Abstract
Crucial to designing angiostatic and vascular targeting agents is the identification of target molecules. Because angiogenesis is not limited to pathologic conditions, careful evaluation of putative therapeutic targets is warranted to prevent adverse effects associated with impaired physiologic angiogenesis. To identify tumor-specific angiogenesis markers, we compared transcriptional profiles of angiogenic endothelial cells isolated from malignant and nonmalignant tissues with those of resting endothelial cells. We identified 17 genes that showed specific overexpression in tumor endothelium but not in angiogenic endothelium of normal tissues, creating a therapeutic window for tumor vasculature-specific targeting. Antibody targeting of 4 cell-surface–expressed or secreted products (vimentin, CD59, HMGB1, IGFBP7) inhibited angiogenesis in vitro and in vivo. Finally, targeting endothelial vimentin in a mouse tumor model significantly inhibited tumor growth and reduced microvessel density. Our results demonstrate the usefulness of the identification and subsequent targeting of specific tumor endothelial markers for anticancer therapy.
Introduction
Tumor progression and the development of distant metastases require the presence of an extensive vasculature.1,2 Active angiogenesis is a hallmark of most malignancies, and inhibition of this process is considered a promising strategy for the treatment of tumors. To develop the most specific and effective antiangiogenic therapies for treating cancer, it is important to have a fundamental understanding of the molecular differences between tumor endothelial cells (ECs) and their normal counterparts. Because angiogenesis is not limited to pathologic conditions, careful evaluation of the putative targets is warranted to prevent adverse effects associated with impaired normal physiologic angiogenesis. Gene-expression profiling techniques are widely used to detect changes in transcript-expression levels to study molecular events in biologic processes. Different cell culture models have been developed to study angiogenesis, but the temporal and spatial complex actions of all factors exerting effects on ECs in vivo may not be accurately reflected in cultured ECs.3 Gene-expression analysis of tumor ECs can be problematic because these cells are embedded in complex tissues and constitute only a small fraction of the cells present in a tumor.
To date, only a limited number of studies have characterized the gene-expression profiles of freshly isolated tumor ECs.4-6 Serial analysis of gene expression-(SAGE) tag repertoires were generated from ECs isolated from tumor and normal tissues and were compared to identify differentially expressed genes for the discovery of tumor endothelial markers. In these studies, however, gene expression associated with physiologic processes was never directly taken into account.4-6 In addition, proof of successful targeting of tumor endothelial markers in vivo for cancer therapy has not yet been provided.
In the present study, we used suppression subtractive hybridization (SSH) to compare gene-expression profiles of isolated ECs from colon carcinoma tissues, nonmalignant angiogenic placental tissues, and nonangiogenic normal tissues. We identified 17 tumor angiogenesis genes (TAGs) overexpressed in tumor endothelium compared with angiogenic and nonangiogenic endothelium. Four of these genes, encoding surface-expressed or secreted proteins, such as vimentin, CD59, high-mobility group box 1 (HMGB1), and insulin-like growth factor binding protein 7 (IGFBP7), were selected for further investigation. Antibodies targeting these proteins inhibited angiogenesis in in vitro and in vivo assays. We demonstrate the success of our approach by targeting one of these markers, vimentin, in a mouse tumor model, resulting in markedly inhibited tumor growth, accompanied by reduced microvessel density. These results validate our approach to target finding and suggest the applicability of the identified TAGs.
Materials and methods
Isolation of endothelial cells from fresh tissues
Colorectal carcinoma tissue (Dukes C; n = 5) and distant normal colon tissue (n = 5) of the same patient were obtained directly after surgery. Fresh placental tissue (n = 5) was obtained from the Department of Obstetrics (University Hospital Maastricht, Maastricht, The Netherlands). ECs were isolated by previously described methods,6 with minor modifications. Tissue was minced with surgical blades and digested for 30 minutes with 1 mg/mL collagenase and 2.5 U/mL dispase (Life Technologies, Bethesda, MD) at 37°C during continuous agitation. DNAse I (Sigma, St Louis, MO) was added to a final concentration of 100 μg/mL, and the cell suspension was incubated for another 30 minutes before Ficoll Paque density gradient centrifugation (Amersham Biosciences, Freiburg, Germany). ECs were immunolabeled with a combination of anti-CD31 (clones JC/70A [DAKO, Carpinteria, CA]; EN4 [Monosan; Sanbio, Uden, The Netherlands]) and anti-CD34 (clone Qbend10; Novocastra, New Castle, United Kingdom) antibodies. In pilot studies we examined the suitability of CD31, CD34, CD105, and CD146 antibodies. A mixture of CD31 and CD34 antibodies gave the most complete coverage of blood vessels, CD105 antibody extensively stained trophoblast in placenta, and CD146 antibody showed unspecific staining (data not shown). ECs were isolated by positive selection using goat anti–mouse IgG–coated paramagnetic beads (Dynal, Oslo, Norway). The purity of the isolated EC fraction was assessed by immunofluorescence staining for endothelium-specific von Willebrand factor (DAKO) and was estimated to be greater than 97%.
Cell culture
Human umbilical vein ECs (HUVECs) were isolated and cultured as previously described.7 HUVECs were activated in RPMI 1640 supplemented with 20% human serum (HS), 10% filter-sterile conditioned medium of the colorectal tumor-cell line LS174T, 10% filter-sterile conditioned medium of the Caco-2 colorectal tumor-cell line, 2 mM l-glutamine (Life Technologies), 50 ng/mL streptomycin and 50 U/mL penicillin (MP Biomedicals, Irvine, CA), 1 ng/mL bFGF, and 10 ng/mL VEGF (ReliaTech, San Pablo, CA) until 80% confluence was reached. Quiescent ECs were prepared by culturing nearly confluent HUVECs in RPMI 1640 supplemented with low amounts (2%) of serum. Cells were cultured for 3 days.
RNA isolation and cDNA synthesis
RNA was isolated with the use of RNeasy Mini reagents (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA samples were pooled per 5 EC fractions, and cDNA (SMART; BD Biosciences, San Jose, CA) was synthesized from the RNA and amplified to be used for SSH.
Suppression subtractive hybridization
SSH was performed with the PCR-Select cDNA subtraction kit (BD Biosciences) according to the manufacturer's instructions. Subtractions were performed to create cDNA repertoires enriched for genes overexpressed in colon tumor ECs (TECs) by subtracting TECs in separate reactions with normal colon ECs (TEC-NECs) and placenta ECs (TEC-PLECs) and for genes differentially expressed in activated and quiescent HUVECs by subtracting activated HUVECs with quiescent HUVECs (forward) and vice versa (reverse).
Subtracted cDNA repertoires were T/A cloned in pCR2.1 (Invitrogen, Carlsbad, CA) and introduced in TOP10 cells according to the manufacturer's instructions. Individual colonies were picked and grown overnight at 37°C in 2xTY bacterial medium (BD Biosciences) supplemented with 10 μg/mL ampicillin (Roche, Mannheim, Germany).
Differential screening
Inserts were amplified using the adaptor-specific primers Nested 1 and Nested 2R (BD Biosciences) using Hot Goldstar Taq polymerase (Eurogentec, Liege, Belgium). PCR products were spotted in duplicate onto nylon membranes and hybridized to 33P-dCTP (Amersham)–labeled cDNA probes derived from colon tumor ECs (TECs), normal colon ECs (NECs), placenta ECs (PLECs), activated HUVECs, and quiescent HUVECs. Membranes were exposed to phosphor screens (Eastman Kodak, Rochester, NY), and images were acquired using the Personal FX Phosphor Imager (Bio-Rad, Hercules, CA). All experiments were performed twice.
Data were processed in MS Excel (Microsoft, Redmond, WA) to identify differentially expressed transcripts. Pairwise comparisons were performed between duplicate filters hybridized with different probes. Duplicate spots showed excellent concordance (R2 > 0.99; data not shown) and were averaged. Average spot intensities were included in the analysis when expression was at least 2.5 times background in any experiment. Spot intensities were normalized for total intensity of the filters under comparison. Gene-expression ratios were calculated using the average normalized intensities for each spotted insert cDNA. Hierarchical clustering analysis was performed with Cluster 3.08 and visualized using Tree View (Michael Eisen, University of California, Berkeley, CA).
Additional detailed information regarding the SSH analysis and associated procedures can be found in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Sequencing and database searching
Inserts of selected clones were sequenced using Big Dye Terminator Cycle Sequencing mix (Applied Biosystems, Foster City, CA) and were analyzed on a 3100 Genetic Analyzer (Genome Center Maastricht). Homology searches were performed using NCBI nucleotide–nucleotide Blast (blastn) algorithm on the combined GenBank/EMBL/DDBJ nonredundant (nr) and expressed sequence tags (est) databases (http://www.ncbi.nlm.nih.gov/blast/). Functional annotation of the genes was determined using public Gene Ontology databases. Chromosomal regions of the identified TAGs and general angiogenesis genes (GAGs) were investigated for cytogenetic abnormalities in colon cancer using the Progenetix database (www.progenetix.de).
Real-time quantitative PCR
SYBR green assays were performed using 10 ng cDNA template per reaction, consisting of 1 × SYBR Green Master Mix (Applied Biosystems) and 200 μM each primer (Sigma Genosys) (Table S1). Reactions were run and analyzed on the ABI7700 (Applied Biosystems). All reactions were performed in triplicate, analyzed using SDS software (Applied Biosystems), and further processed in MS Excel. All experiments were normalized for cyclophilin A transcript expression to account for variations in template input, as previously described.9
Immunohistochemistry
Colon carcinoma and normal colon tissue sections were stained with mouse anti–human CD31 (clone JC70/A; DAKO), mouse anti–human vimentin (clone V9; DAKO), mouse anti–human CD59 (clone MEM-43; Chemicon, Temecula, CA), rabbit anti–human HMGB1 (kind gift of Dr R.G. Roeder, The Rockefeller University, New York, NY), and rabbit anti–human IGFBP7 (kind gift of Dr R. Rosenfeld, Oregon Health and Sciences University, Portland, OR). Primary antibodies were diluted in PBS/0.5% BSA and were detected with peroxidase-conjugated rabbit anti–mouse IgG (DAKO) or goat anti–rabbit IgG (DAKO). Color was developed using DAB according to standard protocols. Images were acquired by using a Leica DMIL inverted microscope equipped with an HC Plan Apo 20×/0.70 objective lens (Leica, Wetzlar, Germany) and a Leica 10C 300FX camera. Images were processed with Qwin Pro3 software (Leica).
Fluorescence-activated cell-sorter analysis
Single-cell suspensions of fresh colorectal carcinoma and normal colon tissue were obtained as described and were fixed in 1% paraformaldehyde. ECs were stained with a PE-labeled anti-CD31 antibody (Serotec, Oxford, United Kingdom) and were separated from other cells by cell sorting (BD FACSAria). CD31+ cells were subsequently stained using the antibodies rabbit antivimentin, rabbit anti-IGFBP7, and rabbit anti-HMGB1, diluted in PBS/0.5% BSA, followed by biotinylated swine anti–rabbit IgG (DAKO) and streptavidin-FITC (DAKO).
In vitro sprouting
Sprouting of ECs was studied in the tube formation assay. Cytodex-3 beads were overgrown with bovine capillary ECs (BCEs) and were transferred to a 3-dimensional gel, as described previously.7 Antibodies dialyzed to PBS were added to the collagen gel and culture medium at the described concentrations. A nonrelevant control anti–c-Myc antibody was included as control (9E10; Roche). Beads with cells were incubated for 48 hours, after which photographs were taken by using a Leica DM5000B microscope equipped with a C Plan 20×/0.50 PH1 objective lens (Leica) and a Basler A101FC-LE camera (Basler, Ahrensburg, Germany). Images were acquired through Research Assistant 5 (RVC, Soest, the Netherlands) and subsequently imported into Microsoft Powerpoint (Microsoft, Seattle, WA). Five concentric rings were projected over the photographs, and the number of intersections of rings and sprouting ECs was determined digitally and used as a measure of in vitro tube formation.
Chick chorioallantoic membrane assay
Fertilized white leghorn chicken eggs were used to monitor vessel development in the chick chorioallantoic membrane (CAM), as described previously.7 Antibodies were dialyzed to 0.9% NaCl and were administered at the indicated concentrations in a volume of 65 μL for 4 consecutive days beginning on day 10. On day 14, the CAMs were visualized by using a Leica MZ8 microscope equipped with an N Plan 3.2×/0.70 objective lens (Leica) and a Basler A101FC-LE camera. Quantification of microvessels was performed as described for the sprouting assay.
Mouse tumor model
Female athymic Swiss nu/nu mice were used and randomly split into 4 groups. All experiments were approved by the University of Minnesota Research Animal Resources ethics committee. Mice (n = 6 per group) were inoculated subcutaneously with 106 LS174T colorectal carcinoma cells in 100 μL RPMI in the right flank. Four days after inoculation, when established and palpable tumors were present, treatment was started. Mice were treated by intraperitoneal injection every third day with purified RV202 antivimentin antibody (10 mg/kg and 1 mg/kg; MUbio Products, Maastricht, The Netherlands), isotype-matched IIB5 anti-BrdU control antibody (10 mg/kg; MUbio), or saline alone. Tumor volume was determined daily by measuring the diameters of tumors using calipers and was calculated as follows: width2 × length × 0.52. Mice were weighed daily to assess possible toxic effects of the treatment.
Cryosections (5 μm) of excised tumors were stained for CD31, and microvessel density was evaluated as described previously.10 Antibody targeted to the tissue in vivo was detected using biotinylated secondary goat antimouse antibodies (DAKO) and were visualized with streptavidin-FITC.
Statistical analysis
Results are expressed as mean ± SEM. Comparisons of values in the in vitro biosassays were assessed using the Student t test. For statistical analysis of the tumor growth curves, 2-way ANOVA was performed.
Results
Identification of tumor angiogenesis genes by SSH
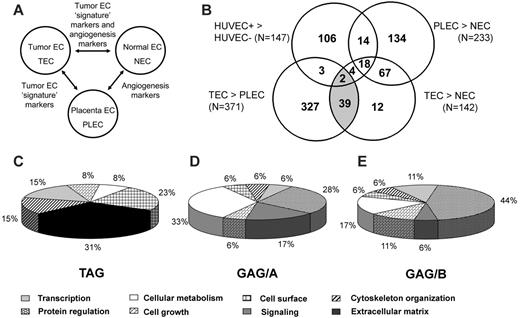
We performed SSH in combination with cDNA array screening to identify novel tumor-specific EC markers in an unbiased manner. We isolated tumor ECs (TECs) from a series of colon carcinoma tissues and patient-matched normal ECs (NECs) from adjacent normal colon tissues and from placental tissues (PLECs). These isolations were greater than 97% pure and, therefore, were perfectly suitable for gene-expression analysis, as has been demonstrated.11 RNA was isolated and used to create subtraction repertoires of genes overexpressed in TECs. In addition, HUVECs were stimulated in vitro with tumor-conditioned medium and were used to create additional subtraction repertoires. Subtracted cDNA libraries consisting of 2746 cDNA sequences were arrayed and screened for differential expression in TECs, NECs, PLECs, and HUVECs. Merely comparing gene-expression differences between TECs and NECs may identify genes involved in angiogenesis in general, in addition to genes that represent a “tumor signature” of the ECs. To distinguish between these 2 groups, a comparison with PLECs—representing angiogenic ECs—was included. Genes overexpressed in TECs, compared with NECs and PLECs, were considered to be tumor angiogenesis specific (Figure 1A).
By comparing expression profiles of TECs with those of NECs, PLECs, and HUVECs, we categorized the TEC-overexpressed genes in 3 different subgroups. First, we identified genes specific for tumor endothelium (TAGs). Forty-one cDNA clones classified as TAGs (Figure 1B; Table 1) and showed overexpression in TECs compared with NECs and in TECs compared with PLECs. Second, 85 cDNA clones were found to be upregulated in TECs compared with NECs and as in PLECs compared with NECs (general angiogenesis genes group A [GAG/A]; Figure 1B; Table 1). Third, we identified 24 upregulated cDNA clones in activated HUVECs compared with quiescent HUVECs and in TECs compared with NECs, which we named GAG/B markers (Figure 1B; Table 1).
Characteristics of EC gene expression identified by differential screening of SSH repertoires
. | Classification of tumor angiogenesis gene . | Spots* . | Gene IDs† . |
|---|---|---|---|
| TAG | TEC > NEC and TEC > PLEC | 41 | 17 |
| GAG/A‡ | TEC > NEC and PLEC > NEC | 85 | 46 |
| GAG/B§ | TEC > NEC and HUVEC+ > HUVEC- | 24 | 22 |
. | Classification of tumor angiogenesis gene . | Spots* . | Gene IDs† . |
|---|---|---|---|
| TAG | TEC > NEC and TEC > PLEC | 41 | 17 |
| GAG/A‡ | TEC > NEC and PLEC > NEC | 85 | 46 |
| GAG/B§ | TEC > NEC and HUVEC+ > HUVEC- | 24 | 22 |
Number of spots that showed at least 2-fold difference in expression in the indicated comparisons.
Number of genes represented differentially expressed spots.
General angiogenesis genes (in vivo markers).
General angiogenesis genes (in vitro and vivo markers).
Sequence analysis revealed that the 41 TAG cDNA clones represented 17 different genes (Table 2). Five of these have previously been identified by SAGE analysis as overexpressed on tumor-derived ECs, validating our approach.4-6 The highly abundant collagen Iα1 and collagen IVα1 are overexpressed on the endothelium of different tumors, including colon carcinoma, breast carcinoma, and glioma,4-6 pointing toward the possible existence of pan-tumor endothelium–specific transcripts. HEYL is a basic helix-loop-helix transcription factor recently reported to be upregulated in breast tumor vasculature.5 SPARC and IGFBP7 have been classified as pan-endothelial markers and showed overexpression in colon carcinoma ECs compared with normal colon ECs.6 We identified 2 more TAG cDNAs with proposed roles in angiogenesis. PPAP2B was upregulated during VEGF-stimulated tube formation of ECs in vitro,12 and HMGB1 promoted angiogenesis in vitro.13 The 10 remaining TAG markers have so far not been reported to functionally contribute to (tumor) angiogenesis, though some, such as vimentin and CD59, are described as expressed on ECs.14,15
Tumor angiogenesis genes
. | . | . | . | . | . | . | Library origin§ . | . | . | |
|---|---|---|---|---|---|---|---|---|---|---|
| TAG . | Gene ID* . | . | GenBank accession no. . | Function† . | Component‡ . | No. spots representing TAG . | HUVEC . | TEC . | CGH∥ . | |
| Previously associated with angiogenesis | ||||||||||
| TAG-5 | Collagen type I, α1 | COL1A1 | NM_000088.2 | Extracellular matrix | E | 4 | — | × | — | |
| TAG-7 | Collagen type IV, α1 | COL4A1 | NM_001845.2 | Extracellular matrix | E | 16 | × | × | G13q34 | |
| TAG-21 | High-mobility group protein-1 | HMGB1 | NM_002128.3 | Transcription | N, E | 1 | — | × | G13q12 | |
| TAG-25 | Hairy/enhancer of split with YRPW motif | HEYL | NM_014571 | Transcription | N | 1 | — | × | — | |
| TAG-29 | Insulin-like growth factor-binding protein 7 | IGFBP7 | NM_001553 | Extracellular matrix | E | 1 | × | — | — | |
| TAG-31 | Phosphatidic acid phosphatase type 2B | PPAP2B | CV337080 | Cell surface | P | 1 | — | × | — | |
| TAG-33 | Secreted protein acidic, rich in cysteine | SPARC | NM_003118.1 | Extracellular matrix | E | 5 | — | × | — | |
| No previous association with angiogenesis | ||||||||||
| TAG-1 | Actin-related protein 2/3 complex | ARPC2 | NM_152862.1 | Cytoskeleton organization | C | 2 | — | × | — | |
| TAG-3 | CD59 antigen p18-20 | CD59 | NM_000611 | Cell surface | P | 1 | — | × | — | |
| TAG-4 | CDK2-associated protein 1 | CDK2AP1 | NM_004642.2 | Cell growth | C, N | 1 | — | × | — | |
| TAG-23 | IMAGE 5299642 | EST | BC041913 | Unknown | Unknown | 1 | × | — | — | |
| TAG-27 | IMAGE 4332094 | EST | NM_017994.1 | Unknown | Unknown | 1 | — | × | G7q11.21 | |
| TAG-28 | Insulin receptor precursor | HSIRPR | X02160 | Cell surface | P | 1 | — | × | — | |
| TAG-30 | Lactate dehydrogenase B | LDHB | NM_002300 | Cellular metabolism | C | 1 | × | — | — | |
| TAG-32 | Polyhomeotic-like 3 | PHC3 | AJ320486 | Unknown | N | 1 | — | × | — | |
| TAG-38 | Voltage-gated K channel β-subunit 4.1 | HSPC014 | AF077200 | Unknown | Unknown | 1 | × | — | G13q12.3 | |
| TAG-39 | Vimentin | VIM | X56134 | Cytoskeleton organization | C | 3 | × | × | — | |
. | . | . | . | . | . | . | Library origin§ . | . | . | |
|---|---|---|---|---|---|---|---|---|---|---|
| TAG . | Gene ID* . | . | GenBank accession no. . | Function† . | Component‡ . | No. spots representing TAG . | HUVEC . | TEC . | CGH∥ . | |
| Previously associated with angiogenesis | ||||||||||
| TAG-5 | Collagen type I, α1 | COL1A1 | NM_000088.2 | Extracellular matrix | E | 4 | — | × | — | |
| TAG-7 | Collagen type IV, α1 | COL4A1 | NM_001845.2 | Extracellular matrix | E | 16 | × | × | G13q34 | |
| TAG-21 | High-mobility group protein-1 | HMGB1 | NM_002128.3 | Transcription | N, E | 1 | — | × | G13q12 | |
| TAG-25 | Hairy/enhancer of split with YRPW motif | HEYL | NM_014571 | Transcription | N | 1 | — | × | — | |
| TAG-29 | Insulin-like growth factor-binding protein 7 | IGFBP7 | NM_001553 | Extracellular matrix | E | 1 | × | — | — | |
| TAG-31 | Phosphatidic acid phosphatase type 2B | PPAP2B | CV337080 | Cell surface | P | 1 | — | × | — | |
| TAG-33 | Secreted protein acidic, rich in cysteine | SPARC | NM_003118.1 | Extracellular matrix | E | 5 | — | × | — | |
| No previous association with angiogenesis | ||||||||||
| TAG-1 | Actin-related protein 2/3 complex | ARPC2 | NM_152862.1 | Cytoskeleton organization | C | 2 | — | × | — | |
| TAG-3 | CD59 antigen p18-20 | CD59 | NM_000611 | Cell surface | P | 1 | — | × | — | |
| TAG-4 | CDK2-associated protein 1 | CDK2AP1 | NM_004642.2 | Cell growth | C, N | 1 | — | × | — | |
| TAG-23 | IMAGE 5299642 | EST | BC041913 | Unknown | Unknown | 1 | × | — | — | |
| TAG-27 | IMAGE 4332094 | EST | NM_017994.1 | Unknown | Unknown | 1 | — | × | G7q11.21 | |
| TAG-28 | Insulin receptor precursor | HSIRPR | X02160 | Cell surface | P | 1 | — | × | — | |
| TAG-30 | Lactate dehydrogenase B | LDHB | NM_002300 | Cellular metabolism | C | 1 | × | — | — | |
| TAG-32 | Polyhomeotic-like 3 | PHC3 | AJ320486 | Unknown | N | 1 | — | × | — | |
| TAG-38 | Voltage-gated K channel β-subunit 4.1 | HSPC014 | AF077200 | Unknown | Unknown | 1 | × | — | G13q12.3 | |
| TAG-39 | Vimentin | VIM | X56134 | Cytoskeleton organization | C | 3 | × | × | — | |
Extracellular matrix indicates ECM organization and biosynthesis; cell surface, cell-surface receptor-linked signal transduction, antigen presentation; transcription, regulation of transcription; cytoskeleton organization, cytoskeleton organization and biogenesis; cell growth, regulation of cell growth, cell cycle, apoptosis; protein regulation, cellular protein metabolism, protein biosynthesis, translation, transport; C, cytoplasm; N, nucleus; P, plasma membrane; E, extracellular; CGH, comparative genomic hybridization; and —, not applicable.
Sequence identity.
Functional classification of the reported TAG, inferred from Gene Ontology Database.
Cellular component of the reported TAG, inferred from Gene Ontology Database.
SSH cDNA repertoire origin of the spots.
Progenetix large intestine database (www.progenetix.de). G indicates gain; L, loss; defined by 15% difference in aberrations per band.
Gene expression of tumor ECs resembles the profile of physiologic angiogenesis
Further gene-expression analysis revealed that most TEC-overexpressed cDNA clones (85 [60%] of 142) are also associated with angiogenesis under physiologic conditions in vivo and are therefore not specific for tumor angiogenesis (Figure 1B). These 85 GAG/A cDNA clones represent 46 different genes (Table 3), including genes that have been associated with angiogenesis such as matrix metalloproteinases (MMPs),16 integrin β1,17 and endothelial cell–specific molecule-1.18
General angiogenesis genes
. | . | GenBank accession no. . | GAG class . | . | . | No. spots representing GAG . | Library origin§ . | . | . | |
|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID* . | . | . | . | Biologic process† . | Component‡ . | . | HUVEC . | TEC . | CGH∥ . | |
| A kinase (PRKA) anchor protein 13 (AKAP13) | AKAP13 | NM_007200 | A | Signaling | C | 1 | × | — | — | |
| Cathepsin B | CTSB | NM_147783.1 | A | Cellular metabolism | C | 11 | — | × | L8p22 | |
| Caveolin 1, caveolae protein, 22 kDa | CAV1 | NM_001753 | A | Protein regulation | P | 2 | × | G7q31.1 | ||
| CD86 antigen | CD86 | NM_175862 | A | Signaling | P | 1 | — | × | — | |
| cDNA FLJ32199 clone PLACE6002710 | EST | AK056761 | A | Unknown | Unknown | 1 | × | — | ||
| Chemokine (C-C motif) ligand 2 | CCL2 | NM_002982 | A | Signaling | E | 1 | × | — | — | |
| dCMP deaminase | DCTD | NM_001921.1 | A | Cellular metabolism | 1 | × | L4q35.1 | |||
| Defender against cell death 1 | DAD1 | NM_001344 | A | Cellular metabolism | C | 1 | × | — | — | |
| EST10870 HUVEC | EST | AA296386 | A | Unknown | 2 | × | — | — | ||
| Eukaryotic translation initiation factor 4A, isoform 1 | EIF4A1 | NM_001416 | A | Protein regulation | C | 1 | × | — | — | |
| FK506-binding protein 1A, 12 kDa | FKBP1A | NM_000801 | A | Signaling | C | 1 | × | — | G20p13 | |
| FLJ37490 | EST | AK094809.1 | A | Unknown | Unknown | 2 | — | × | — | |
| Heme-binding protein 1 | HEBP1 | NM_015987.2 | A | Cellular metabolism | C | 1 | — | × | — | |
| Hypothetical protein MGC 7036 | MGC7036 | NM_145058 | A | Unknown | Unknown | 1 | — | × | — | |
| IMAGE 2028956 | EST | AI793182 | A | Unknown | Unknown | 1 | × | — | ||
| IMAGE 2096486 | EST | AI422919 | A | Unknown | Unknown | 1 | × | — | ||
| IMAGE 2816112 | EST | AW269823 | A | Unknown | Unknown | 1 | × | — | ||
| Integrin β1 | ITGB1 | NM_002211 | A | Cell surface | P | 1 | × | — | ||
| Isoprenylcysteine carboxyl methyltransferase | ICMT | NM_170705.1 | A | Cellular metabolism | C | 1 | — | × | — | |
| Lung cancer oncogene 5 | HLC5 | AY117690.1 | A | Unknown | Unknown | 1 | × | — | ||
| Major histocompatibility complex, class II, DR α | HLA-DRA | BC032350 | A | Cell surface | P | 24 | × | — | ||
| Matrix metalloproteinase 1, interstitial collagenase | MMP1 | NM_002421 | A | Cellular metabolism | E | 1 | × | — | — | |
| Matrix metalloproteinase 3, stromelysin 1 | MMP3 | NM_002422 | A | Cellular metabolism | E | 1 | — | × | — | |
| Pituitary tumor-transforming 1 interacting protein | PTTG1IP | NM_004339.2 | A | Protein regulation | N | 1 | — | × | — | |
| Rad51-associated protein | RAD51AP1 | NM_006479 | A | Cellular metabolism | N | 1 | × | — | — | |
| Ribosomal protein L22 | RPL22 | NM_000983 | A | Cellular metabolism | C | 1 | × | — | — | |
| Tubulin, α3 | TUBA3 | BC050637 | A | Cytoskeleton organization | C | 1 | × | — | — | |
| v-ral simian leukemia viral oncogene homolog B | RALB | BC018163 | A | Signaling | P | 1 | — | × | — | |
| Androgen-induced 1 | AIG-1 | BC025278 | A,B | Unknown | Unknown | 1 | × | — | — | |
| ATP synthase H+ transporting complex, subunit c | ATP5G1 | NM_001575 | A,B | Protein regulation | C | 1 | × | — | — | |
| Diazepam-binding inhibitor | DBI | M15887 | A,B | Protein regulation | C | 1 | × | — | — | |
| Ectonucleoside triphosphate diphosphohydrolase 1 | ENTPD1 | BC047664 | A,B | Signaling | P | 1 | — | × | — | |
| Endothelial cell-specific molecule 1 | ESM1 | NM_007036 | A,B | Cell growth | E | 1 | × | — | — | |
| Eukaryotic translation elongation factor 1 ϵ1 | EEF1E1 | NM_004280.2 | A,B | Protein regulation | C, N | 2 | × | — | — | |
| Heterogeneous nuclear ribonucleoprotein C (C1/C2) | HNRPC | BC003394 | A,B | Cellular metabolism | N | 1 | × | — | — | |
| HSP90 α | HSP90AA1 | NM_005348 | A,B | Cellular metabolism | Unknown | 1 | × | — | — | |
| IMAGE 757234 | EST | BX115183 | A,B | Unknown | Unknown | 1 | × | — | — | |
| IMAGE 4182080, CARD8 homolog | CARD8 | BC046136 | A,B | Cell growth | N | 1 | × | — | — | |
| Ubiquitin protein ligase E3 component n-recognin 2 | UBR2 | AB002347 | A,B | Protein regulation | N | 1 | — | × | — | |
| Matrix metalloproteinase 10, stromelysin 2 | MMP10 | NM_002425 | A,B | Cellular metabolism | E | 2 | × | — | — | |
| Mitochondrial ribosomal protein S27 | MRPS27 | BC011818 | A,B | Protein regulation | C | 1 | × | — | — | |
| Split hand/foot malformation (ectrodactyly) type 1 | SHFM1 | NM_006304 | A,B | Protein regulation | C | 1 | × | — | G7q21.3 | |
| SRY (sex-determining region Y)-box 4 | SOX4 | NM_003107 | A,B | Transcription | N | 2 | × | — | — | |
| Thymosin, β4, X-linked | TMSB4X | NM_021109 | A,B | Cytoskeleton organization | C | 1 | × | — | — | |
| Ubiquitin-conjugating enzyme E2L3 | UBE2L3 | NM_003347 | A,B | Protein regulation | C | 1 | × | — | — | |
| Zinc finger motif enhancer-binding protein 2 | ZNF644 | NM_032186 | A,B | Transcription | N | 1 | × | — | — | |
| Platelet/endothelial cell adhesion molecule 1 | PECAM1 | NM_000442.2 | B | Cell surface | P | 1 | × | — | — | |
| Ribosomal protein L21 | RPL21 | NM_000982.2 | B | Protein regulation | C | 2 | × | — | G13q12.2 | |
. | . | GenBank accession no. . | GAG class . | . | . | No. spots representing GAG . | Library origin§ . | . | . | |
|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID* . | . | . | . | Biologic process† . | Component‡ . | . | HUVEC . | TEC . | CGH∥ . | |
| A kinase (PRKA) anchor protein 13 (AKAP13) | AKAP13 | NM_007200 | A | Signaling | C | 1 | × | — | — | |
| Cathepsin B | CTSB | NM_147783.1 | A | Cellular metabolism | C | 11 | — | × | L8p22 | |
| Caveolin 1, caveolae protein, 22 kDa | CAV1 | NM_001753 | A | Protein regulation | P | 2 | × | G7q31.1 | ||
| CD86 antigen | CD86 | NM_175862 | A | Signaling | P | 1 | — | × | — | |
| cDNA FLJ32199 clone PLACE6002710 | EST | AK056761 | A | Unknown | Unknown | 1 | × | — | ||
| Chemokine (C-C motif) ligand 2 | CCL2 | NM_002982 | A | Signaling | E | 1 | × | — | — | |
| dCMP deaminase | DCTD | NM_001921.1 | A | Cellular metabolism | 1 | × | L4q35.1 | |||
| Defender against cell death 1 | DAD1 | NM_001344 | A | Cellular metabolism | C | 1 | × | — | — | |
| EST10870 HUVEC | EST | AA296386 | A | Unknown | 2 | × | — | — | ||
| Eukaryotic translation initiation factor 4A, isoform 1 | EIF4A1 | NM_001416 | A | Protein regulation | C | 1 | × | — | — | |
| FK506-binding protein 1A, 12 kDa | FKBP1A | NM_000801 | A | Signaling | C | 1 | × | — | G20p13 | |
| FLJ37490 | EST | AK094809.1 | A | Unknown | Unknown | 2 | — | × | — | |
| Heme-binding protein 1 | HEBP1 | NM_015987.2 | A | Cellular metabolism | C | 1 | — | × | — | |
| Hypothetical protein MGC 7036 | MGC7036 | NM_145058 | A | Unknown | Unknown | 1 | — | × | — | |
| IMAGE 2028956 | EST | AI793182 | A | Unknown | Unknown | 1 | × | — | ||
| IMAGE 2096486 | EST | AI422919 | A | Unknown | Unknown | 1 | × | — | ||
| IMAGE 2816112 | EST | AW269823 | A | Unknown | Unknown | 1 | × | — | ||
| Integrin β1 | ITGB1 | NM_002211 | A | Cell surface | P | 1 | × | — | ||
| Isoprenylcysteine carboxyl methyltransferase | ICMT | NM_170705.1 | A | Cellular metabolism | C | 1 | — | × | — | |
| Lung cancer oncogene 5 | HLC5 | AY117690.1 | A | Unknown | Unknown | 1 | × | — | ||
| Major histocompatibility complex, class II, DR α | HLA-DRA | BC032350 | A | Cell surface | P | 24 | × | — | ||
| Matrix metalloproteinase 1, interstitial collagenase | MMP1 | NM_002421 | A | Cellular metabolism | E | 1 | × | — | — | |
| Matrix metalloproteinase 3, stromelysin 1 | MMP3 | NM_002422 | A | Cellular metabolism | E | 1 | — | × | — | |
| Pituitary tumor-transforming 1 interacting protein | PTTG1IP | NM_004339.2 | A | Protein regulation | N | 1 | — | × | — | |
| Rad51-associated protein | RAD51AP1 | NM_006479 | A | Cellular metabolism | N | 1 | × | — | — | |
| Ribosomal protein L22 | RPL22 | NM_000983 | A | Cellular metabolism | C | 1 | × | — | — | |
| Tubulin, α3 | TUBA3 | BC050637 | A | Cytoskeleton organization | C | 1 | × | — | — | |
| v-ral simian leukemia viral oncogene homolog B | RALB | BC018163 | A | Signaling | P | 1 | — | × | — | |
| Androgen-induced 1 | AIG-1 | BC025278 | A,B | Unknown | Unknown | 1 | × | — | — | |
| ATP synthase H+ transporting complex, subunit c | ATP5G1 | NM_001575 | A,B | Protein regulation | C | 1 | × | — | — | |
| Diazepam-binding inhibitor | DBI | M15887 | A,B | Protein regulation | C | 1 | × | — | — | |
| Ectonucleoside triphosphate diphosphohydrolase 1 | ENTPD1 | BC047664 | A,B | Signaling | P | 1 | — | × | — | |
| Endothelial cell-specific molecule 1 | ESM1 | NM_007036 | A,B | Cell growth | E | 1 | × | — | — | |
| Eukaryotic translation elongation factor 1 ϵ1 | EEF1E1 | NM_004280.2 | A,B | Protein regulation | C, N | 2 | × | — | — | |
| Heterogeneous nuclear ribonucleoprotein C (C1/C2) | HNRPC | BC003394 | A,B | Cellular metabolism | N | 1 | × | — | — | |
| HSP90 α | HSP90AA1 | NM_005348 | A,B | Cellular metabolism | Unknown | 1 | × | — | — | |
| IMAGE 757234 | EST | BX115183 | A,B | Unknown | Unknown | 1 | × | — | — | |
| IMAGE 4182080, CARD8 homolog | CARD8 | BC046136 | A,B | Cell growth | N | 1 | × | — | — | |
| Ubiquitin protein ligase E3 component n-recognin 2 | UBR2 | AB002347 | A,B | Protein regulation | N | 1 | — | × | — | |
| Matrix metalloproteinase 10, stromelysin 2 | MMP10 | NM_002425 | A,B | Cellular metabolism | E | 2 | × | — | — | |
| Mitochondrial ribosomal protein S27 | MRPS27 | BC011818 | A,B | Protein regulation | C | 1 | × | — | — | |
| Split hand/foot malformation (ectrodactyly) type 1 | SHFM1 | NM_006304 | A,B | Protein regulation | C | 1 | × | — | G7q21.3 | |
| SRY (sex-determining region Y)-box 4 | SOX4 | NM_003107 | A,B | Transcription | N | 2 | × | — | — | |
| Thymosin, β4, X-linked | TMSB4X | NM_021109 | A,B | Cytoskeleton organization | C | 1 | × | — | — | |
| Ubiquitin-conjugating enzyme E2L3 | UBE2L3 | NM_003347 | A,B | Protein regulation | C | 1 | × | — | — | |
| Zinc finger motif enhancer-binding protein 2 | ZNF644 | NM_032186 | A,B | Transcription | N | 1 | × | — | — | |
| Platelet/endothelial cell adhesion molecule 1 | PECAM1 | NM_000442.2 | B | Cell surface | P | 1 | × | — | — | |
| Ribosomal protein L21 | RPL21 | NM_000982.2 | B | Protein regulation | C | 2 | × | — | G13q12.2 | |
Extracellular matrix indicates ECM organization and biosynthesis; cell surface, cell-surface receptor-linked signal transduction, antigen presentation; transcription, regulation of transcription; cytoskeleton organization, cytoskeleton organization and biogenesis; cell growth, regulation of cell growth, cell cycle, apoptosis; protein regulation, cellular protein metabolism, protein biosynthesis, translation, transport; C, cytoplasm; N, nucleus; P, plasma membrane; E, extracellular; CGH, comparative genomic hybridization; and —, not applicable.
Sequence identity.
Functional classification of the reported GAG, inferred from Gene Ontology Database.
Cellular component of the reported GAG, inferred from Gene Ontology Database.
SSH cDNA repertoire origin of the spots.
Progenetix large intestine database (www.progenetix.de). G indicates gain; L, loss; defined by 15% difference in aberrations per band.
In vitro endothelial-cell activation has limited value for the study of tumor angiogenesis
Based on our gene-expression analysis, it is obvious that only a limited number of genes upregulated in TECs compared with NECs overlapped with genes overexpressed in tumor-conditioned HUVECs in vitro (GAG/B [Figure 1B; Table 1]). Most of the GAG/B markers overlapped with genes associated with physiologic angiogenesis (GAG/A [Figure 1B; Table 3]). In addition, hierarchical clustering analysis suggested that the overall expression pattern in the HUVECs model related most to that emerging from physiologic angiogenesis (ie, the comparison between PLECs and NECs; Figure S1).
Endothelial gene-expression analysis. (A) Interrelationship between ECs of different sources and their gene-expression signatures. ECs from a malignant and proangiogenic environment (TEC) are compared with ECs from organ-matched and patient-matched nonmalignant sources (NEC) and with nonmalignant proangiogenic microenvironment (placenta)–derived ECs (PLEC) to identify the subset of genes that showed expression induced by the tumor microenvironment specifically (tumor EC signature markers). (B) Venn diagram representation of subtraction repertoire screening. Four pairwise comparisons were performed by cDNA array screening of SSH repertoires: tumor-conditioned (HUVEC+) compared with quiescent HUVECs (HUVEC–), colorectal carcinoma ECs compared with normal colon ECs (TEC compared with NEC), colorectal carcinoma ECs compared with placental ECs (TEC compared with PLEC), and placental ECs compared with normal colon ECs (PLEC compared with NEC). Included are clones that showed at least a 2-fold difference in expression. The shaded area represents the TAGs (overexpressed in TECs compared with NECs and in TECs compared with PLECs). (C) TAG markers are strongly biased toward genes associated with extracellular matrix remodeling. (D) GAG/A markers (overexpressed in TECs and PLECs compared with NECs) show a diverse functional profile. (E) GAG/B markers (overexpressed in TECs and in activated HUVECs) are strongly biased to protein regulation. Functional distributions among the 3 different classes were significantly different (P < .001), as determined by χ2 test. TAG functional classification differed from that of GAG/A (P < .001) and GAG/B (P = .006), though GAG/A functional distribution did not differ significantly from that of GAG/B (P = .630). Classifications were obtained using the Gene Ontology Database and were graphically represented according to the data presented in Tables 2 and 3.
Endothelial gene-expression analysis. (A) Interrelationship between ECs of different sources and their gene-expression signatures. ECs from a malignant and proangiogenic environment (TEC) are compared with ECs from organ-matched and patient-matched nonmalignant sources (NEC) and with nonmalignant proangiogenic microenvironment (placenta)–derived ECs (PLEC) to identify the subset of genes that showed expression induced by the tumor microenvironment specifically (tumor EC signature markers). (B) Venn diagram representation of subtraction repertoire screening. Four pairwise comparisons were performed by cDNA array screening of SSH repertoires: tumor-conditioned (HUVEC+) compared with quiescent HUVECs (HUVEC–), colorectal carcinoma ECs compared with normal colon ECs (TEC compared with NEC), colorectal carcinoma ECs compared with placental ECs (TEC compared with PLEC), and placental ECs compared with normal colon ECs (PLEC compared with NEC). Included are clones that showed at least a 2-fold difference in expression. The shaded area represents the TAGs (overexpressed in TECs compared with NECs and in TECs compared with PLECs). (C) TAG markers are strongly biased toward genes associated with extracellular matrix remodeling. (D) GAG/A markers (overexpressed in TECs and PLECs compared with NECs) show a diverse functional profile. (E) GAG/B markers (overexpressed in TECs and in activated HUVECs) are strongly biased to protein regulation. Functional distributions among the 3 different classes were significantly different (P < .001), as determined by χ2 test. TAG functional classification differed from that of GAG/A (P < .001) and GAG/B (P = .006), though GAG/A functional distribution did not differ significantly from that of GAG/B (P = .630). Classifications were obtained using the Gene Ontology Database and were graphically represented according to the data presented in Tables 2 and 3.
TAG markers are functionally classified as associated with late events of angiogenesis
We assigned a functional annotation to every gene and analyzed the distribution of genes within different functional classes. TAG markers were predominantly biased toward genes associated with cytoskeleton organization and extracellular matrix remodeling, indicative of late events in the process of tumor angiogenesis. Genes associated with protein regulation and cell growth, indicative of early events in the angiogenesis cascade, are underrepresented within TAG (Figure 1C). In contrast, genes functioning in cellular metabolism and protein regulation are dominantly present in the GAG/A class (Figure 1D). Similarly, GAG/B molecules are mainly involved in these early stages in the angiogenesis process, indicated by the predominant presence of genes associated with protein regulation (Figure 1E).
Validation of TAGs
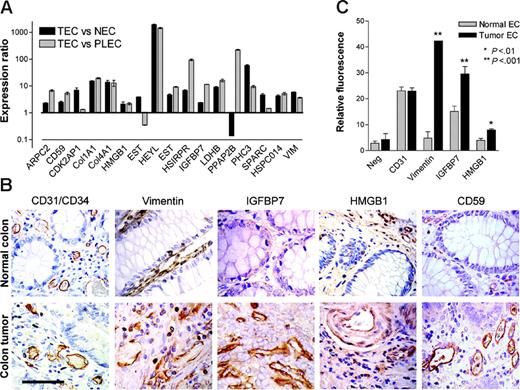
Overexpression of the 17 TAGs was confirmed using real-time quantitative PCR (RTQ-PCR) as a second independent validation technique. For 16 genes, overexpression in TECs compared with NECs was confirmed, as was overexpression in TECs compared with PLECs (Figure 2A). Taken together, 15 (88%) of 17 genes were validated by RTQ-PCR to be TAG markers.
For subsequent studies of TAG markers at the protein level, we selected 4 different membrane-associated or secreted molecules that may be feasible drug targets: (1) CD59 (TAG-3), a GPI-membrane–anchored inhibitor of complement activation19 ; (2) HMGB1 (TAG-21), a secreted cytokine and a nonhistone DNA-binding protein20,21 ; (3) insulin-like growth factor binding protein 7 (IGFBP7, TAG-29), a secreted molecule with growth factor modulating function22 ; and (4) vimentin (TAG-39), an intermediate filament protein recently demonstrated in macrophages to be actively secreted.23
Immunohistochemical analysis in colorectal carcinoma and normal colon indicated that all 4 proteins were overexpressed on the tumor vasculature (Figure 2B). Although vimentin, IGFBP7, and CD59 were predominantly expressed in the endothelial compartment, HMGB1 was also found to be expressed in stromal and epithelial cells (Figure 2B). Vimentin expression was detected in ECs of tumor and normal colon tissue, although it was heavily overexpressed on tumor endothelium. IGFBP7 expression in normal colon tissue was hardly detected, whereas tumor blood vessels showed abundant expression of IGFBP7. CD59 expression was mainly localized to vasculature, in particular to the luminal-cell membrane (Figure 2B). This is in line with its reported expression as a membrane protein with a role in protecting ECs from complement-mediated lysis by binding the complement proteins C8 and C9 to prevent the formation of the membrane-attack complex.19 HMGB1 staining was detected in ECs as cytoplasmic protein and in epithelial cells, where the localization was predominantly nuclear. In addition, diffuse stromal staining was observed. Protein expression was more abundant in colorectal tumor tissue than in normal colon tissue, predominantly in the stromal compartment, consistent with a secretion product (Figure 2B).24
Because immunohistochemistry is a qualitative rather than a quantitative technique, we determined the expression of these TAGs on freshly isolated ECs of tumor and normal tissues by flow cytometry. We were able to quantitatively confirm the overexpression of vimentin, IGFBP7, and HMGB1 protein on TECs compared with NECs (Figure 2C). Antibody species cross-reactivity prevented CD59 protein detection on freshly isolated ECs by this method. CD31 expression did not differ between TECs and NECs (Figure 2C). These observations further support the value of these TAG proteins as tumor EC markers.
Interference with TAG proteins inhibits angiogenesis in vitro and in vivo
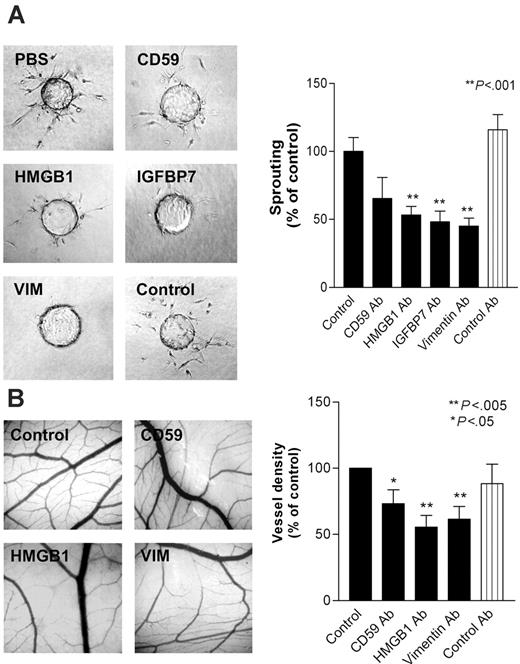
To investigate whether overexpression of the selected TAG markers is causally related to the process of angiogenesis, in vitro bioassays were performed. Antibodies directed against CD59, HMGB1, IGFBP7, and vimentin were tested for their effect on endothelial tube formation in an in vitro collagen gel–based sprout formation assay. Antibodies directed against the latter 3 showed significant inhibitory effects on sprout formation in vitro, whereas antibodies directed against CD59 had a limited effect. The control antibody did not show any effect (Figure 3A). These observations suggest that the targeted proteins are actively involved in the process of capillary tube formation in vitro.
To investigate whether these TAGs are involved in angiogenesis in vivo, antibodies were tested in the chorioallantoic membrane (CAM) of the chick embryo. All 4 antibodies were checked for reactivity with chick embryo endothelium by immunohistochemistry. Three of the antibodies (directed against CD59, HMGB1, and vimentin) positively stained chicken blood vessels and were therefore usable in the CAM assay (data not shown). Similar to the sprout formation assay, antibodies against CD59, HMGB1, and vimentin inhibited angiogenesis by 27%, 45%, and 40%, respectively, whereas a control antibody did not show any activity (Figure 3B). These results strongly suggest a role for these molecules in the process of angiogenesis and, together with the overexpression on tumor endothelium, support their potential for use in targeting of tumor vasculature as therapy against cancer.
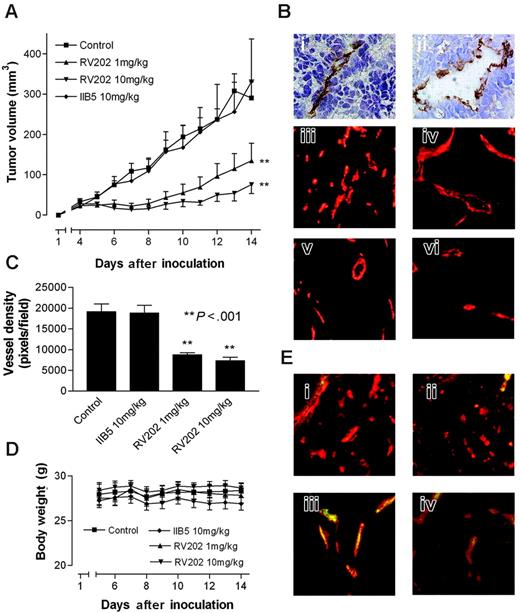
In a subsequent experiment, we provided evidence for the validity of our approach to identify endothelial targets that can be used for the treatment of cancer. Athymic mice bearing human LS174T colon carcinoma tumors were treated with the mouse-reactive RV202 mouse antivimentin antibody. This treatment resulted in a significant dose-dependent inhibition of tumor growth (Figure 4A). Because LS174T tumor cells do not express vimentin (Figure 4Bi,ii), this effect was the result of targeting endothelial vimentin. The latter was supported by the reduced microvessel density (MVD) in the RV202-treated tumors (Figure 4Bv,vi) compared with MVD in mice treated with control antibody or saline (Figures 4Biii,iv, C). Furthermore, we showed the binding of injected RV202 antibody to the tumor vasculature (Figure 4E), suggesting that activity is mediated through EC vimentin. Treatment with antivimentin antibody did not induce toxicity, as determined by normal weight gain (Figure 4D) and behavior.
Expression validation of TAGs. (A) Transcriptional validation of TAG markers by quantitative real-time PCR. Expression ratios (± SEM) in TECs compared with NECs (▪) and TECs compared with PLECs (▦) normalized for cyclophilin A are shown. (B) Immunohistochemical staining of CD31/CD34 and different TAGs in colorectal tumor and normal colon tissue sections. Vimentin expression is detected in ECs of tumor and normal colon tissue, although heavily overexpressed on tumor endothelium. IGFBP7 and CD59 show predominant endothelial expression that is more abundant in tumor tissue. HMGB1 is also localized to stromal and epithelial cells, suggestive of secretion. Scale bar = 50 μm. (C) Protein-expression levels (mean fluorescence intensity, ± SEM) of HMGB1, IGFBP7, and vimentin on tumor ECs compared with normal ECs, assessed by flow cytometry (*P < .01; **P < .001).
Expression validation of TAGs. (A) Transcriptional validation of TAG markers by quantitative real-time PCR. Expression ratios (± SEM) in TECs compared with NECs (▪) and TECs compared with PLECs (▦) normalized for cyclophilin A are shown. (B) Immunohistochemical staining of CD31/CD34 and different TAGs in colorectal tumor and normal colon tissue sections. Vimentin expression is detected in ECs of tumor and normal colon tissue, although heavily overexpressed on tumor endothelium. IGFBP7 and CD59 show predominant endothelial expression that is more abundant in tumor tissue. HMGB1 is also localized to stromal and epithelial cells, suggestive of secretion. Scale bar = 50 μm. (C) Protein-expression levels (mean fluorescence intensity, ± SEM) of HMGB1, IGFBP7, and vimentin on tumor ECs compared with normal ECs, assessed by flow cytometry (*P < .01; **P < .001).
Discussion
In the present study, we describe the identification of tumor angiogenesis genes (TAGs) that are overexpressed in tumor ECs compared with physiologically activated placental ECs and normal ECs. From the series of 17 TAGs, we identified several genes that encoded membrane-bound or secreted proteins. Four of these were selected so that their role in angiogenesis could be investigated and they could serve as a tumor endothelial target for therapeutic applications. We demonstrated that all 4 proteins are necessary in the process of angiogenesis and provided proof that TAGs can be used for intervention in angiogenesis and tumor growth with antibodies as a treatment opportunity. A crucial element in designing antiangiogenic and vascular targeting approaches is the identification of specific target molecules. Some tumor endothelial cell– associated markers have been identified,4-6,25,26 but translation to the clinic remains an obstacle. Because angiogenesis is not limited to pathologic conditions, careful evaluation of putative targets is warranted to prevent adverse effects associated with impaired physiologic angiogenesis. The subtraction strategy used provided us with genes that were overexpressed on tumor endothelium, but this did not exclude the possibility of expression on other endothelial cells. We demonstrated in a mouse tumor model that overexpression on tumor ECs provides a therapeutic window that can be used to treat cancer. Alternatively, the approach used did not exclude the identification of markers expressed by tumor cells, although this may even be considered an advantage for therapeutic application. Finally, it may be argued that a target can be expressed by other (vascular) cells (eg, pericytes). However, we demonstrated in the mouse model that injected antivimentin antibody definitely targets the tumor ECs (Figure 4E).
Inhibition of in vitro and in vivo angiogenesis by targeting of TAG proteins. (A) Sprout formation (± SEM) of bovine capillary ECs (BCEs) in a 3-dimensional collagen gel is inhibited by the addition of antibodies (10 μg/mL) directed against cell-surface and secreted TAGs (**P < .001). (B) Angiogenesis in the chick chorioallantoic membrane is inhibited by treatment with antibodies (10 μg/mL) directed against the indicated TAGs. Control antibody used is 9E10 anti-myc antibody (*P < .05; **P < .005).
Inhibition of in vitro and in vivo angiogenesis by targeting of TAG proteins. (A) Sprout formation (± SEM) of bovine capillary ECs (BCEs) in a 3-dimensional collagen gel is inhibited by the addition of antibodies (10 μg/mL) directed against cell-surface and secreted TAGs (**P < .001). (B) Angiogenesis in the chick chorioallantoic membrane is inhibited by treatment with antibodies (10 μg/mL) directed against the indicated TAGs. Control antibody used is 9E10 anti-myc antibody (*P < .05; **P < .005).
Vimentin is an extensively studied intermediate filament protein. In different tumor-cell lines, vimentin expression has been linked to increased invasiveness.27,28 We have shown quantitative data on the overexpression of vimentin on ECs in colon tumor samples compared with normal colon samples, both at the transcriptional level (Figure 2A) and the protein level (Figures 2B-C), suggesting a contribution of this protein to the tumor endothelial phenotype. We present evidence that targeting of vimentin by means of antibodies clearly inhibits angiogenesis in vitro and in vivo. In a mouse tumor model, an almost full abrogation of tumor growth was observed. Because of the absence of vimentin on tumor cells in this model (Figure 4Bi) and the demonstration of homing of injected antibody to the tumor ECs (Figure 4Eiii,iv), it was suggested that this resulted from a direct effect on tumor angiogenesis. The latter was supported by the marked inhibition of microvessel density. The absence of toxicity suggested that the effect of the antivimentin antibody in healthy tissues did give rise to concerns regarding the targeting of other vasculature. An interesting and as yet not fully elucidated aspect of vimentin is its presumed extracellular localization, which makes it targetable with antibodies. Vimentin is actively secreted in macrophages and functionally involved in the phagocytosis of bacteria.23 It may be argued that the targeting of tumor-associated macrophages is an alternative explanation for the observed antitumor effect. This production of vimentin by macrophages was reported from in vitro studies; to our knowledge, no evidence indicates that vimentin is secreted by macrophages in tumors. Therefore, without further evidence, we favor the view that the observed effects are mainly through inhibition of angiogenesis. Vimentin is furthermore detected on the surfaces of apoptotic and activated T-lymphocytes and apoptotic neutrophils.29-31 One might speculate that in tumor endothelium, vimentin is associated with integrins32,33 that mediate its externalization. This cell-biology aspect of vimentin awaits further study.
Although HMGB1 was originally identified as a nonhistone DNA-binding molecule,20 focus has shifted toward its role as a secreted cytokine. As an extracellular protein, it has been involved in the regulation of cell migration,34 tumorigenesis,35 cell activation,21 inflammation,36 and sprouting of ECs.13 Given its role in positive feedback mechanisms in endothelial-cell activation, cell migration, and invasion, targeting HMGB1 as a means of antiangiogenic therapy is promising. Here we demonstrated that an antibody directed against HMGB1 was effective at inhibiting endothelial-cell sprouting in vitro and angiogenesis in vivo.
CD59 is a GPI-anchored membrane protein and an inhibitor of complement activation.19 Cells overexpressing CD59 show resistance to antibody-mediated immunotherapy when complement activation is compromised.19 Neutralizing CD59 function can improve tumor immunotherapy.37 Therefore, theoretically, anti-CD59 antibodies may have dual efficacy in vivo by inducing complement on antibody binding and by inhibiting CD59 as a complement inhibitory protein. Complement activation does not apply in vitro, which may explain our result that antibodies directed against CD59 were not readily effective in our in vitro assays. It may well be that targeting of CD59 in vivo is more successful.38
IGFBP7 is a secreted protein that accumulates in the basement membrane, where it can bind collagen types II, IV, and V, heparan sulfates, and cytokines.22,39 By binding these collagens, it supports the organization of ECs into tubelike structures. In addition, IGFBP7 induces prostacyclin secretion from ECs, potent vasodilators that may mediate metastatic spread of tumors.22 In summary, there is ample evidence of an important function of IGFBP7 in tumor blood vessels. Indeed, we showed that the overexpression of IGFBP7 in tumor endothelium was evident at the transcriptional level and at the protein level. In addition, targeting IGFBP7 with an antibody clearly inhibited endothelial sprouting in vitro, possibly caused by inhibition of the interaction between IGFBP7 and collagens in the 3-dimensional culture matrix.
Inhibiting tumor growth by targeting tumor endothelial vimentin. (A) Tumor growth curves of LS174T human colon carcinoma tumor xenografts in nude mice, treated with vehicle, IIB5 anti-BrdU isotype-matched control antibody (10 mg/kg), or RV202 antivimentin antibody (10 mg/kg and 1 mg/kg). Antibodies were administered intraperitoneally every 3 days for a period of 12 days. A dose-dependent inhibition of tumor growth is evident in mice treated with antivimentin antibody (1 mg/kg, **P < .001; 10 mg/kg, **P < .001), whereas treatment with the isotype control antibody did not show inhibition of tumor growth (IIB5; 10 mg/kg, P = .661). (B) Immunohistochemical staining of LS174T tumor xenografts in mice with CD31 (i) and antivimentin antibody RV202(ii) show that vimentin expression is restricted to the endothelium. Microvessel staining with phycoerythrin-labeled CD31 antibody in control mice (iii), isotype control antibody–treated mice (iv), RV202 (1 mg/kg per treatment) (v), and RV202 (10 mg/kg per treatment)–treated mice (vi). (C) Quantification of microvessel density (±SEM) was assessed digitally (**P < .001; Student t test). (D) Body weight of mice during treatment, indicating absence of toxicity. (E) Detection of treatment antibodies targeted to the tumor endothelium. Mouse antibodies were detected (green fluorescence) in mice treated with saline (i), isotype control antibody (ii), RV202 (1 mg/kg per treatment) (iii), and RV202 (10 mg/kg per treatment) (iv). Endothelial cells are stained with phycoerythrin-labeled anti-CD31 antibody in red. Yellow indicates colocalization.
Inhibiting tumor growth by targeting tumor endothelial vimentin. (A) Tumor growth curves of LS174T human colon carcinoma tumor xenografts in nude mice, treated with vehicle, IIB5 anti-BrdU isotype-matched control antibody (10 mg/kg), or RV202 antivimentin antibody (10 mg/kg and 1 mg/kg). Antibodies were administered intraperitoneally every 3 days for a period of 12 days. A dose-dependent inhibition of tumor growth is evident in mice treated with antivimentin antibody (1 mg/kg, **P < .001; 10 mg/kg, **P < .001), whereas treatment with the isotype control antibody did not show inhibition of tumor growth (IIB5; 10 mg/kg, P = .661). (B) Immunohistochemical staining of LS174T tumor xenografts in mice with CD31 (i) and antivimentin antibody RV202(ii) show that vimentin expression is restricted to the endothelium. Microvessel staining with phycoerythrin-labeled CD31 antibody in control mice (iii), isotype control antibody–treated mice (iv), RV202 (1 mg/kg per treatment) (v), and RV202 (10 mg/kg per treatment)–treated mice (vi). (C) Quantification of microvessel density (±SEM) was assessed digitally (**P < .001; Student t test). (D) Body weight of mice during treatment, indicating absence of toxicity. (E) Detection of treatment antibodies targeted to the tumor endothelium. Mouse antibodies were detected (green fluorescence) in mice treated with saline (i), isotype control antibody (ii), RV202 (1 mg/kg per treatment) (iii), and RV202 (10 mg/kg per treatment) (iv). Endothelial cells are stained with phycoerythrin-labeled anti-CD31 antibody in red. Yellow indicates colocalization.
Some genes resided in chromosomal locations associated with colorectal cancer (Tables 2, 3), in particular the gain of loci containing HMGB1, HSPC014, COL4A1, and RPL21 on chromosome 13. Although this is an interesting observation, evidence is limited that chromosomal abnormalities contribute to the phenotype of tumor ECs.40,41
The extensive bias toward genes functioning in extracellular matrix remodeling among the TAG markers (Figure 1C) was also evident in published SAGE data sets of isolated tumor ECs that were only compared with normal ECs. Genes supposedly playing a role in the initiation of angiogenesis are rarely identified in gene-expression profiling of ECs derived from tumors.3-6 In contrast, gene-expression profiling of growth factor–stimulated ECs and of 3-dimensional EC cultures show biases to early events such as cell turnover and cell adhesion, respectively (for a review, see van Beijnum and Griffioen3 ), and can be paralleled with the functional gene distribution in GAG/B and GAG/A, respectively (Figure 1D-E). These differences in global gene-expression profiles in ECs cultured in vitro compared with isolated ECs are likely influenced by differences in the microenvironment and the duration of stimuli in vitro compared with in vivo. Therefore, extrapolation of data generated by in vitro experiments to the in vivo situation may be limited because it appears difficult to accurately mimic in vitro the complex temporal and spatial actions present in vivo, emphasizing the importance of approaches that make use of more relevant cell sources such as tissue-derived cells.
The data presented here demonstrate the presence of genes that show differential expression in tumor angiogenesis compared with physiologic angiogenesis, which may be sufficient to create a therapeutic window for cancer treatment without inducing significant adverse effects associated with interference in physiologic angiogenesis. The successful targeting of tumor endothelium with antibodies directed at one such TAG, vimentin, and the subsequent inhibition of tumor growth underscores the power of our approach and holds promise for the therapeutic development of the remaining TAGs.
Prepublished online as Blood First Edition Paper, June 22, 2006; DOI 10.1182/blood-2006-02-004291.
Supported by the sixth EU Framework Programme (Integrated Project Angiotargeting; contract no. 504743; “Life Sciences, Genomics, and Biotechnology for Health”) and by a grant from GROW, Maastricht University.
J.R.v.B. and A.W.G. designed the research and wrote the paper. J.R.v.B., E.v.d.L., R.P.M.D., and B.M.M.Z. performed the research and analyzed the data. F.C.S.R. and K.H.M. provided vital reagents and tools.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Susan Joosten and Mat Rousch for technical assistance with endothelial-cell selection.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal