Abstract
We have previously associated high natural killer (NK)–cell activity and protection against HIV-1 infection in Vietnamese exposed uninfected intravascular drug users (EUs). Considering that activating and inhibitory signals sensed by NK-cell receptors regulate NK-cell activation, we performed phenotypic and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) transcript analyses of the NK-cell receptor (NKR) repertoire in 25 EUs, 19 HIV+ intravenous drug users, and 26 uninfected blood donors. Although NK-cell activation was not linked to a unique NKR repertoire in EUs, various patterns consistent with NK-cell activation were detected in EUs: high KIR3DS1/KIR3DL1 ratio associated with down-regulated KIR3DL1 transcript levels, KIR2DL3+ low-affinity receptor expansion associated to group HLA-C1 ligand in 2DS2−/2DL2− EUs, enhanced NKG2C/NKG2A ratio, and increased CD69 expression. Remarkably, EUs exhibited high constitutive degranulation activity in the absence of exogenous stimulation, as shown by the CD107a assay. Furthermore, CD161 expression was increased within the CD107a+ NK-cell compartment. Our results suggest that in response to viral exposition, particular genetic or regulated features of the NKR repertoire of EUs contribute to their high constitutive NK-cell potential. This might allow NK cells to generate a more rapid and effective immune response to HIV-1, thereby contributing to prevention toward infection.
Introduction
Natural killer (NK) cells are key actors in innate host immune responses to a variety of pathologic challenges. NK-cell activation results from integration of signals induced by the interaction of a highly diversified repertoire of inhibitory and activating NK-cell receptors (NKRs) with constitutive or inducible ligands expressed on target cells, including virus-infected cells.1–5 Inhibitory receptors essentially recognize specific subsets of classical and nonclassical HLA class I molecules, and include KIR-L, ILT2/CD85j, and heterodimeric CD94/NKG2A NKRs. Inhibitory KIR2DL1 recognizes HLA-C group 2 alleles (HLA-C2: Asn77Lys80), whereas KIR2DL2 and KIR2DL3 both recognize HLA-C group 1 alleles (HLA-C1: Ser77Asn80) but with different affinity.2,6 KIR3DL1 recognizes the HLA-Bw4 group of alleles7 and KIR3DL2 HLA-A11/HLA-A3. Recognition of the relevant HLA class I molecule by KIR-L receptors delivers NK inhibitory signals, whereas nonrecognition allows stimulatory signals to activate NK cells. MHC-KIR interactions shape the NK-cell repertoire and calibrate NK-cell potential to control infectious, tumoral, or alloreactive challenges.5,8,9 In contrast to KIR-L, activating KIR-S counterparts can deliver activating signals to NK cells, but their physiologic ligands remain mostly uncharacterized. Recent evidence suggests that KIR2DS1 and KIR2DS2, respectively, share the capacity to bind HLA-C2 or HLA-C1 ligands with their inhibitory counterparts in vitro, but with lower affinity.10 Receptors delivering activating signaling include NK-specific natural cytotoxicity receptors (NCRs), in addition to receptors more broadly distributed among T-lymphocyte subsets, such as activating KIR-S, CD94/NKG2C, NKG2D, CD226/DNAM, and CD161/NKRP1. Activated NK cells mediate various effector mechanisms, including antibody-dependent and natural cytotoxicity, and secretion of chemokines and cytokines with antiviral activity.1,2,4,5 In addition to these effector functions, crosstalk between NK cells and antigen-presenting cells, and cytokine secretion by NK cells influence the efficiency of adaptive immune responses, thereby constituting a major link between innate and adaptive immunity.6,7
NK cells therefore have the potential to contribute to the prevention and control of viral infections, including HIV-1 infection. The NK-cell phenotype and functions are altered in HIV+ patients.7–14 Up-regulation of inhibitory receptors in viremic patients has been proposed as a basis for the inhibition of NK-cell activity observed in such subjects.12,15,16 Decreased expression of NKp30 and NKp46 NCR-activating receptors has also been linked to impaired cytolytic potential in viremic patients relative to highly activated antiretroviral therapy (HAART)–treated aviremic subjects and to healthy individuals.11–13 Partial quantitative and functional reconstitution of NK cells can be obtained during HAART, thus linking NK-cell suppression to HIV replication.14–17 A role of NK cells in controlling HIV disease progression is suggested by the association between delayed progression to AIDS and concomitant presence in the host genome of genes that contribute to the HLA-Bw4 allelic group and activating KIR3DS1 NK receptor genes.18
The rare individuals who are naturally resistant to HIV-1 infection represent a unique model in which to examine the role of innate immunity in the control of HIV-1, with potential implications for preventive and therapeutic strategies. We have previously observed enhanced NK-cell cytotoxic activity and cytokine and β-chemokine production in Vietnamese intravascular drug users (IDUs) who remained uninfected despite several years of high-risk exposure to HIV-1, when compared with HIV+ drug users IDUs and low-risk uninfected blood donors.19 In contrast, NK-cell activity was low in some drug users who seroconverted during the study.19 These results suggested that NK cells contribute to the protection of these exposed uninfected individuals (EUs). A recent study analyzing genomic HLA-KIR combinations in African female sex workers suggests that resistance to HIV-1 infection associates with inhibitory KIR in absence of their HLA ligand.20 The present study was designed to investigate whether specific patterns of NK-cell receptor expression associate with higher NK cytotoxic and secretory activities in Vietnamese EU. We therefore conducted a comparative genomic, transcriptional, and phenotypic analysis of the NK-cell repertoire, in association with the functional assay of the CD107a degranulation potential of NK cells, in EUs, HIV+, and uninfected low-risk blood donors.
Patients, materials, and methods
Study population
Twenty-five exposed uninfected (EU) intravascular drug users (IDUs) from a previously described cohort recruited in the Binh Trieu Hospital in Ho Chi Minh City19,21 were included in this study. Risk factors for HIV exposure, the high seroprevalence of other blood-borne viruses such HBV, HCV, and HTLV, and the follow-up for this substudy were previously described.19 Enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (PCR) confirmed the HIV-1− status of EUs as described.21 Nineteen HIV+ IDUs were studied (HIV+ IDUs) and offered free diagnosis, medical follow-up, and treatment for opportunistic infections. Twenty-six low-risk uninfected controls (Cs) were recruited from the Blood Bank of Ho Chi Minh City. Informed consent was obtained from each participant at recruitment, in accordance with the Declaration of Helsinki. All the subjects were men, and there was no age difference among the study groups. The Ethics Committee of Binh Trieu Hospital, Ho Chi Minh City, approved the protocol. Peripheral blood mononuclear cells (PBMCs) were recovered from blood samples after Ficoll-Hypaque gradient and cryopreserved in 90% FCS, 10% DMSO. When PBMC or NK-cell counts were a limiting factor, in particular in HIV+ individuals, functional, phenotypic, and transcriptional analysis of NK cells were privileged over DNA extraction as a source of informative data.
HLA and KIR genotyping
HLA genotyping was performed with a reverse dot blot assay using a commercial kit and according the manufacturer's instructions (Innogenetics, Gent, Belgium). Due to the limited availability of biologic samples, KIR genotyping could only be performed on 14 EUs, 8 HIV+ IDUs, and 12 controls. A reverse hybridization assay read using Luminex technology was used for KIR genotyping according to manufacturer's instructions (One Lambda, Canoga Park, CA).
RNA purification and real time RT-PCR analysis of KIR transcript levels
Total RNA was isolated from PBMCs using RNeasy mini kits (Qiagen, Valencia, CA) including a DNase I digestion step removing gDNA. Total RNA (5 μg) was converted to cDNA using 200 U Supercript II reverse transcriptase (Invitrogen, Cergy Pontoise, France). Real-time PCR amplification was performed with the FastStart DNA MasterPLUS SYBR Green I kit as recommended by the manufacturer, on a LightCycler instrument (Roche, Meylan, France). Cycling conditions were 10 minutes at 95°C (hot-start PCR), followed by 40 cycles, 10 seconds at 95°C (denaturation), 10 seconds at 62°C (annealing), and 20 seconds at 72°C (elongation). Melting curve analysis was performed to check the specificity of amplification. Reported values are relative numbers of specific transcripts detected per 106GAPDH transcripts. KIR expression was analyzed with primers specifically designed to discriminate KIR locus, as follows (forward and reverse sequences): KIR2DL3 5′-CTCCTTCATCGCTGGTGCTG-3′ and 5′-GGCAGGAGACAACTTTGGATCA-3′; KIR2DL4 5′-CTCCCAGAGCTCCTTTGACATC-3′ and 5′-GAGCCGAAGCATCTGTAGGTCT-3′; KIR3DS1 5′-CCAAGCTCCAAATCTGGTAACCT-3′ and 5′-TCAGAATCCTCGCTGTTCACTTCT-3′; KIR3DL1 5′-GGACATCGTGGTCACAGGTCC-3′ and 5′-CACTGAGGTCCCAATCAGAATG-3′; KIR3DL2 5′-CCAACTTCTCCATCGGTCCCT-3′ and 5′-GTTGACCTTGGGCACTGCAC-3′; GAPDH 5′-GGTGGTCTCCTCTGACTTCAACA-3′ and 5′-GTTGCTGTAGCCAAATTCGTTGT-3′.
Phenotypic analysis
The percentage of CD3−CD56+CD16+ NK cells among total lymphocytes and the size of each NKR+ NK-cell subset were determined by 3-color flow cytometry (Beckman Coulter, Villepinte, France). Eleven distinct NK-cell subsets were defined as NK cells (CD3−CD56+) expressing specific NKRs. The following antibodies were used: FITC-conjugated anti-CD3 (IgG1, UCHT1); PC5-conjugated anti-CD56 (IgG1, N901); PE-conjugated: anti-CD158a/h (clone EB6, IgG1 recognizing KIR2DS1/2DL1), anti-CD158b1/b2/j (clone GL183, IgG1 recognizing KIR2DS2/2DL2/2DL3), anti-CD158e1/e2 (clone Z27.3.7, IgG1, recognizing KIR3DL1/KIR3DS1), anti-CD158i (clone FES172, IgG2a, recognizing KIR2DS4), anti-CD25 (IgG2a, B.1.49.9), anti-CD161 (IgG1, 191B8), anti-NKp30 (IgG1, Z25), anti-NKp46 (IgG1, BAB281), anti-CD85j (IgG1, HP-F1), anti-CD159a (IgG2b, Z199), all from Beckman Coulter; anti-CD69 (IgG1, FN50, BD PharMingen, San Diego, CA); and anti-NKG2C (IgG1, 134591, R&D Systems, Minneapolis, MN).
Flow cytometer analysis of NK function using the degranulation marker CD107a/LAMP-1
CD107a lysosome-associated membrane protein-1 (LAMP-1) expression was used to measure NK-cell degranulation, as described.22,23 PBMCs were incubated with or without HLA class I− K562 target cells (American Type Culture Collection, Manassas, VA) at an effector-target (E/T) ratio of 2:1 for 3 hours. CD107a-PC5 antibody (IgG1, H4A3) and GolgiStop reagent diluted at 1:150 (BD PharMingen) were added directly to the cultures. Cells were then stained with anti-CD3–FITC (IgG1, UCHT1), CD56-PC7 (IgG1, N901) prior to fluorescence-activated cell sorting (FACS) analysis. Additional NKR-PE staining was performed with anti-CD85j, anti-CD159a, anti-CD161 (Beckman Coulter), anti-NKG2C (R&D Systems), and anti-CD69 (BD PharMingen) antibodies. All samples were analyzed by means of 4-color flow cytometry and CXP software (Beckman Coulter).
Statistical analysis
Analyses were performed with Prism software (GraphPad 4.0b, GraphPad, San Diego, CA) implementing the nonparametric Kruskal-Wallis test followed by the Dunn post-test to compare 3 or more continuous variables, the Mann-Whitney test to compare 2 unpaired groups, and the Wilcoxon matched pairs test for the CD107a degranulation assay results.
Results
Distinctive features of the KIR repertoire in EU
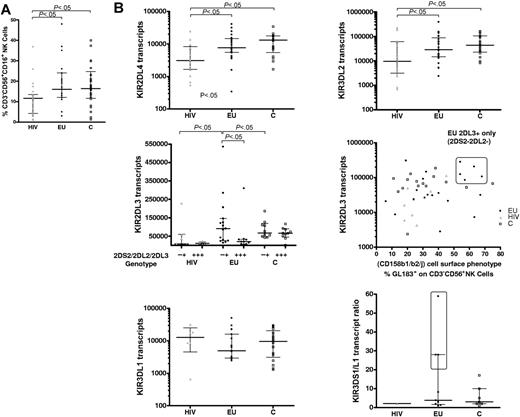
The size of the NK-cell compartment did not differ between EUs and controls (median 16% and 16.4%, respectively), whereas NK-cell percentages were lower in HIV+ IDUs (HIV; median 11.6%, P < .05, Figure 1A).
Genomic and real-time PCR transcriptional analysis of the KIR repertoire. (A) The size of the NK-cell compartment was analyzed by flow cytometry (% CD3−CD56+CD16+ cells) in exposed HIV-infected IDU (HIV, ▴) exposed HIV-uninfected IDU (EU, •), and unexposed control blood donors (C, □). (B) Real-time PCR analysis of altered KIR transcript patterns in HIV and EUs. KIR2DL4, KIR3DL2, KIR2DL3, and KIR3DL1 mRNA transcript levels are represented as relative copy numbers per 106GAPDH transcripts. KIR2DL3 transcript levels where examined according to the alternative KIR2DL3+KIR2DS2−KIR2DL2− or KIR2DL3+KIR2DS2+KIR2DL2+ genomic gene combinations (middle left) and also plotted according to the size of the phenotypically defined GL183+ NK-cell subset (middle right). A high percentage of GL183+ NK cells (> 50% of GL183+ NK cells within CD3−CD56+ NK cells) identified a specific subgroup of KIR2DL3+KIR2DS2−KIR2DL2− EU individuals who exhibit higher KIR2DL3 transcript levels in the absence of KIR2DS2/KIR2DL2 transcription (boxed area). Activating KIR3DS1/inhibitory KIR3DL1 transcript ratio is represented (bottom right). EUs with higher KIR3DS1/KIR3DL1 ratios as compared with C and HIV+ are boxed. Horizontal bars represent the median and the 25th and 75th percentiles.
Genomic and real-time PCR transcriptional analysis of the KIR repertoire. (A) The size of the NK-cell compartment was analyzed by flow cytometry (% CD3−CD56+CD16+ cells) in exposed HIV-infected IDU (HIV, ▴) exposed HIV-uninfected IDU (EU, •), and unexposed control blood donors (C, □). (B) Real-time PCR analysis of altered KIR transcript patterns in HIV and EUs. KIR2DL4, KIR3DL2, KIR2DL3, and KIR3DL1 mRNA transcript levels are represented as relative copy numbers per 106GAPDH transcripts. KIR2DL3 transcript levels where examined according to the alternative KIR2DL3+KIR2DS2−KIR2DL2− or KIR2DL3+KIR2DS2+KIR2DL2+ genomic gene combinations (middle left) and also plotted according to the size of the phenotypically defined GL183+ NK-cell subset (middle right). A high percentage of GL183+ NK cells (> 50% of GL183+ NK cells within CD3−CD56+ NK cells) identified a specific subgroup of KIR2DL3+KIR2DS2−KIR2DL2− EU individuals who exhibit higher KIR2DL3 transcript levels in the absence of KIR2DS2/KIR2DL2 transcription (boxed area). Activating KIR3DS1/inhibitory KIR3DL1 transcript ratio is represented (bottom right). EUs with higher KIR3DS1/KIR3DL1 ratios as compared with C and HIV+ are boxed. Horizontal bars represent the median and the 25th and 75th percentiles.
KIR+ NK-cell subset distribution was analyzed by flow cytometry in 25 EUs (Table 1), 19 HIV+, and 26 unexposed Vietnamese control donors. Four KIR+CD3−CD56+ NK-cell subsets were evaluated using antibodies EB6 (CD158a/h or KIR2DL1/2DS1), GL183 (CD158b1/b2/j or KIR2DL2/2DL3/2DS2), Z27.3.7 (CD158e1/e2 or KIR3DL1/3DS1), and FES172 (CD158i or KIR2DS4). As previously described in southeast France,24 high interindividual heterogeneity was observed in the distribution of each KIR+ NK-cell subset. Still, no significant differences in the size of these KIR+ NK-cell phenotype subsets were observed among the 3 Vietnamese groups (data not shown).
Genomic, transcriptional, and phenotypic analysis of the KIR repertoire in EUs
| EU . | HLA-C . | HLA-B . | KIR2DS1/KIR2DL1 . | KIR2DS2/2DL2/2DL3 . | KIR3DS1/KIR3DL1 . | KIR2DS4 . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype, 2DS1/2DL1 . | Transcripts, 2DS1/2DL1 (ratio) . | Phenotype, % EB6+ . | Genotype, 2DS2/2DL2/2DL3 . | Transcripts, 2DS2/2DL2/2DL3 . | Phenotype, % GL183+ . | Genotype, 3DS1/3DL1 . | Transcripts, 3DS1/3DL1 (ratio) . | Phenotype, % Z27.3.7+ . | Genotype, 2DS4 . | Transcripts, 2DS4/1D . | Phenotype, % FES172+ . | |||
| Individual values for each EU | ||||||||||||||
| 1 | C1 | Bw4 Bw6 | −/+ | −/+ | 15 | −/−/+ | −/−/+ | 24 | −/+ | −/+ | 30 | + | +/− | 53 |
| 2 | C1 | Bw4 Bw6 | −/+ | −/+ | 21 | −/−/+ | −/−/+ | 57* | −/+ | −/+ | 7 | + | +/− | 81 |
| 3 | C1 | Bw4 Bw6 | ND | −/+ | ND | ND | −/−/+ | ND | ND | −/+ | ND | ND | +/− | ND |
| 4 | C1 | Bw4 Bw6 | −/+ | −/+ | 6 | +/+/+ | +/+/+ | 63* | −/+ | −/+ | 13 | + | −/+ | 38 |
| 5 | C1 | Bw6 | −/+ | −/+ | 32 | −/−/+ | −/−/+ | 68* | −/+ | −/− | 0 | + | −/− | 0 |
| 6 | C1 | Bw6 | +/+ | +/+ (2.8) | 29 | −/−/+ | −/−/+ | 26 | +/+ | +/+ (28)* | 6 | + | −/+ | 0 |
| 7 | C1 | Bw6 | +/+ | +/+ (61) | 4 | +/+/+ | +/+/+ | ND | +/+ | +/+ (59)* | 1 | + | −/+ | 0 |
| 8 | C1 | Bw6 | ND | +/+ (0.9) | 8 | ND | +/+/+ | 33 | ND | −/+ | 22 | ND | +/+ | 45 |
| 9 | C1 | Bw6 | ND | +/+ (3.7) | ND | ND | −/−/+ | ND | ND | +/+(1.3) | ND | ND | −/+ | ND |
| 10 | C1 C2 | Bw4 Bw6 | +/+ | +/− | 13 | +/+/+ | +/+/+ | 67* | +/+ | +/− | 6 | + | +/+ | 24 |
| 11 | C1 C2 | Bw4 Bw6 | ND | +/+ (4.0) | 50 | ND | −/−/+ | 54* | ND | +/+ (3.0) | 27 | ND | +/+ | 44 |
| 12 | C1 C2 | Bw4 Bw6 | +/+ | +/+ (1.0) | 83 | +/+/+ | +/+/+ | 6 | +/+ | +/+ (1.9) | 6 | + | +/− | 37 |
| 13 | C1 C2 | Bw4 Bw6 | +/+ | +/+ (2.3) | 21 | −/−/+ | −/−/+ | 36 | +/+ | +/+ (0.9) | 26 | + | −/+ | 0 |
| 14 | C1 C2 | Bw4 Bw6 | ND | −/+ | ND | ND | −/−/+ | ND | ND | −/− | ND | ND | +/+ | ND |
| 15 | C1 C2 | Bw6 | −/+ | −/− | 10 | +/+/+ | −/+/+ | 41 | +/+ | +/− | ND | + | +/+ | 16 |
| 16 | C1 C2 | Bw6 | +/+ | +/+ (1.0) | 38 | −/−/+ | −/−/+ | 32 | +/+ | +/+ (28)* | 8 | + | −/+ | 0 |
| 17 | C1 C2 | Bw6 | −/+ | −/+ | 9 | +/+/+ | +/+/+ | 12 | −/+ | −/+ | 5 | + | +/− | 30 |
| 18 | C1 C2 | Bw6 | +/+ | +/+ (0.1) | 47 | −/−/+ | −/−/+ | 63* | −/+ | −/+ | 17 | + | +/+ | 22 |
| 19 | C2 | Bw4 Bw6 | ND | −/+ | ND | ND | −/−/+ | 31 | ND | −/+ | ND | ND | +/− | ND |
| 20 | C2 | Bw6 | −/+ | −/+ | ND | −/−/+ | −/−/+ | ND | −/+ | −/+ | ND | + | −/+ | ND |
| 21 | ND | ND | ND | +/+ (1.0) | 25 | ND | −/−/+ | 54* | ND | +/+ (8.1) | 10 | ND | +/− | 40 |
| 22 | ND | ND | ND | +/+ (3.45) | 31 | ND | +/+/+ | 46 | ND | +/+ (3.8) | 21 | ND | +/− | 45 |
| 23 | ND | ND | ND | −/+ | 5 | ND | +/+/+ | 19 | ND | −/+ | 1 | ND | −/+ | 0 |
| 24 | ND | ND | ND | +/+ (47) | 20 | ND | +/+/+ | 40 | ND | ND | 8 | ND | −/− | ND |
| 25 | ND | ND | ND | −/+ | ND | ND | −/−/+ | 30 | ND | −/+ | ND | ND | −/+ | ND |
| Summary of values by group, median (25th-75th percentile ranges) | ||||||||||||||
| HIV | — | — | — | 1.4 (1.4-1.4) | 11.3 (7.6-22.5) | — | — | 28.4 (19.5-44.1) | — | 2.1 (2.1-2.1) | 11.3 (6.3-15.7) | — | — | 33.7 (18.1-37.7) |
| Total EUs | — | — | — | 1.7 (0.5-3.6) | 15 (9-31) | — | — | 36 (23-56.7) | — | 4.7 (1.5-28.0) | 8.6 (6.9-17.5) | — | — | 30.5 (20.9-42.0) |
| C | — | — | — | 4.1 (3.1-4.8) | 14.5 (7-26) | — | — | 35.2 (20-48.3) | — | 3.0 (2.0-10.0) | 12 (8.8-21.5) | — | — | 32.6 (21.7-65.0) |
| EU . | HLA-C . | HLA-B . | KIR2DS1/KIR2DL1 . | KIR2DS2/2DL2/2DL3 . | KIR3DS1/KIR3DL1 . | KIR2DS4 . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype, 2DS1/2DL1 . | Transcripts, 2DS1/2DL1 (ratio) . | Phenotype, % EB6+ . | Genotype, 2DS2/2DL2/2DL3 . | Transcripts, 2DS2/2DL2/2DL3 . | Phenotype, % GL183+ . | Genotype, 3DS1/3DL1 . | Transcripts, 3DS1/3DL1 (ratio) . | Phenotype, % Z27.3.7+ . | Genotype, 2DS4 . | Transcripts, 2DS4/1D . | Phenotype, % FES172+ . | |||
| Individual values for each EU | ||||||||||||||
| 1 | C1 | Bw4 Bw6 | −/+ | −/+ | 15 | −/−/+ | −/−/+ | 24 | −/+ | −/+ | 30 | + | +/− | 53 |
| 2 | C1 | Bw4 Bw6 | −/+ | −/+ | 21 | −/−/+ | −/−/+ | 57* | −/+ | −/+ | 7 | + | +/− | 81 |
| 3 | C1 | Bw4 Bw6 | ND | −/+ | ND | ND | −/−/+ | ND | ND | −/+ | ND | ND | +/− | ND |
| 4 | C1 | Bw4 Bw6 | −/+ | −/+ | 6 | +/+/+ | +/+/+ | 63* | −/+ | −/+ | 13 | + | −/+ | 38 |
| 5 | C1 | Bw6 | −/+ | −/+ | 32 | −/−/+ | −/−/+ | 68* | −/+ | −/− | 0 | + | −/− | 0 |
| 6 | C1 | Bw6 | +/+ | +/+ (2.8) | 29 | −/−/+ | −/−/+ | 26 | +/+ | +/+ (28)* | 6 | + | −/+ | 0 |
| 7 | C1 | Bw6 | +/+ | +/+ (61) | 4 | +/+/+ | +/+/+ | ND | +/+ | +/+ (59)* | 1 | + | −/+ | 0 |
| 8 | C1 | Bw6 | ND | +/+ (0.9) | 8 | ND | +/+/+ | 33 | ND | −/+ | 22 | ND | +/+ | 45 |
| 9 | C1 | Bw6 | ND | +/+ (3.7) | ND | ND | −/−/+ | ND | ND | +/+(1.3) | ND | ND | −/+ | ND |
| 10 | C1 C2 | Bw4 Bw6 | +/+ | +/− | 13 | +/+/+ | +/+/+ | 67* | +/+ | +/− | 6 | + | +/+ | 24 |
| 11 | C1 C2 | Bw4 Bw6 | ND | +/+ (4.0) | 50 | ND | −/−/+ | 54* | ND | +/+ (3.0) | 27 | ND | +/+ | 44 |
| 12 | C1 C2 | Bw4 Bw6 | +/+ | +/+ (1.0) | 83 | +/+/+ | +/+/+ | 6 | +/+ | +/+ (1.9) | 6 | + | +/− | 37 |
| 13 | C1 C2 | Bw4 Bw6 | +/+ | +/+ (2.3) | 21 | −/−/+ | −/−/+ | 36 | +/+ | +/+ (0.9) | 26 | + | −/+ | 0 |
| 14 | C1 C2 | Bw4 Bw6 | ND | −/+ | ND | ND | −/−/+ | ND | ND | −/− | ND | ND | +/+ | ND |
| 15 | C1 C2 | Bw6 | −/+ | −/− | 10 | +/+/+ | −/+/+ | 41 | +/+ | +/− | ND | + | +/+ | 16 |
| 16 | C1 C2 | Bw6 | +/+ | +/+ (1.0) | 38 | −/−/+ | −/−/+ | 32 | +/+ | +/+ (28)* | 8 | + | −/+ | 0 |
| 17 | C1 C2 | Bw6 | −/+ | −/+ | 9 | +/+/+ | +/+/+ | 12 | −/+ | −/+ | 5 | + | +/− | 30 |
| 18 | C1 C2 | Bw6 | +/+ | +/+ (0.1) | 47 | −/−/+ | −/−/+ | 63* | −/+ | −/+ | 17 | + | +/+ | 22 |
| 19 | C2 | Bw4 Bw6 | ND | −/+ | ND | ND | −/−/+ | 31 | ND | −/+ | ND | ND | +/− | ND |
| 20 | C2 | Bw6 | −/+ | −/+ | ND | −/−/+ | −/−/+ | ND | −/+ | −/+ | ND | + | −/+ | ND |
| 21 | ND | ND | ND | +/+ (1.0) | 25 | ND | −/−/+ | 54* | ND | +/+ (8.1) | 10 | ND | +/− | 40 |
| 22 | ND | ND | ND | +/+ (3.45) | 31 | ND | +/+/+ | 46 | ND | +/+ (3.8) | 21 | ND | +/− | 45 |
| 23 | ND | ND | ND | −/+ | 5 | ND | +/+/+ | 19 | ND | −/+ | 1 | ND | −/+ | 0 |
| 24 | ND | ND | ND | +/+ (47) | 20 | ND | +/+/+ | 40 | ND | ND | 8 | ND | −/− | ND |
| 25 | ND | ND | ND | −/+ | ND | ND | −/−/+ | 30 | ND | −/+ | ND | ND | −/+ | ND |
| Summary of values by group, median (25th-75th percentile ranges) | ||||||||||||||
| HIV | — | — | — | 1.4 (1.4-1.4) | 11.3 (7.6-22.5) | — | — | 28.4 (19.5-44.1) | — | 2.1 (2.1-2.1) | 11.3 (6.3-15.7) | — | — | 33.7 (18.1-37.7) |
| Total EUs | — | — | — | 1.7 (0.5-3.6) | 15 (9-31) | — | — | 36 (23-56.7) | — | 4.7 (1.5-28.0) | 8.6 (6.9-17.5) | — | — | 30.5 (20.9-42.0) |
| C | — | — | — | 4.1 (3.1-4.8) | 14.5 (7-26) | — | — | 35.2 (20-48.3) | — | 3.0 (2.0-10.0) | 12 (8.8-21.5) | — | — | 32.6 (21.7-65.0) |
HLA-B and -C typing was performed and grouped as HLA-C1 (Asn80), HLA-C2 (Lys80), and Bw4/Bw6 groups, according to KIR recognition pattern. Real-time PCR evaluation of each KIR transcript level was performed in 25 exposed uninfected individuals (EUs) and associated to KIR genotyping in 14 EUs. When KIR transcripts were detected (+), the ratio between activating and inhibitory counterparts was indicated (in parentheses). Phenotypic expression of KIR receptors is indicated as the percentage of KIR+ cells among CD3−CD56+ NK cells using EB6+, GL183+, Z27.3.7+, and FES172† mAbs. Data from total EUs were analyzed in reference to median values and 25th to 75th percentiles (in parentheses) in low-risk Vietnamese unexposed control blood donors (C) and HIV-infected individuals (HIV).
ND indicates not determined; —, not applicable.
Values over the 75th percentile observed in control donors.
Because phenotypic analysis of the KIR repertoire with currently available monoclonal antibodies (mAbs) cannot discriminate between inhibitory and activating KIR-expressing NK-cell subsets, we further performed real-time PCR analysis of the KIR repertoire to quantify the mRNA transcripts corresponding to each of the 5 activating KIR-S and 6 inhibitory KIR-L receptors. In addition, HLA and KIR genotyping was also performed in 14 EUs, 8 HIV+ IDUs, and 12 controls for whom enough cells were available for DNA analysis (Table 1).
Overall, the KIR gene frequencies observed in our control group (Table 2) were comparable to those previously reported in Vietnamese blood donors.25 Moreover, we show that KIR transcripts frequencies were mostly similar in EUs and controls (Table 2). In contrast, transcript frequencies of given KIRs were selectively altered within the HIV+ group (Table 2).
Frequency of KIR transcript detection
| . | KIR Transcript Frequency . | KIR Genotype C (Toneva et al25 ) . | ||
|---|---|---|---|---|
| HIV+ . | EUs . | C . | ||
| KIR2DS1 | 27 | 52 | 48 | 50 (37) |
| KIR2DS2 | 39 | 36 | 48 | 50 (41) |
| KIR2DS3 | 0 | 8 | 24 | 58 (31) |
| Expressed KIR2DS4 | 77* | 60 | 67 | NT (NT) |
| KIR2DS5 | 47 | 28 | 33 | 42 (NT) |
| KIR2DL1 | 73 | 92 | 100 | 100 (98) |
| KIR2DL2 | 33 | 40 | 52 | 50 (45) |
| KIR2DL3 | 76* | 100 | 95 | 92 (66) |
| KIR2DL4 | 88 | 100 | 100 | 100 (NT) |
| KIR2DL5 | 22 | 40 | 52 | 50 (NT) |
| KIR3DS1 | 18 | 48 | 43 | 50 (41) |
| KIR3DL1 | 35† | 76 | 86 | 92 (88) |
| KIR3DL2 | 94 | 100 | 100 | 100 (100) |
| KIR3DL3 | 78 | 60 | 71 | 100 (NT) |
| 2DL3+2DS2−2DL2− | 47 | 60 | 38 | NT (NT) |
| 2DL3+2DS2+/−2DL2+/− | 53 | 40 | 62 | NT (NT) |
| . | KIR Transcript Frequency . | KIR Genotype C (Toneva et al25 ) . | ||
|---|---|---|---|---|
| HIV+ . | EUs . | C . | ||
| KIR2DS1 | 27 | 52 | 48 | 50 (37) |
| KIR2DS2 | 39 | 36 | 48 | 50 (41) |
| KIR2DS3 | 0 | 8 | 24 | 58 (31) |
| Expressed KIR2DS4 | 77* | 60 | 67 | NT (NT) |
| KIR2DS5 | 47 | 28 | 33 | 42 (NT) |
| KIR2DL1 | 73 | 92 | 100 | 100 (98) |
| KIR2DL2 | 33 | 40 | 52 | 50 (45) |
| KIR2DL3 | 76* | 100 | 95 | 92 (66) |
| KIR2DL4 | 88 | 100 | 100 | 100 (NT) |
| KIR2DL5 | 22 | 40 | 52 | 50 (NT) |
| KIR3DS1 | 18 | 48 | 43 | 50 (41) |
| KIR3DL1 | 35† | 76 | 86 | 92 (88) |
| KIR3DL2 | 94 | 100 | 100 | 100 (100) |
| KIR3DL3 | 78 | 60 | 71 | 100 (NT) |
| 2DL3+2DS2−2DL2− | 47 | 60 | 38 | NT (NT) |
| 2DL3+2DS2+/−2DL2+/− | 53 | 40 | 62 | NT (NT) |
The frequency of KIR transcript detection was analyzed in 17 HIV-infected individuals (HIV), 25 EUs, and 21 unexposed controls (C) (and compared to genomic KIR frequencies reported by Toneva et al25 in 59 Vietnamese subjects [in parentheses]). HIV+ individuals show lower KIR transcript frequencies for KIR3DS1 (P < .10) and KIR3DL1 (P < .001). Genomic analysis confirmed the weak KIR3DS1 frequency in HIV+ (18%). Dissociation between KIR3DL1 gene and transcript detection was detected in 3 (21.4%) of 14 EUs tested and 3 (37.5%) of 8 HIV+ individuals, but was never observed in tested controls (n = 12). Expressed KIR2DS4 transcript frequencies refer to transcription alleles encoding the expressed KIR2DS4 receptor (exclusive of individuals only expressing the KIR1D null allele).
Italics in the data field indicate statistical trends (P < .1).
P < .05.
P < .001.
Remarkably, KIR2DL4 and KIR3DL2 transcripts, corresponding to core KIR genes found at the genomic level in all individuals, were found in all EUs and controls, but were undetectable in some HIV+ individuals (Table 2). Similarly, the core KIR3DL3 gene was detected in 100% of controls but the corresponding transcripts were undetectable in 22% of HIV+, 40% of EUs, and 29% of controls tested. The dissociation between KIR3DL3 gene content and transcript frequency (P < .05) was thus not specific to a given group (Table 2). Transcript frequencies of other KIRs showed significant differences in HIV+ subjects in comparison with EUs and controls (Table 2). First, the frequency of individuals with detectable KIR3DL1 and KIR2DL3 transcripts were lowered in HIV+ IDUs (P < .001 and P = .017, respectively). Similarly, frequency of individuals expressing KIR3DS1 transcripts tended to be lower in HIV+ individuals (P = .099). Our transcriptional analysis included design of specific primers that discriminate transcripts corresponding either to the KIR1D null allele that is not expressed as an NK receptor or to the group of KIR2DS4 alleles that contribute to cell surface expression of the KIR2DS4/CD158i receptor.26 Using this approach we showed that frequency of KIR2DS4 transcripts coding for KIR2DS4 receptor was higher in HIV+ IDUs than in EUs and controls (P = .035; Table 2).
When allowed by cell or DNA availability, we further investigated whether the absence of detectable KIR3DL1 or KIR3DS1 transcripts was associated with the absence of the gene or whether it may correspond to lack of transcription in individuals who bear the gene. For KIR3DS1, a good correlation between gene content and transcript detection was observed in all groups. In particular, HIV+ IDUs in whom KIR3DS1 transcripts were undetectable were also typed as negative for KIR3DS1 at the gene level. In contrast, whereas the presence of the KIR3DL1 gene was always associated to KIR3DL1 transcripts in controls, 3 of 8 HIV+ IDUs and 3 of 14 EUs genotyped were KIR3DL1+ at the gene level but lacked the corresponding transcripts.
We further analyzed quantitative differences in the levels of KIR transcript expression in the 3 groups (Figure 1B and results not shown). KIR2DL4 and KIR3DL2 transcripts levels were lower in HIV+ subjects than in controls or EUs (P < .05; Figure 1B). On the other hand, KIR3DL3 transcript levels were also lower in EUs than in HIV+ and controls (P < .05; data not shown).
In contrast, in HIV+ subjects, KIR2DL3 transcript levels were lowered in reference to EUs and controls (P < .05 and P < .001, respectively). Combined KIR2DS2−/2DL2−/2DL3+ transcript patterns tended to be more frequent in EUs than in HIV+ individuals or controls (P = .10; Table 2). We further observed higher KIR2DL3 transcript levels in these KIR2DS2−/2DL2−/2DL3+ EU individuals (P < .05; Figure 1B). Notably, 5 EUs exhibiting more than 50% GL183+-stained NK cells were further identified as KIR2DL3+and 2DL2−/2DS2−, both at the gene and transcript levels (Figure 1B). KIR2DL3 was thus the only KIR gene associated to the GL183+ phenotype in these individuals.
Further analysis of HLA groups associated with the KIR repertoire in EUs showed that groups HLA-C1/C2 and HLA-C1/C1 were equally represented and that only 2 EUs were homozygous HLA-C2/C2 (Table 1). The highly represented GL183+ NK-cell subsets in KIR2DL3+/2DL2−/2DS2− EUs were always associated with presence of the HLA-C1 ligand in these individuals.
KIR3DL1 transcript levels also tended to be lower in EUs than in HIV+ subjects and controls (Figure 1B), although statistical significance was not reached. Accordingly, and given the high KIR3DS1 transcript levels in some EUs, the ratio between activating KIR3DS1 and inhibitory KIR3DL1 transcript levels was higher in some EUs, and particularly elevated at values never observed in tested controls in 3 EUs (Figure 1B; nos. 6, 7, and 16 in Table 1).
Increased expression of NK-cell activation markers in EUs
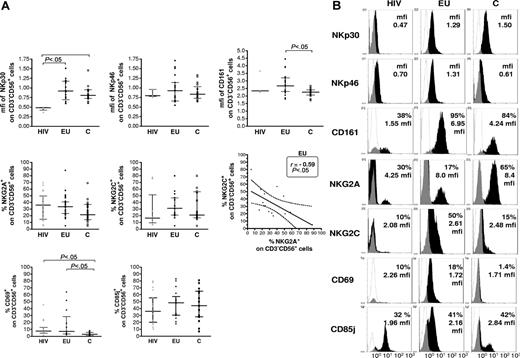
The NK-cell repertoire was further characterized by phenotypic analysis of NK-cell subsets expressing activating and inhibitory NKRs, CD25 and CD69 activation markers (Figure 2A-B and data not shown). NKp30 expression was found on all NK cells and at comparable levels in EUs and controls. The level of NKp30, assayed by mean fluorescence intensity (mfi) of NKp30 staining, was lower in HIV+ than in EUs and controls (P < .05; Figure 2A). In contrast, no significant difference in NKp46 (Figure 2A) nor NKG2D and DNAM expression (data not shown) was observed among the 3 groups. Although the size of the CD161+ NK-cell subset was not different among the 3 groups, CD161 mean fluorescence in CD161+CD3−CD56+ cells was significantly higher in EUs than in controls (P < .05; Figure 2A). Whereas the NK-cell surface expression of the CD25 activation marker was comparable in all tested groups (data not shown), a higher percentage of CD69+ NK cells was observed in EUs and HIV+ as compared to controls (P < .05; Figure 2A). Overall, CD85j, NKG2A, and NKG2C expression on NK cells did not differ significantly among the groups. Nevertheless, expression of the activating NKG2C subunit of the CD94/NKG2 receptor was inversely correlated to that of its inhibitory NKG2A counterpart in EUs, but not in the other 2 groups (P < .05; Figure 2A).
Flow cytometry analysis of NKR expression on CD3−CD56+ NK cells. (A) The size of NKR+ NK-cell subsets expressing NKp30, NKp46, CD161, NKG2A and NKG2C, CD69, and CD85j were evaluated after gating of positive CD3−CD56+ NK cells in exposed HIV-infected IDU (HIV, ▴), exposed HIV-uninfected IDU (EU, •), and unexposed control blood donors (C, □, Horizontal bars represent the median and the 25th-75th percentiles. NKp30 and NKp46 were expressed on all NK cells (100% CD3−CD56+), and comparison of expression levels where thus analyzed by analysis of mean fluorescence intensity (mfi). NK-cell surface expression of NKG2A inhibitory subunit of the CD94/NKG2A heterodimeric receptor was inversely correlated to that of the stimulatory NKG2C subunit among CD3−CD56+ NK cells in EU subjects (Spearman correlation test), whereas correlation was nonsignificant in C and HIV groups. (B) Representative flow cytometer analyses of NKR staining in exposed HIV-infected IDU (HIV), exposed uninfected IDU (EU) and unexposed controls (C) (white: isotypic control staining; black, NKR specific staining).
Flow cytometry analysis of NKR expression on CD3−CD56+ NK cells. (A) The size of NKR+ NK-cell subsets expressing NKp30, NKp46, CD161, NKG2A and NKG2C, CD69, and CD85j were evaluated after gating of positive CD3−CD56+ NK cells in exposed HIV-infected IDU (HIV, ▴), exposed HIV-uninfected IDU (EU, •), and unexposed control blood donors (C, □, Horizontal bars represent the median and the 25th-75th percentiles. NKp30 and NKp46 were expressed on all NK cells (100% CD3−CD56+), and comparison of expression levels where thus analyzed by analysis of mean fluorescence intensity (mfi). NK-cell surface expression of NKG2A inhibitory subunit of the CD94/NKG2A heterodimeric receptor was inversely correlated to that of the stimulatory NKG2C subunit among CD3−CD56+ NK cells in EU subjects (Spearman correlation test), whereas correlation was nonsignificant in C and HIV groups. (B) Representative flow cytometer analyses of NKR staining in exposed HIV-infected IDU (HIV), exposed uninfected IDU (EU) and unexposed controls (C) (white: isotypic control staining; black, NKR specific staining).
Strong constitutive activation of NK cells in EUs
We then investigated the granule exocytosis pathway characterizing NK-cell activation by evaluating CD107a+ expression within CD3−CD56+ NK cells,23 both without stimulation and in the presence of stimulatory K562 cells (Figure 3).
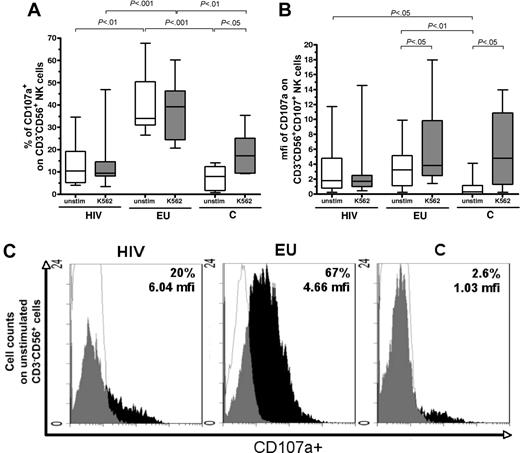
Constitutive and inducible activation of NK cells is revealed by the CD107 degranulation assay. (A) Comparative analysis of CD107a expression on CD3−CD56+ NK cells was performed in the 3 study groups, in the absence of stimulation (unstim) and after K562 target cells stimulation (K562). Boxes represent CD3−CD56+ cells expressing CD107a in exposed HIV-infected IDU (HIV), exposed HIV-uninfected IDU (EU), and unexposed control donors (C), y-axis refers to 25th percentile, median, 75th percentile values. (B) Comparative analysis of mfi of CD107a staining among CD107+CD3−CD56+ NK cells in exposed HIV-infected IDU (HIV), exposed uninfected IDU (EU), and controls (C) in the absence of stimulation (unstim) and on stimulation with K562 target cells (K562). Boxes represent the median mfi of CD107a staining among CD3−CD56+ NK cells (25th percentile, median, 75th percentile). Horizontal bars indicate minimum and maximum values. (C) Examples of flow cytometer analyses of CD107a staining on CD3−CD56+ NK cells in the absence of stimulation (white, isotypic control staining; black, NKR specific staining).
Constitutive and inducible activation of NK cells is revealed by the CD107 degranulation assay. (A) Comparative analysis of CD107a expression on CD3−CD56+ NK cells was performed in the 3 study groups, in the absence of stimulation (unstim) and after K562 target cells stimulation (K562). Boxes represent CD3−CD56+ cells expressing CD107a in exposed HIV-infected IDU (HIV), exposed HIV-uninfected IDU (EU), and unexposed control donors (C), y-axis refers to 25th percentile, median, 75th percentile values. (B) Comparative analysis of mfi of CD107a staining among CD107+CD3−CD56+ NK cells in exposed HIV-infected IDU (HIV), exposed uninfected IDU (EU), and controls (C) in the absence of stimulation (unstim) and on stimulation with K562 target cells (K562). Boxes represent the median mfi of CD107a staining among CD3−CD56+ NK cells (25th percentile, median, 75th percentile). Horizontal bars indicate minimum and maximum values. (C) Examples of flow cytometer analyses of CD107a staining on CD3−CD56+ NK cells in the absence of stimulation (white, isotypic control staining; black, NKR specific staining).
Without stimulation, the size of the activated CD107a+ NK-cell subset was much larger in EUs (median: 34%; 25%-75% interquartile ranges: 31%-50%) than in HIV+ individuals (10%; 5%-19%; P < .01) and controls (8%; 1.7%-12%; P < .001; Figure 3A,C). Kinetic analysis of CD107a degranulation at 10 minutes, 1 hour, 2 hours, and 3 hours showed that degranulation of unstimulated EU NK cells occurred between 10 minute and 1 hour, in contrast to 2 hours in cells from controls (data not shown). Median percentage of CD107a+ cells among control CD3−CD56+ NK cells was increased by K562 stimulation (from 8% to 17%; P < .05), but remained unchanged in HIV+ individuals (Figure 3A).
On stimulation by K562 cells, median percentage of CD107a+ NK cells remains significantly higher in EUs (39%; 24%-46%) than in HIV+ persons (9.5%; 8%-15%; P < .001) and stimulated controls (17%; 9%-25%; P < .01; Figure 3A).
We further analyzed variations in intensity of CD107a expression, as assayed by mfi of CD107 staining within the activated CD3−CD56+ NK-cell subset. Stimulation with K562 cells induced increased CD107a mean fluorescence staining intensity within CD107+ NK cells, in controls as well as in EUs (Figure 3B). In contrast, the mfi of CD107a+ NK cells from HIV+ individuals was unchanged after K562 stimulation (Figure 3B).
Increased CD161 expression is associated with the CD107a+ activated NK compartment
Finally, we examined the NK-cell surface expression of CD69, CD85j, NKG2A, NKG2C, and CD161 within the activated CD107+CD3−CD56+ NK-cell compartment, both in the presence and absence of stimulatory K562 target cells. Overall, the distribution of CD85j, NKG2A, and NKG2C expression among CD107a+CD3−CD56+ NK cells did not differ significantly among the 3 study groups (data not shown). Median percentages of CD69 expression among CD107a+ NK cells tended to be higher in EUs (40%) than in HIV+ (20%) and controls (28%) in the absence of K562 stimulation and were induced to similar levels in the 3 groups after K562 stimulation (data not shown).
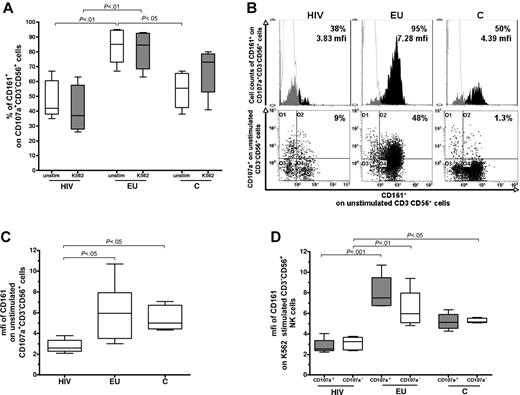
In contrast, CD161 expression differed significantly between EUs and the other groups (Figure 4). In the absence of K562 stimulation, the percentage of CD161+ cells among CD107+CD3−CD56+ NK cells were higher in EUs than in controls (P < .05) and HIV+ subjects (P < .01; Figure 4A-B). In addition, the intensity of CD161 staining in unstimulated CD161+CD107+CD3−CD56+ NK cells (mfi) was comparable in EUs and controls and significantly lowered in HIV+ individuals (P < .05; Figure 4B-C), but not affected by K562 stimulation in any of the 3 groups (data not shown). Among CD161+ NK cells, intensity of CD161 staining (mfi) in EUs tended to be higher in the CD107a+ NK subset than in the CD107a− NK subset (P = .12; Figure 4D). In contrast, CD161 expression levels were similar in the CD107a+ and CD107− NK-cell compartments in HIV+ and controls subjects (Figure 4D).
Analysis of CD161 expression by the activated CD107a+CD3-CD56+ NK-cell subset. (A) Cell surface expression of CD161 was evaluated by 4-color flow cytometry after gating on CD107a+CD3−CD56+ NK cells. The percentage of CD161+ stained cells was analyzed within CD107a+CD3−CD56+ NK cells in absence of stimulation (unstim) and after 3 hours of K562 cell stimulation (K562) in exposed HIV-infected IDUs (HIV), exposed uninfected IDUs (EU), and unexposed controls (C). The y-axis values boxed represent 25th percentile, median, and 75th percentile. Horizontal bars indicate minimum and maximum values. (B) Representative flow cytometer analysis of percentage and mfi of CD161 staining among the CD107a+CD3−CD56+ NK-cell subset before exogenous stimulation (white: isotypic control staining, black, CD161 specific staining). (C) mfi of CD161 staining among the CD161+ unstimulated CD107a+CD3−CD56+ NK-cell subset. The y-axis values boxed represent 25th percentile, median, and 75th percentile. (D) Evaluation of mfi of CD161 expression within the CD107a+ and CD107a−CD3−CD56+NK-cell subsets after K562 cell stimulation. The y-axis values boxed represent 25th percentile, median, and 75th percentile.
Analysis of CD161 expression by the activated CD107a+CD3-CD56+ NK-cell subset. (A) Cell surface expression of CD161 was evaluated by 4-color flow cytometry after gating on CD107a+CD3−CD56+ NK cells. The percentage of CD161+ stained cells was analyzed within CD107a+CD3−CD56+ NK cells in absence of stimulation (unstim) and after 3 hours of K562 cell stimulation (K562) in exposed HIV-infected IDUs (HIV), exposed uninfected IDUs (EU), and unexposed controls (C). The y-axis values boxed represent 25th percentile, median, and 75th percentile. Horizontal bars indicate minimum and maximum values. (B) Representative flow cytometer analysis of percentage and mfi of CD161 staining among the CD107a+CD3−CD56+ NK-cell subset before exogenous stimulation (white: isotypic control staining, black, CD161 specific staining). (C) mfi of CD161 staining among the CD161+ unstimulated CD107a+CD3−CD56+ NK-cell subset. The y-axis values boxed represent 25th percentile, median, and 75th percentile. (D) Evaluation of mfi of CD161 expression within the CD107a+ and CD107a−CD3−CD56+NK-cell subsets after K562 cell stimulation. The y-axis values boxed represent 25th percentile, median, and 75th percentile.
Discussion
The role of NK cells in controlling HIV infection has mainly been investigated in infected individuals. Thus, although considerable information has been gained on NK-cell dysfunctions associated with HIV infection and viremia, the role of NK cells in protection against HIV infection remains to be clarified. Natural resistance to infection among exposed uninfected individuals may be due partly to efficient early innate antiviral responses. Indeed, we previously showed that highly exposed uninfected Vietnamese IDUs have higher NK cytokine/chemokine secretion capacities and lytic activity.19 More recently, Montoya et al reported increased interferon-γ (IFN-γ) production by NK cells from uninfected individuals sexually exposed to HIV-1.27
The present study was designed to further characterize the NK effector potential of Vietnamese EU IDUs using a recently described degranulation functional assay22 and to investigate whether increased levels of activation could be associated with peculiar features of the NK-cell receptor repertoire in EUs. In addition to the phenotypic analysis of KIR+ NK-cell subsets, our study is, to our knowledge, the first to provide a quantitative analysis discriminating KIR transcripts in virally exposed IDUs in reference to low-risk control donors.
Using the CD107a NK functional assay,22 we found that EU NK cells exhibit a high constitutive degranulation potential. Indeed, degranulation occurred rapidly in vitro, without the need for exogenous target cell stimulation. This high constitutive activity, which likely reflects a strong in vivo activation status of the NK-cell compartment in EUs, was not observed in HIV+ individuals or in low-risk controls recruited in the same environment. In other studies, high levels of CD107a expression on NK cells from HIV viremic subjects was associated with a decreased number of CD3−CD56+CD161+ NK cells.28 Up-regulation of CD107a has been shown to correlate with NK cytokine secretion and cytolytic activity.22,23,28 This is in line with our previous observation that EUs exhibit higher NK cytolytic potential cells toward Daudi and K562 targets and an enhanced NK-mediated production of IFN-γ, TNF-α, and CCL3-5.19 The additional functional readout of NK cells provided in this study, therefore, stresses the fact that NK cells in EUs have a particularly high constitutive potential to display immediate effector functions on infectious challenge, which possibly contributes to the protection of EUs against HIV infection.
The phenotypic analysis of the NK-cell repertoire identified particular NK-cell receptor patterns that may sustain constitutive NK-cell activation in EUs, such as enhanced CD69 and CD161 expression. In particular, intensity of CD161 staining on NK cells was higher in EUs than in HIV+ individuals and controls. Furthermore, following stimulation, CD161 was preferentially expressed by the activated CD107a+ NK-cell subset. The number of CD161+ NK cells was reduced in HIV-infected viremic subjects,29 but no correlation with NK functional defects observed in HIV+ patients has yet been established. CD161 engagement appears to have a complex role in the modulation of immune functions. Both activating and inhibitory effects have been described in mice after CD161 receptor engagement.30 In humans, LLT1, an inhibitory ligand of human CD161, was shown to modulate NK-cell activity.31,32 CD161 expression was also involved in the triggering of NK-cell cytotoxicity toward the Daudi tumor cell line.33 Interestingly, we have previously shown that EU NK cells display substantial cytotoxic activity against Daudi cells.19 Although direct evidence implicating CD161 in enhanced NK-cell activation is still lacking, these observations suggest that CD161 expression may contribute to the NK-cell activation status, at least in HIV-exposed uninfected subjects.
Although down-regulation of NKp30 was confirmed in HIV+ individuals,12 no major alteration of NCRs was found in EUs in comparison to low-risk controls, thus suggesting that NK activation in EUs was not associated with increased expression of activating NKp30 or NKp46 receptors. Other features of the NK-cell repertoire that senses virally induced modification of HLA class I expression may sustain NK-cell activation in EUs. Cell activity against HIV-1–infected blasts was shown to be partly dependent on NK-cell recognition of HIV-modulated HLA-E expression.34 This recognition is mediated by the heterodimeric receptor CD94, in association with either its inhibitory NKG2A subunit or its activating NKG2C subunit. In some EUs the stimulatory NKG2C-to-inhibitory NKG2A ratio was inverted, in favor of the activating subunit. Whereas no significant change in the relative size of the CD94+ NK subset was found in EUs, this suggests that HLA-E interaction with NK cells expressing the CD94/NKG2C activating receptor may be dominant in some EUs. Such a switch from inhibitory to activating NKG2 subunit expression of the CD94/NKG2 receptor for HLA-E has been linked to concomitant infections in HIV+ individuals and may be driven in EUs by exposure to viral infections other than HIV.35
Although the phenotypic distribution of NK cells expressing KIR and CD85j MHC-class I receptors was not profoundly affected in EUs, further discrimination of the activating and inhibitory KIR genes that contribute to the phenotype of KIR+ NK-cell subsets, by locus-specific real-time PCR transcript quantification, revealed qualitative or quantitative differences between EUs and the other 2 groups of subjects. In contrast to the other 2 study groups, high percentages of GL183+ NK cells in EUs correlate with the expansion of KIR2DL3 transcripts in the absence of KIR2DS2 and KIR2DL2 transcripts. In addition, this expansion of KIR2DL3 transcripts was observed in EUs displaying either HLA-C1/C1 or C1/C2 allelic specificities. Because the inhibitory KIR2DL3 receptor binds to its ligand HLA-C1 with lower affinity than KIR2DL2, KIR2DL3 expression in the absence of KIR2DL2 expression may favor NK-cell activation in EUs, by a mechanism similar to that proposed for KIR2DL3-HLA-C1 in the resolution of HCV infection.36 Indeed, NK-cell interactions involving KIR2DL3-HLA-C1 have been shown to directly influence the resolution of HCV infection in whites and African Americans.36 This led to the concept that not only activation but also lowered NK inhibitory responses may be an advantage in the control of viral infection. KIR2DL2/2DL3 heterozygosity in absence of HLA-C1 ligand has been recently reported to be associated to the seronegative status of exposed African sex workers.20 In our study, all KIR2DL2/2DL3 heterozygous EUs were homozygous HLA-C1/C1 or heterozygous HLA-C1/C2. Moreover, the 2 C2/C2 EUs were typed as homozygous for KIR2DL3. Therefore, although our sample is clearly too limited to establish any firm statistics on HLA-KIR combinations in EUs, the KIR2DL2/2DL3 heterozygosity in absence of HLA-C1 does not appear to contribute to resistance in Vietnamese EU IDUs.
Resistance to HIV-1 infection among African sex workers was also associated with KIR3DL1 homozygosity in the absence of its cognate HLA-Bw4 ligand.20 No such trend was identified in Vietnamese EUs, because homozygous HLA-Bw6 and Bw4/Bw6 were equally represented in KIR3DL1 homozygous EUs (Table 1). Interestingly, in some EUs, concomitant expression of lowered inhibitory KIR3DL1 transcript levels and high activating KIR3DS1 levels resulted in a high KIR3DS1/KIR3DL1 ratio that may confer an enhanced activating NK-cell repertoire profile to these EUs. Indeed, very recent data suggest that KIR3DS1 associates with DAP12 and triggers both cytolysis and IFN-γ production.37 Previous genetic studies have linked KIR3DS1 and HLA-Bw4 expression to slow progression toward AIDS.18,38 However, in our study, strongest KIR3DS1 expansion in EUs occurred in HLA-Bw6 individuals rather than in homozygous HLA-Bw4 EUs who were not found in our study population (Table 1). Furthermore, it was recently suggested that activating KIR3DS1 receptor does not recognize HLA-Bw4.37,39
Although the number of individuals analyzed was too small to draw statistically powerful conclusions, combined transcriptional and genomic analysis suggested that KIR3DL1 and core KIR2DL4 and KIR3DL2 transcription might be specifically abolished in some HIV-exposed individuals. Indeed, KIR3DL1 transcripts reflected the KIR3DL1 gene content in controls, whereas the absence of KIR3DL1 transcripts was observed in the presence of the KIR3DL1 gene in some HIV+ and EU IDUs. Although the possibility that KIR3DL1 gene polymorphisms specific to HIV-exposed Vietnamese were not detected by our reverse transcription PCR (RT-PCR) is not excluded because PCR primers designed to specifically detect KIR3DL1 transcripts did not allow their use for gene amplification, this is unlikely considering that KIR3DL1 genotype was confirmed in these individuals by commercially available kits. Our transcriptional approach of the KIR repertoire, although limited by the number of individuals studied, thus suggests that genomic KIR content is not always associated with transcript detection in HIV-exposed populations, and that mechanisms regulating KIR transcripts may occur in exposed individuals. Representation of KIR transcripts might not only represent a down- or up-regulation but a selection at infection. Interestingly, the frequency of individuals who exhibit KIR2DS4 transcripts encoding the KIR2DS4 receptor was significantly increased in HIV+ IDUs, suggesting that the KIR2DS4-bearing haplotype, often associated with lack of other activating KIR-S gene content,8 might be more prevalent in HIV+ individuals. A challenging issue to be addressed in wider population studies would be to investigate whether KIR regulation is selected at infection and to what extent regulatory mechanisms can be specifically related to HIV or to concomitant coinfection occurring in exposed individuals.
In conclusion, our results and data from other studies of HIV+ patients suggest that the NK-cell receptor repertoires in EUs may be quite different from those of HIV-1–infected individuals, whether they are viremic or nonprogressors.13,18,28,29,40 Several mechanisms, including cellular restriction of HIV replication, T-cell activity, and specific IgA responses have been proposed to explain resistance to HIV infection,41 but it is more likely that complex genetic or selected mechanisms operate independently or in synergy to confer individual resistance to HIV-1. HIV-exposed uninfected individuals are a heterogeneous population, and it is therefore not surprising that the high level of NK-cell activation that characterizes the Vietnamese EUs studied here was associated with diverse NK receptor repertoires. Yet, in some EUs, no particular pattern of NK receptors could be identified. The constitutive highly activated status of NK cells from EUs may also be dependent on the HLA-NK receptor genetic background or on selective induction of NK receptors that endow NK cells with the capacity to mount an effective defense against HIV-1. The increased NK-cell activity that we found in Vietnamese EUs may reflect more general immune activation resulting from several factors including exposure to multiple viruses.42 However, HIV-1–infected IDUs exposed to similar environmental conditions and to multiple viral infections19,43 have different NK-cell receptor patterns. Therefore, our data strongly suggest that the increased NK-cell activity in virally exposed EUs is linked to particular genetic backgrounds that adapt the regulation of diverse NK-cell receptors on viral challenge, thereby shaping a constitutively highly activated NK-cell status that may contribute to protection of EUs toward HIV-1 infection.
Authorship
Contribution: P.P., F.B.-S., D.S.-A., and G.P. conceived the study; P.P., D.S.-A., and G.P. contributed to the experimental design and coordination of the study; L.X.T. and N.N. recruited the study subjects and coordinated the study in Vietnam; S.R., E.B., and H.K.T. performed the transcriptional, phenotypic, and functional analyses; I.T. and T.T. contributed the HLA and KIR genotyping; and P.P., S.R., G.P., and D.S.-A. wrote the manuscript, with I.T. performing its critical correction. All authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pascale Paul, Laboratoire Exploration NK, Hematology Unit, Hôpital de la Conception, 147 Bd Baille, 13005 Marseille, France; e-mail: pascale.paul@ap-hm.fr.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from Agence Nationale de Recherche sur le Sida, Assistance publique Hôpitaux de Marseille (AP-HM), INSERM (action thématique concertée ATC Biothérapies), and Ministère de la Recherche Réseau Genhomme.
We thank all the physicians, family practitioners, and medical staff from the Binh Trieu Hospital and the Pasteur Institute of Ho Chi Minh City who took part in this study. We thank Denis Reviron, Christophe Picard, and Coralie Frassati for their helpful advice in KIR genotyping and members of the Hematology Unit; AP-HM for support and availability of flow cytometer facilities; and Françoise Dignat-George for providing helpful comments on the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal