden Blood Bank in The Netherlands, and stored for 72 to 96 hours, 4°C, without shaking to allow maturation of reticulocytes to RBCs. Although it has recently been suggested that the maturation of reticulocytes in vitro is limited at 4°C,39 our analysis of the final material (see “Purity assessment of RBC samples”) clearly shows that the procedures taken to eliminate reticulocytes in our analyses were effective. We also eliminated the top RBC layers after centrifugation.
White-cell filters (Plasmodipur, Euro-diagnostica, Arnhem, The Netherlands) were used according to the manufacturer's instructions, and RBCs were pelleted at 1734g for 5 minutes in an Allegra X-22R centrifuge (Beckman Coulter, Mijdrecht, The Netherlands). Supernatant was discarded, and RBCs were resuspended before being layered on 30 mL CL5020 (Cederlane Laboratories, Hornby, ON, Canada) in a 50-mL Falcon tube, with centrifugation as described. The remaining RBC fraction was passed through nylon nets to further eliminate granulocytes and was washed 5 times with ice-cold RPMI 1640 (Sigma-Aldrich, Zwijndrecht, The Netherlands). At each step, along with supernatant, the upper RBC layer was removed. Remaining RBCs were immediately used to prepare membrane and cytoplasmic fractions.
Purity assessment of RBC samples
The packed RBC samples from 3 separate purifications were diluted with RPMI 1640 medium to the following packed RBCs–medium ratios: 1:0.3, 1:0.5, 1:1, and 1:2. These samples were counted for white blood cells (WBCs), granulocytes, monocytes, platelets, and RBCs using a Sysmex SF-3000 system and for reticulocytes using a Sysmex R-500 system (Goffin Meyvis Medical & Analytical Systems, Etten Leur, The Netherlands). For the 3 highest dilutions, RBC counts consistent with human hematology reference data were measured (4.27 × 1012-5.87 × 1012 cells/L). Only mature RBCs were detected. At the lowest dilution, 8.07 × 1012 cells/L RBCs and 0.01 × 109/L WBCs were found. Again, no reticulocytes or other cell types were found.
Protein determinations
Proteins were measured using the MicroBCA assay using BSA as a standard (Pierce, Rockford, IL), per the manufacturer's instructions.
Membrane preparation
Packed RBCs (10 mL) were suspended in 50 mL ice-cold 5 mM phosphate buffer, pH 8, and centrifuged (9000g, 20 minutes, 4°C). Hemolysate was discarded and the operation repeated (at least 5 times) until the supernatant appeared colorless. Centrifugation was then increased to 20 000g and washing was repeated until the ghost membranes appeared yellow-whitish. Membranes were stored at –80°C.
Soluble protein preparation
Washed, packed RBCs suspended in RPMI 1640 were subjected to 5 successive freeze/thaw cycles (liquid nitrogen),40 after which suspensions were centrifuged (50 000g Sorval RC M150 GX, 4°C, for 90 minutes). The resulting supernatant was aliquoted and stored at –80°C prior to analysis.
Membrane extractions
By Na2CO3. RBC membranes (10 μL; 80 μg protein) were resuspended in 1 mL 100 mM Na2CO3 (pH 11) and passed 5 times through a 25-gauge needle, mixed by rotation (30 minutes, 4°C), and pelleted (90 minutes, 245 000g), and the supernatant was removed. This process of suspension, rotation, pelleting, and washing was repeated twice more. The pellet was either digested directly or treated with EtOH.
By EtOH. RBC membrane pellets were diluted with 4 volumes absolute EtOH and brought to 50 mM sodium acetate, using 2.5 M sodium acetate, pH 5.0. Final pH was approximately 7.5. Twenty micrograms of glycogen per mL of original sample was added, the suspension was mixed at room temperature (RT) for 90 minutes and centrifuged (10 minutes, RT), and the supernatant was discarded.
Cytoskeleton extraction. Membranes isolated from 10 mL packed RBCs were washed (10 volumes of ice-cold 0.1 mM EDTA, pH 8.0), resuspended to 40 mL in the same buffer, and incubated (30 minutes, 37°C) prior to centrifugation (30 minutes, 250 000g, 4°C). The pellet was immediately digested.
In-solution digestion. Membrane samples were supplemented with a 1:1 or 1:2 volume ratio of either 8 M urea, 6 M urea/2 M thiourea, or 8 M guanidine-HCl (pH 1.5, 10 minutes, RT) and centrifuged (10 minutes, 9300g). The supernatant was removed and its pH was checked to confirm it was approximately 8.0. One microgram DTT was added per 50 μg protein and samples were incubated (30 minutes, RT). Reduced proteins were alkylated by supplementing with 5 μg iodoacetamide per 50 μg protein (20 minutes, RT). Lys C (endoproteinase Lys C from Lysobacter enzymogenes; Sigma-Aldrich) was added (1 μg/50 μg protein), the mixture was incubated (3 hours, RT) and diluted (4 volumes 50 mM ammonium carbonate), and digestion was initiated by adding 1 μg trypsin/50 μg protein and incubation (15 hours, RT).
SDS-PAGE. For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), 10 μL of 10% (wt/vol) lithium dodecyl sulfate was added to 10-μL samples, the mixture was heated (10 minutes, 70°C), and 10 μL of this was run on a precast 4% to 12% polyacrylamide gel (NuPAGE, Invitrogen, Breda, The Netherlands) in MOPS buffer supplemented with 0.025% (vol/vol) reducing agent (Invitrogen) in the inner chamber to prevent sample reoxidation.
In-gel digestion and preparation for MS. SDS-PAGE track lengths (top to tracking dye) averaged 7 cm and were cut into 15 slices, which were individually digested as previously reported.41 Aliquots of trypsin-digested material were diluted 1:5 with 0.5% glacial acidic acid, 1% trifluoroacetic acid (vol/vol). Samples were loaded on a Stage tip42 to desalt and stored for maximally 12 hours, 4°C. Peptides were eluted (3 times 10 μL buffer B [80% acetonitrile, 20% MilliQ water, 0.5% glacial acidic acid vol/vol]) directly into 96-well plates (ABgene; AB-0800, Courtaboeuf, France) that were centrifuged under vacuum until volumes were 4 to 6 μL, which was brought to 10 μL with buffer A containing 0.3% trifluoroacetic acid.
MS. Trypsin-digested samples were analyzed by capillary liquid chromatography coupled on-line with tandem mass-spectrometry (LC-MS/MS) using an Agilent 1100 series system and a Q-STAR (Q-STAR pulsar from PE Sciex; 17 runs) or LTQ-FT (Hybrid-2D-Linear Quadrupole Ion Trap-Fourier Transform Ion Cyclotron Resonance [FTICR] Mass Spectrometer, Thermo Electron, Bremen, Germany, 3 runs). Samples from 3 μg protein were separated by reverse-phase chromatography (3 μm Reposil C18, 75 μm × 12-cm column) using a gradient from 98% MS buffer A and 2% MS buffer B solution at 0.5-μL/min flow rate. MS buffer A was 0.5% glacial acidic acid vol/vol; MS buffer B, was 80% acetonitrile, 0.5% glacial acidic acid vol/vol. At 24 minutes the flow was decreased to 0.25 μL/min and the amount of buffer B was increased to 7% (27 minutes), 13% (35 minutes), 33% (95 minutes), 50% (112 minutes), 60% (117 minutes), and finally to 80% (123 minutes). Eluted peptides were ionized to charge states 1+,2+, or higher by the electrospray source and peptides that were at least doubly charged were analyzed in data-dependent MS experiments with dynamic exclusion. Two FT methods were used, the 3 most intense ions method43 and the 5 mass ranges method, the methods differing in the mode in which the ion trap is filled. For the Q-STAR, raw spectra were converted to a detected peak list of 1+, 2+, and 3+ ions, providing an exclusion list that was iteratively applied, thereby progressively excluding the most abundant ions from sequencing and enabling progressively less abundant peptides to be sequenced.
Database search. Acquired MS/MS spectra were searched against the nonredundant International Protein Index (IPI) human sequence database (www.ebi.ac.uk)44 using Mascot software.45 To determine levels of false-positive peptide identifications, spectra were also searched against the corresponding reverse database. The estimated rate of peptide false positives from reverse database analysis was 0.3%. Search parameters for initial peptide and fragment mass tolerance were, respectively, ± 0.2 Da and ± 0.8 Da (Q-STAR) and ± 5 ppm and ± 0.6 Da (LTQ-FT), with allowance made for one missed trypsin, fixed modification of cysteine through carbamidomethylation, acetylation and methionine oxidation. Only fully tryptic peptide matches were allowed.
Data obtained from annotation databases require critical evaluation to ascribe the most likely functions when several are available. For example, moesin is reported to be involved in shape regulation, in MAPK kinase activation, and in photoreceptor cell differentiation. The latter role may be relevant in other cell types, but can reasonably be excluded in RBCs. As a further example Ras-related C3 botulinum toxin substrate 4 has been assigned roles in signal transduction (small GTPase), cell adhesion, and dendrite morphogenesis. Again, for RBCs, involvement in the latter process can be excluded. Thus, annotation data presented in Table S1 (available on the Blood website; see the Supplemental Tables link at the top of the online article) should be viewed as an effort to assemble all available information via Uniprot (comprising Swiss-Prot and Tembl)46 and Ensembl databases47 using protein accession numbers, but, because databases are incomplete, should not be taken as absolute. To reduce uncertainty, in case of doubt, the protein description was submitted to PubMed48 and cross-checked for literature relevant to RBCs (Table S1, column AG). Relevant papers were often identified (not all of which could be cited) although findings were often contradictory, and results were often obtained for RBCs from species other than humans.
Validation. Validation was based on MSQuant (open source software developed by our laboratory, http://msquant.sourceforge.net/), enabling manual score and spectrum evaluation of each peptide that led to the identification of a given protein. Stringent protein identification criteria were imposed: each protein required minimally a unique, 7-amino acid peptide, with a Mascot score more than 35 (corresponding to 99.9% identification confidence) and an MS/MS spectrum featuring a continuous series of at least 3 y-ions in the area equal to or more than y5 (Q-STAR) or a continuous series of 3 y- or b-ions (LTQ-FT). Protein identifications by single peptides were allowed only if the protein in question was identified at least in 2 runs. Proteins were then blasted all versus all with a cutoff of 95% to remove redundancy.
Annotation. Swiss-Prot/TrEMBL (http://www.expasy.org),46 Ensembl (http://www.ensembl.org),47 and the Gene Ontology databases49 were used for annotation. Unique Swiss-Prot/TrEMBL/Ensembl numbers provided access to sequence, isoform, family, localization, and function data for identified proteins and in parallel IPI numbers were queried against the Gene Ontology database49 using GoMinerH (http://discover.nci.nih.gov/gominer/), enabling grouping by class, function, or localization and quantitation within a grouping—for example, within the different signal transduction pathways.
Results
RBC preparations for MS were analyzed for their purity using automatic counting of WBCs and reticulocytes to have a measure of the extent of contamination. The automatic counts gave a maximum of 1 WBC in 1 million RBCs, and no granulocytes, monocytes, platelets, or reticulocytes. This suggests that the combined use of different purification procedures (see “Materials and methods”) reduced contaminant blood cells by 1000-fold such that we deal with an essentially pure RBC proteome. Nevertheless, it should be borne in mind that the nature of the study means that despite the extensive purification protocols and relatively stringent protein identification criteria used, we cannot rule out inclusion of a small number of false-positive identifications nor the occasional presence of contaminating proteins wrongly assigned to the RBC.
Membrane proteins in the proteome
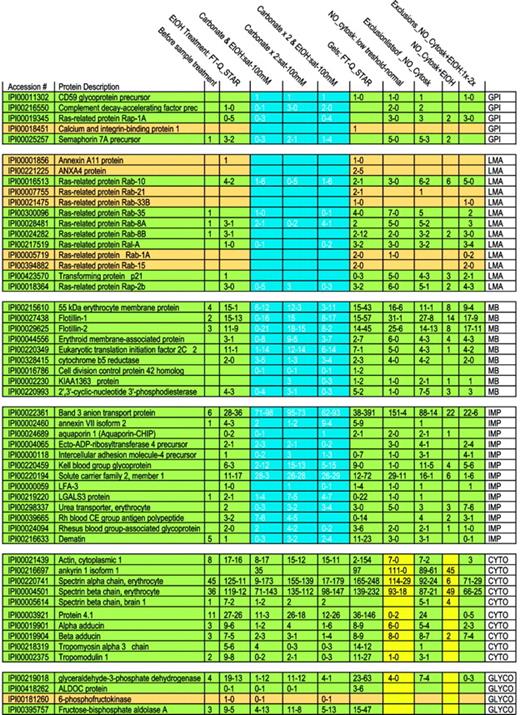
Membrane proteins are often partially shielded from tryptic digestion by lipids and may occur as either integral or membrane-associated proteins. To begin analysis of membrane proteins, we therefore evaluated biochemical procedures that might increase the number of detectable proteins. Initially 35 proteins were identified on MS analysis of digested crude RBC membranes (Table 1). An ethanol solubilization/precipitation protocol was used to remove lipids.50 Following this, 95 proteins were identified; band 3 peptide hits increased from 6 to 36, whereas spectrin α and β peptide hits decreased from 45 to 11 and from 36 to 12, respectively. To detect more proteins and to help differentiate between membrane-associated and integral membrane proteins, we combined the ethanol treatment with sodium carbonate extraction at 2 concentrations.51 Three procedures were compared: (1) ethanol solubilization followed by calcium carbonate extraction, (2) 2 extractions with calcium carbonate alone, and (3) ethanol solubilization, followed by 2 calcium carbonate extractions. A trend is evident showing the progressive disappearance of loosely associated membrane proteins (such as Rab and annexins A11 and A4) as more intense carbonate extraction of the samples was used (Table 2 and Figure 1). At the same time, detection of integral membrane proteins remained stable. Tightly membrane-associated proteins, such as membrane protein 55 kDa, showed a trend similar to that of loosely associated membrane proteins (the number of peptides after the carbonate wash decreased when compared with that of the combined ethanol/carbonate treatment) but some peptides were always present, reflecting strong association with the membrane and high abundance. We did not observe a marked difference in behavior toward a carbonate treatment for membrane-bound and GPI-anchored proteins. Only CD59 glycoprotein showed a constant peptide number throughout all carbonate extractions. On the other hand, protein dissociation in a saturated calcium carbonate solution appeared to be more efficient in that loosely associated membrane proteins such as ras-related Rab proteins almost completely disappeared, whereas membrane protein peptide numbers increased, probably through counter-ion and ionic strength effects. Thus, whereas a 100-mM carbonate solution alone decreased the number of identified proteins in comparison with a combined ethanol/carbonate treatment, a saturated carbonate solution revealed more proteins.
Specifications of the MS detection systems used and summary of the number of membrane proteins identified by using different extraction methods and 2 different databases
. | IPI Human, no. . | NCBI Mammalia, no. . |
|---|---|---|
| Not treated | 35 | ND |
| EtOH | 95 | 93 |
| Carbonate saturated | ||
| Na2CO3 + EtOH | 48 | 36 |
| Na2CO3 × 2 | 97 | 87 |
| Na2CO3 × 2 + EtOH | 69 | 61 |
| Carbonate | ||
| Na2CO3 + EtOH | 108 | 89 |
| Na2CO3 × 2 | 76 | 75 |
| Na2CO3 × 2 + EtOH | 117 | 106 |
| Gel (combo) | 144 | ND |
. | IPI Human, no. . | NCBI Mammalia, no. . |
|---|---|---|
| Not treated | 35 | ND |
| EtOH | 95 | 93 |
| Carbonate saturated | ||
| Na2CO3 + EtOH | 48 | 36 |
| Na2CO3 × 2 | 97 | 87 |
| Na2CO3 × 2 + EtOH | 69 | 61 |
| Carbonate | ||
| Na2CO3 + EtOH | 108 | 89 |
| Na2CO3 × 2 | 76 | 75 |
| Na2CO3 × 2 + EtOH | 117 | 106 |
| Gel (combo) | 144 | ND |
Number of identified peptides by using different extraction methods and 2 different databases for annotation.
ND indicates not determined.
Details of extraction procedures and MS methods used
Before sample treatment . | Crude membrane digest . |
|---|---|
| EtOH treatment: FT-Q-STAR | EtOH-treated crude membrane analyzed by LTQ-FT (1st peptide count) versus Q-STAR (2nd peptide count) |
| Carbonate and EtOH:sat-100 mM | Crude membrane treated with carbonate solution followed by EtOH: saturated carbonate solution versus 100 mM |
| Carbonate × 2:sat-100 mM | Crude membrane treated twice with carbonate: saturated solution versus 100 mM |
| Carbonate × 2 and EtOH:sat-100 mM | Crude membrane treated twice with carbonate solution followed by EtOH: saturated carbonate solution versus 100 mM |
| Gels: FT-Q-STAR | Gel analyzed with FT versus Q-STAR |
| NO_Cytosk: normal-low threshold | Cytoskeletal removal on crude membranes analyzed by normal versus low threshold |
| Exclusion lists of_NO_Cytosk | Exclusion list of the above samples |
| NO_Cytosk + EtOH | Cytoskeletal removal on crude membranes followed by EtOH treatment |
| Exclusions_NO_Cytosk + EtOH: 1 × -2 × | Exclusion list of the above samples run once versus run twice |
Before sample treatment . | Crude membrane digest . |
|---|---|
| EtOH treatment: FT-Q-STAR | EtOH-treated crude membrane analyzed by LTQ-FT (1st peptide count) versus Q-STAR (2nd peptide count) |
| Carbonate and EtOH:sat-100 mM | Crude membrane treated with carbonate solution followed by EtOH: saturated carbonate solution versus 100 mM |
| Carbonate × 2:sat-100 mM | Crude membrane treated twice with carbonate: saturated solution versus 100 mM |
| Carbonate × 2 and EtOH:sat-100 mM | Crude membrane treated twice with carbonate solution followed by EtOH: saturated carbonate solution versus 100 mM |
| Gels: FT-Q-STAR | Gel analyzed with FT versus Q-STAR |
| NO_Cytosk: normal-low threshold | Cytoskeletal removal on crude membranes analyzed by normal versus low threshold |
| Exclusion lists of_NO_Cytosk | Exclusion list of the above samples |
| NO_Cytosk + EtOH | Cytoskeletal removal on crude membranes followed by EtOH treatment |
| Exclusions_NO_Cytosk + EtOH: 1 × -2 × | Exclusion list of the above samples run once versus run twice |
The table lists and explains the headings of Figure 1, detailing the extraction procedures and MS methods used. The first number in each column always refers to the amount of peptides detected with the first method and is compared with the second number, which is the amount of peptides detected with the second method.
Interestingly, the use of a cytoskeleton-removal protocol not only resulted in a reduction in peptides from cytoskeletal and cytoskeleton-associated protein, but also reduced the number of hits for almost all glycolytic proteins. This phenomenon became even more marked when the proteome was studied in the greatest possible depth (by the use of exclusion lists as described in “Materials and methods”). This suggests that glycolytic proteins in the RBC, with the exception of glyceraldehyde dehydrogenase, may be as loosely associated with the membrane as the membrane skeleton to ankyrin. This finding agrees with earlier observations that binding is sensitive to band 3 tyrosine phosphorylation and oxidation.3,52 Alternatively, it may point to a direct association of glycolytic enzymes with cytoskeleton proteins.
Number of peptides identified following different extraction procedures and different MS methods. In each rectangle, the figure lists for a particular extraction procedure a range of the number of detected peptides. Because the number of detected peptides differed for the 2 types of MS techniques applied, we list a range. The investigated proteins are grouped, as abbreviated in the right column: GPI, GPI-anchored proteins; LMA, loosely membrane-associated proteins; TMA, tightly membrane-associated proteins; IMP, integral membrane protein; CYTO, cytoskeletal proteins; and GLYCO, glycolytic proteins. Proteins are shown in green or dark yellow to maintain the consistent statistical color-coding adopted in the supplementary tables, where green proteins are detected in almost all runs and tan in a high number of runs. To illustrate the most dramatic differences among “membrane proteins” following extractions, an area is highlighted in blue.
Number of peptides identified following different extraction procedures and different MS methods. In each rectangle, the figure lists for a particular extraction procedure a range of the number of detected peptides. Because the number of detected peptides differed for the 2 types of MS techniques applied, we list a range. The investigated proteins are grouped, as abbreviated in the right column: GPI, GPI-anchored proteins; LMA, loosely membrane-associated proteins; TMA, tightly membrane-associated proteins; IMP, integral membrane protein; CYTO, cytoskeletal proteins; and GLYCO, glycolytic proteins. Proteins are shown in green or dark yellow to maintain the consistent statistical color-coding adopted in the supplementary tables, where green proteins are detected in almost all runs and tan in a high number of runs. To illustrate the most dramatic differences among “membrane proteins” following extractions, an area is highlighted in blue.
Identifying low-abundance RBC membrane proteins: maximizing the depth of proteome coverage. After evaluating extraction protocols, as described, samples were routinely analyzed by MS after SDS-PAGE and gel slicing. In comparison with MS analysis without prior SDS-PAGE fractionation, this approach reduced the complexity of the individual sample, improved detection of low-abundance proteins, and also provided a molecular size window for the proteins analyzed. In this way, the most effective carbonate/ethanol protocol was further improved to increase identified proteins from 117 to 144 (Table 1). The presence of abundant proteins in a particular sample results in repeated detection of peptides from this protein and thereby lowers the probability of detecting rare proteins (dynamic range challenge). This problem was most pronounced for band 3 protein (anion channel, 1 million copies/cell) that comprises approximately 30% of the membrane proteome53 and spectrin tetramer (100 000 copies/cell) that makes up 75% of the cytoskeleton.54 Two additional strategies were therefore used to minimize this problem, namely, the use of a highly sensitive mass spectrometer with very fast sequencing cycles (LTQ-FT) and the use of exclusion lists on the Q-STAR instrument (see “Materials and methods”).
The Q-STAR offers better mass accuracy than ion trap MS (with which the first human RBC proteome was obtained).34 The LTQ-FT offers several important advantages over Q-STAR, in that it provides high mass accuracy from the FT, high fragmentation speed from the LTQ, and further provides the opportunity for an additional step of fragmentation (MS3 ) in the ion trap part of the instrument (LTQ). In this way, the LTQ-FT made it possible to identify extracellular proteins that bind to RBCs, which are generally expected to be of very low abundance. Nonetheless, the Q-STAR still offers advantages, such as better quantitation statistics than LTQ-FT.
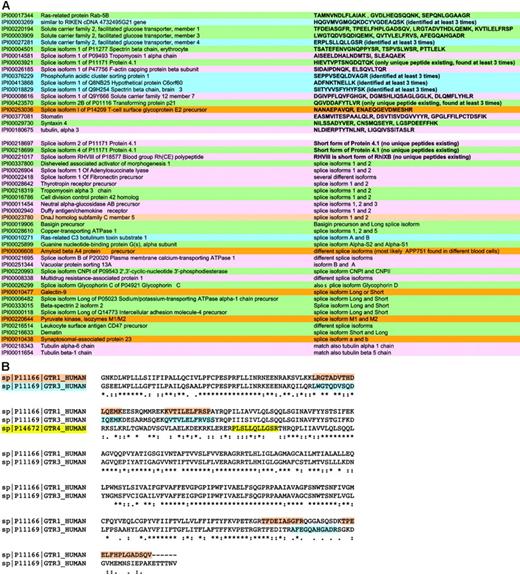
Detection and validation of proteins found including isoforms and protein families. These strategies allowed detection of more proteins than previously possible and in many cases the sequencing of peptides specific for protein isoforms (Figure 2A-B). Members of the same protein family, such as the glucose transporters, were differentiated wherever possible by identification of unique peptides. This enabled confirmation that more than one family member was present in the sample. Peptide spectra (only scores over 30 were accepted) were checked for correct attribution of the amino acid sequence.
As an example of a protein family, 3 different members of the glucose transporter family were identified, namely, member 1 for which several unique peptides (eg, FLLINRNEENR) were found, member 3 for which the unique peptide LWGTQDVSQDIQEMK was sequenced, and member 4, featuring the unique peptide ERPLSLLQLLGSR (Figure 2B).
Sometimes it was also possible to discriminate between different splice isoforms, as in the case of the splice isoform 1 of P09493 tropomyosin 1 α chain (accession no. IPI 00014581) and the splice isoform 1 of P47756 F-actin capping protein β subunit. These were AISEELDHALNDMTSI and SIDAIPDNQK, respectively, and are signature peptides specific for the isoform 1 of these proteins.
In some cases, though, it was not possible to discriminate between isoforms. One example is the ankyrin family. Three proteins were detected: ankyrin 1 isoform 1, alt. ankyrin, and ankyrin 1 isoform 4. Alignment of these proteins shows that ankyrin isoform 4 is the long form, the others 2 shorter splice forms. In this case, because the sequences of the 2 short splice isoforms completely overlap with that of isoform 4, we cannot discriminate between these proteins. However, ankyrin 1 isoform 4 is present in any case as specific peptides for the long form were detected. All proteins were validated as described in “Materials and methods.” An all-versus-all blast of validated proteins eliminated redundancy, providing the final membrane/soluble protein lists (Table S1).
When proteins were identified in only one run, further information was sought in annotation databases and the literature, allowing us to confirm their likely presence in the RBC or to define them as probable contaminants from other blood sources.
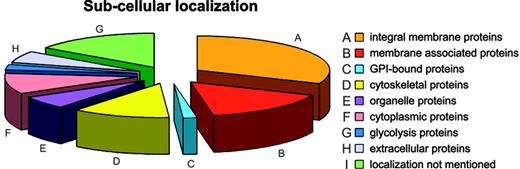
Annotation: the membrane components. The membrane protein set was analyzed for subcellular localization; of 340 membrane proteins found, 105 were annotated as integral membrane proteins, 54 as membrane associated or bound, 5 as GPI anchored, 40 as cytoskeletal, 21 as organelle proteins, 41 as cytoplasmic (8 of which play a role in glycolysis), and 20 as extracellular. For 54 proteins the subcellular localization was not available (Figure 3).
The final list of membrane proteins was evaluated in depth against the available literature and databases. Such sources are, of course, incomplete and changing. Nevertheless, GoMiner recognized 222 of 314 proteins submitted. Annotation can further be complicated when multiple functions and locations are attributed to a protein (eg, Lutheran blood group glycoprotein, involved in adhesion and signal transduction). Limited or no information may be available or the database may not recognize the submitted query (different accession numbers or protein descriptions).
Broadly, proteins were categorized with regard to molecular function and biologic process. Most proteins are involved in binding (115 proteins) or possess a catalytic activity (98 proteins). Many show a transporter (47 proteins), signal transducer (29 proteins), or structural activity (24 proteins). Transport in RBCs is known to be diverse, involving among other substrates carbohydrates, fluids, gases, ions, and proteins. The large number of transporters was, therefore, expected. However, intracellular transport proteins of the Golgi network (eg, Rab 14, vesicle trafficking protein SEC22b, TPM 1, and syntaxin 7), the endoplasmic reticulum (protein disulfide isomerase A3), and the mitochondrion (heat shock protein 90 kDa), were also detected, probably as reticulocyte legacies. Because many catalytic proteins were identified, it is not surprising that 10 enzyme regulators were also found.
RBCs are exposed to membrane oxidation and accordingly we detected antioxidant proteins (5). Although RBCs are devoid of nuclei, proteins annotated to have transcription regulator activity (1), translation regulator activity (2), and chaperone regulatory activity (7), were identified. Some of these proteins (elongation factor 1 α 2, eukaryotic translation initiation factor 2C 2) are most probably in an inactive state as they are no longer needed in the absence of the nucleus and organelles and may be polyubiquitinylated, which could account for the increased observed molecular weight (Figure 5). Alternatively, some of these proteins may have roles in mature RBCs other than those so far ascribed in the literature. Other proteins include 2 with motor activity, one with nutrient reservoir activity, and 8 for which a molecular function is unknown.
The proteins identified are involved in the following biologic processes: 174 in cellular, 170 in physiologic, and 38 in regulatory processes. Some proteins are annotated as involved in development (22), but it cannot be excluded that some of these proteins may have as yet undefined roles in differentiated cells as well.
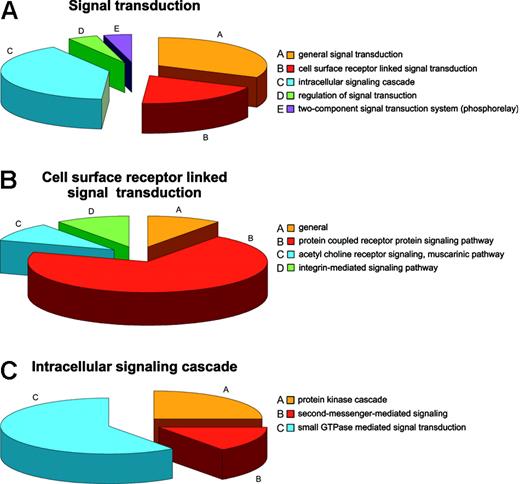
Regulatory mechanisms in the RBCs can be viewed from the standpoint of regulation of cellular processes, physiologic processes, and regulation of enzyme activities. The most common cellular regulatory activity is involvement in programmed cell death, presumably mostly a legacy of prior development,55 followed by involvement in regulation of transport and signal transduction. Fifteen proteins are involved in general signal transduction, 9 are linked to surface receptor signal transducers, 19 proteins have roles in the intracellular signaling cascade, and 2 in regulation thereof. Two proteins are ascribed by databases as being part of the so-called “phosphorelay” (2-component signal transduction system; Figure 4A). Of the proteins involved in surface receptor-linked signal transduction, 7 belong to the G protein-coupled receptor protein signaling pathway, one to the acetylcholine receptor signaling muscarinic pathway, and one to the integrin-mediated signaling pathway (Figure 4B), whereas the intracellular signaling cascade is itself divided into protein kinases (5), second messengers (3), and small GTPases (11), which could further be defined as cell surface and intracellular activities (Figure 4C), respectively. Such a high representation of signal transduction proteins is unlikely to be purely a reticulocyte legacy. Beside the traditional protein kinases PKC and PKA, 3 additional kinases were found, namely, the PRKAR2 protein and the serine-threonine protein kinases VRK1 and WNK1, none of which have previously been reported as RBC constituents. Although for PRKAR2 a role in mediating membrane association by binding to anchoring proteins similar to that of PKA could be envisaged, WNK1 is more likely to be involved in the regulation of ion transport pathways.56 A possible role for VRK157 is less obvious although it seems to be a protein kinase diverged from the caseine kinase 1 branch, a protein constitutively present in RBCs. Small GTPases were the most numerous intracellular signaling proteins in RBCs. Proteins regulating cell shape (guanine nucleotide-binding protein, α-13 subunit, moesin, phospholipid scramblase 4) and proteins annotated to be involved in cell proliferation (amyloid β A4 protein) were also identified.
Protein isoforms in RBC membranes. (A) The splice isoforms of membrane proteins are summarized with the relevant peptides that permitted unequivocal attribution to a specific isoform when such attribution was possible. (B) The partial of a Clustal W alignment of sister proteins is shown for the glucose transporters of RBCs as an example of the way by which we ensured the presence of specific peptides for each protein in the final data set. The evidenced peptides were found (among others) and are clearly unique to each protein.
Protein isoforms in RBC membranes. (A) The splice isoforms of membrane proteins are summarized with the relevant peptides that permitted unequivocal attribution to a specific isoform when such attribution was possible. (B) The partial of a Clustal W alignment of sister proteins is shown for the glucose transporters of RBCs as an example of the way by which we ensured the presence of specific peptides for each protein in the final data set. The evidenced peptides were found (among others) and are clearly unique to each protein.
Most proteins involved in physiologic processes are related to regulation of metabolism (7); a few are involved in vascular processes (guanine nucleotide-binding protein G(i), α-2 subunit, TPM1, guanine nucleotide-binding protein, α-inhibiting activity polypeptide 3) through their involvement in the modulation of nitric oxide,58,59 blood vessels size (vasodilatation brought about by serum albumin), and coagulation (annexin VII isoform 2).60
Negative regulators predominate over positive regulators of enzyme activity, and more proteins were present that are involved in the regulation of adenylate cyclase (guanine nucleotide-binding protein G(i), α-2 subunit, guanine nucleotide-binding protein, α-inhibiting activity polypeptide 3) than in the regulation of GTPase activity (Rab GDP dissociation inhibitor α). In agreement with previous findings, these proteins also appear to be mostly involved in the inhibition of the activity of the GTPase-adenylate cyclase complex (guanine nucleotide-binding protein G(i), α-2 subunit, α-inhibiting activity polypeptide 3, and Rab GDP dissociation inhibitor α) than in its activation (guanine nucleotide binding protein Gs). This confirms the findings of Ikeda et al,61 who identified the 2 α subunits of Gs and Gi and the rab protein by biochemical means together with an unknown G protein, which we suggest could be the α-inhibiting activity polypeptide 3.
Annotation: the example of signal transduction. (A) Distribution of proteins involved in signal transduction in the RBC. (B) Distribution of proteins linked to the cellular surface receptor-mediated signal transduction. (C) Distribution of proteins linked to intracellular signal transduction.
Annotation: the example of signal transduction. (A) Distribution of proteins involved in signal transduction in the RBC. (B) Distribution of proteins linked to the cellular surface receptor-mediated signal transduction. (C) Distribution of proteins linked to intracellular signal transduction.
Furthermore, the inhibitory G proteins (guanine nucleotide-binding protein G(i) and α-inhibiting activity polypeptide 3) seem to be involved in the regulation of the AQP1 water channel62 and inhibition of the nitric oxide-mediated ATP release from RBCs mediated by nitric oxide.63
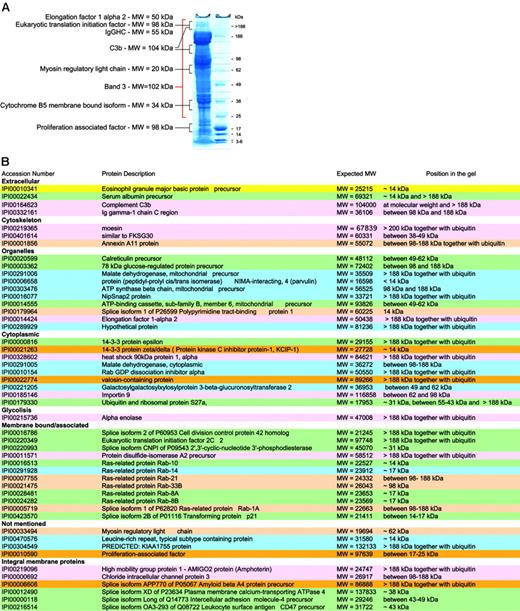
Not all proteins are present in their active form. Although our preparation methods focused on purification of mature RBCs, the protein content of these is likely to vary with cell age. Thus, RBCs may be considered as “in development,” because degradation of organelles occurs during maturation and chemical and enzymatic modifications during cellular aging.”Left over” proteins may therefore be detected and it becomes important to know in which state they exist. It is possible to speculate on the status of a protein based on comparison of the expected and observed molecular weights (MWs) of the proteins in question. Proteins giving unexpected subcellular localization, unexpected MWs, or having functions not attributable to mature RBCs were studied together with control proteins known to be present in mature RBCs. Control proteins were found in gel bands corresponding to their expected MW. Of the 193 proteins identified after in-gel separation and digestion, 49 had apparent MWs at variance with those expected of which 32 migrated at a MW higher than expected in a gel fraction that also contained ubiquitin, and 17 at a lower MW (Figure 5).
For example, the presence of proteins of internal organelles in the final membrane protein list was unexpected, as mature RBCs are devoid of both nucleus and internal organelles. Hence, peptides assignable to such proteins could have originated from partially degraded proteins or from RBC proteins that were modified during RBC maturation. Many of these proteins indeed had lower MWs than expected (14-3-3 protein ζ-δ, similar to FKSG30, ATP-binding cassette subfamily B, member 6, mitochondrial precursor) suggesting that they represented degradation products. However, others, including 78-kDa glucose regulated protein, elongation factor 1 α 2, and eukaryotic translation initiation factor 2C 2, showed increased MWs and were found in bands also containing peptides from ubiquitin/polyubiquitin. Although this may be attributable in part to high MW complexes resulting from reduction-insensitive protein-protein interactions, the presence of ubiquitins within the same gel region makes it attractive to speculate that some proteins not required in the mature RBC underwent modifications by ubiquitin for degradation by the proteasome. Band 3, the spectrins, and other cytoskeletal proteins were present at several MWs in the gels as previously reported.64 A higher MW may be due to ubiquitinylation as is known for spectrin65 or due to oligomer formation (SDS and reduction insensitive) in aging RBCs.66
Extracellular proteins associated with RBCs. Among the membrane proteins we also identified 20 proteins that were most probably of extracellular origin and remained associated with RBC membranes.
Serum albumin was one of them and is known to bind also to white cells.67-69 Cathepsin G is involved in the degradation of RBC glycophorins and plasmodial antigens by human neutrophils.70 Clusterin appears to be involved in complement inhibition.71 It normally forms complexes with liberated C5b-7/8 complement proteins in the fluid phase and thereby prevents their interaction with nearby cells and thus prevents their lysis.72,73 Even though the latter components were not observed in our set, evidence indicates that clusterin is able to promote RBC aggregation and it may therefore not be surprising that we find it in this context.74 Galectin 3 has a role in the binding of IgE and surface carbohydrates, with preference for carbohydrates linked to blood groups Aand B compared with H75 and is able to elicit selected cell responses.76 Lactoferrin, another component apparently associated with RBCs, seems to have a role in the regulation of RBC glycolysis77 although its potential role in the partial inhibition of anti–band 3 naturally occurring antibodies to ageing RBCs is still a matter of debate.78-80 Prolactin binds to RBCs through specific receptors,81 which appear to be more abundant in cord blood than in blood from adult healthy volunteers. Although the importance of prolactin during erythropoiesis is widely understood, its role in mature RBCs is unknown.82 Two components of the innate immune system, C3 and autologous IgG, were also identified among RBC-associated proteins. Interestingly, IgG and C3 protein were identified not only at the MW of their reduced components following in-gel digestion, but also in gel fractions originating from MWs higher than 188 kDa. It is likely that the concomitant presence of both proteins at such a MW may have originated from partially inactivated covalent C3b2-IgG complexes. Such complexes are generated during opsonization of oxidatively damaged and aging RBCs by naturally occurring IgG antibodies to band 3 protein.83 The complexes contain 2 sequentially ester-bonded C3b on one heavy chain of IgG, yielding an apparent MW of 263 kDa in their reduced and intact form and one of 185 kDa in their iC3b2-IgG form.84 Thus, our results provide independent evidence for such high MW adducts on RBC.
RBC membrane proteins with unexpected MWs. (A) Typical SDS-PAGE with the relative size markers and a few proteins that were found not only at their expected MW. (B) Summary lists all proteins that were not found at their expected MWs.
RBC membrane proteins with unexpected MWs. (A) Typical SDS-PAGE with the relative size markers and a few proteins that were found not only at their expected MW. (B) Summary lists all proteins that were not found at their expected MWs.
Soluble RBC proteins
Soluble proteins were analyzed twice by a hybrid linear ion trap Fourier transform mass spectrometer (LTQ-FT) following SDS-PAGE fractionation. To maximize the number of peptide hits for low-abundance proteins, the standard MS method was used, except that it was rendered more sensitive by selectively filling the ion trap to capacity with 5 mass ranges in turn. In this way, dynamic range is maximized and dominant peptides from abundant proteins are excluded from most mass ranges. Proteins were validated as described in “Validation,” and again unique peptides were sought to identify isoforms and to distinguish between members of protein families (Table S2).
Annotation: the cellular component. A total of 252 unique soluble proteins were identified and annotated (Table S3). Of these, 195 were annotated as cytoplasmic, 15 cytoplasmic and nuclear, 5 Golgi or endoplasmic reticulum, 5 nuclear, and 4 mitochondrial or cytoplasmic with only one integral membrane protein; for 27 proteins a subcellular localization was not available. Thus, most proteins could unequivocally be attributed to the cytoplasm. Those annotated both as cytoplasmic and organellar, appeared to have a transporter role, accounting for this distribution.
Are proteins present in their active form? Thirty-nine proteins were classified as unexpected based on their annotation, of which 26 migrated with higher apparent MWs in a fraction that also contained ubiquitin, 9 were found at the expected MW, and 4 at a lower molecular weight (Table S4).
It will be interesting to determine biochemically whether such proteins are indeed ubiquitinylated.
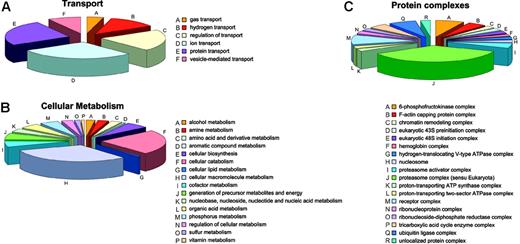
Annotation: cellular process and molecular function. Most proteins identified among the soluble proteins are implicated in cellular metabolism or in cellular transport. Unsurprisingly, given the RBC history that requires degradation of organelles, nucleic acid, and the like, most proteins involved in metabolic processes are catabolic (34) or are involved in macromolecule metabolism (44), whereas only a few proteins are involved in cellular biosynthesis (8) (Figure 6B). The complex with most members is the proteasome (15) followed by ubiquitin ligases (3) (Figure 6C). Other proteins are related to metabolism of amines (4), amino acids and derivatives (4), aromatic compounds (1), lipids (1), cofactors (8), nucleic acids (9), organic acids (7), phosphorous (7), sulfur (2), and vitamins (1) and with generation of precursor metabolites and cellular energy (9).
Involvement of soluble proteins. Soluble proteins involved in transport (A), cellular metabolism (B), and being comprised in protein complexes (C).
Involvement of soluble proteins. Soluble proteins involved in transport (A), cellular metabolism (B), and being comprised in protein complexes (C).
Alongside the proteasome and the ubiquitin ligase complexes, single proteins known to be associated with several other types of protein complexes are present, most probably representing the remains of incomplete degradation processes for complexes no longer needed by the mature RBC (chromatin remodeling complex, eukaryotic 43S preinitiation complex, eukaryotic 48S initiation complex, nucleosome, ribonucleoprotein complex, and ribonucleoside-diphosphate reductase complex). These proteins did not have the expected MWs on SDS-PAGE as defined by Uniprot. Thus, it is likely that most of these functional complexes were partially degraded by incomplete cellular catabolic processes. In contrast, enzymes likely to also be relevant for mature RBCs,85 like the 3 proton-transporting ATPase complexes and proteins belonging to such complexes, were identified at their expected MW in the gel.
Transport processes mainly involve ion (7) and protein transport (6) or the regulation thereof (5) (Figure 6A). Hemoglobin is the sole gas transporter, and there are 3 different hydrogen transporters.
Comparison with other studies of the proteome of blood cells. A comparison of the proteins found in our study and those found in platelets (“secretomes”)86-88 shows that only proteins that are known to belong to both platelets and RBCs or to bind to both cell types are found in both proteomes, whereas platelet-specific proteins such as coagulation factors, fibrinogen, and different platelet glycoproteins are not found in our RBC data set. Worthy of mention is thrombospondin 1, which is known to bind to different blood cells89 and was identified with a high number of peptides in platelet proteomes. Interestingly, the 14-3-3 ζ-δ protein, which was until now attributed solely to platelets by the Uniprot database,90 appears to be present also in RBCs, because it was found in both the membrane and the soluble protein fraction thereof. Likewise, cyclophilin A, which is found in the platelets secretome, may also be present in the RBC most probably as legacies of the reticulocytes, where members of the cyclophilin family have previously been identified.91 A special case is that of the amyloid A4 protein, which does not appear to be present in RBCs, but seems to be able to bind to them—for example, in the central nervous system92 and in the plasma.93 The leukocyte-specific protein, CD45, was not found in our data set.94 The transferrin receptor, a key protein in reticulocytes, which is lost from them in exosomes when they become mature RBCs,95 was also not found in our final protein list. The same is true for the ferritin receptor.96
Further information
A detailed summary of all proteins found is available as supplemental material at the Blood website; click on the Supplemental Tables link at the top of the online article. A database is also being created at http://proteome.biochem.mpg.de/RBC.
Discussion
The most advanced LC-MS technology coupled with different biochemical procedures for sample preparation, and new bioinformatics tools, developed in house, have been used to investigate the proteome of the normal RBC, 72 to 96 hours after collection from a blood donor. The stringent purification procedures applied to the starting RBCs, with only 1 WBC/106 RBCs, and the detection limit of about 500 copies of protein per cell (reported for CR1 on RBCs) allowed us to present the most complete analysis of the RBC proteome performed so far. After validation, a total of 314 membrane and 252 soluble proteins were found and further scanned for their most likely physiologic role in the RBC, for their subcellular localization, and molecular function. Isoforms were critically screened for unique peptides and an all-versus-all Blast was performed to ensure that all proteins in the final list were actually present in the RBCs. All unexpected membrane proteins, membrane-associated proteins, and soluble proteins have been further evaluated for their likely metabolic status by confronting their known MW with their in-gel migration. A protein's appearance at a MW lower than expected was considered a degraded protein, a remainder from reticulocytes. Comigration with ubiquitins at elevated MW was considered evidence for a nonfunctional protein, a remainder from reticulocytes, which was incompletely proteolysed by proteasomes. These and several other aspects clearly revealed that maturation of RBCs from reticulocytes and aging of RBCs are ongoing processes that most probably endure the whole 120 days of their life span. This makes the RBC a highly dynamic blood component and suggests that many aspects of its physiologic role and on its interplay with other cells21-25 and plasma proteins still remain to be discovered.
Prepublished online as Blood First Edition Paper, April 18, 2006; DOI 10.1182/blood-2005-11-007799.
Supported by the Netherlands Organization for Scientific Research (NWO Genomics, grant no. 050-10-053), by the Danish National Research Foundation, and by the BioMalPar Contract CT 2004-503578.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal