Abstract
The 8;21 translocation is a major contributor to acute myeloid leukemia (AML) of the M2 classification occurring in approximately 40% of these cases. Multiple mouse models using this fusion protein demonstrate that AML1-ETO requires secondary mutagenic events to promote leukemogenesis. Here, we show that the negative cell cycle regulator p21WAF1 gene is up-regulated by AML1-ETO at the protein, RNA, and promoter levels. Retroviral transduction and hematopoietic cell transplantation experiments with p21WAF1-deficient cells show that AML1-ETO is able to promote leukemogenesis in the absence of p21WAF1. Thus, loss of p21WAF1 facilitates AML1-ETO–induced leukemogenesis, suggesting that mutagenic events in the p21WAF1 pathway to bypass the growth inhibitory effect from AML1-ETO–induced p21WAF1 expression can be a significant factor in AML1-ETO–associated acute myeloid leukemia.
Introduction
AML1 (RUNX1) is a member of the Runt family of transcription factors that heterodimerize with CBFβ to form a stable DNA-binding complex.1,2 AML1 is also the major gene involved in chromosomal translocations in leukemia,3,4 seen translocated in both lymphoid and myeloid leukemia, such as t(8;21) (AML1-ETO),5 t(3;21) (AML1-Evi1),6 t(12;21) (TEL-AML1),7 t(16;21) (AML1-MTG16),8 t(12;21) (AML1-copine VIII),9 t(X;21) (AML1-Fog2),10 and t(7;21) (AML1-USP42).11 Specifically the AML1-ETO–associated translocation is observed in approximately 40% of cases of acute myeloid leukemia of the M2 classification (AML-M2), while present in approximately 12% of AMLs. This translocation protein contains the N-terminal portion of AML1 up to its Runt homology DNA-binding domain fused to most of the ETO (MTG8) protein.12 The ETO gene is one of the family of ETO/MTG repressor proteins that have homology to the Drosophila Nervy repressor proteins.13,14 Nonconditional AML1-ETO knock-in mice15,16 are similar to AML1-deficient mice17,18 in embryonic lethality and in displaying a block in embryonic definitive hematopoiesis, suggestive of a dominant-negative function. Furthermore, directed controlled expression of AML1-ETO in mice demonstrated that AML1-ETO does not promote leukemia under these circumstances.19–22 However, following the treatment of AML1-ETO–expressing mice with a DNA-alkylating agent, the mice developed AML.20,21 Furthermore, various retroviral transduction models have shown again that it does not promote AML.23,24 These studies suggest that AML1-ETO requires secondary mutagenic events to promote development of AML. Indeed, AML1-ETO promotes AML when combined with the loss of the ICSBP gene, overexpression of WT1, the Flt3-activating mutation, or cotransduction with another translocation fusion protein TEL-PDGFR.24–27
The biologic characterization of AML1-ETO in various cell lines has shown that it has both positive and negative effects on leukemia development. Its positive influence comes from its ability to block differentiation and to expand hematopoietic stem cells23,24,28–32 ; however, its negative effects are that it promotes growth arrest and apoptosis.28,33,34 Therefore, it is reasonable to propose that the additional events to promote AML1-ETO–associated leukemogenesis are mutations that oppose the negative effects of AML1-ETO on cell proliferation and apoptosis. In our investigations to understand the mechanism by which AML1-ETO promotes growth arrest, we found increased expression of p21WAF1 protein in AML1-ETO+ hematopoietic cells.35 p21WAF1 is known to block the activity of cell cycle-dependent kinases.36 We have now further characterized AML1-ETO ability to up-regulate p21WAF1 and analyzed the role of p21WAF1 in AML1-ETO–associated leukemogenesis. We show that AML1-ETO enhances the expression of p21WAF1 at the protein, RNA, and promoter levels. Furthermore, although AML1-ETO alone cannot promote leukemogenesis, its expression in p21WAF1-deficient hematopoietic cells leads to the development of AML, indicating that AML1-ETO is able to promote leukemogenesis by cooperating with the loss of p21WAF1. This result suggests that bypass of the p21WAF1 functional pathways is one of the molecular mechanisms of AML1-ETO–associated leukemogenesis.
Materials and methods
Cell culture and mice
K562, Kasumi-1, and Jurkat cells were maintained in RPMI (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS; Omega Scientific, Tarzana, CA), 2 mM glutamine (Invitrogen). 293T cells were maintained in DMEM (Invitrogen) containing 10% FBS and 2 mM glutamine. The p21WAF1 knockout mice were kindly provided by Dr Dwight Kono at the Scripps Research Institute (originally from Dr Phillip Leder at Harvard Medical School).37 Mice of C57BL/6 strain and C57BL/6 congenic strain B6.SJL-PtprcaPep3b/BoyJ were purchased from Jackson Laboratories (Bar Harbor, ME). Mice were housed in a pathogen-free facility and procedures were approved by the Institutional Animal Care and Use Committee of The Scripps Research Institute. The fetal liver was harvested from embryos at 17.5 days postcoitus (dpc),35 and frozen down in 10% FBS-DMEM-7% DMSO (Fisher Scientific, Hampton, NH).
Expression vectors
MigR1 and HA-AML1-ETO expression retroviral construct Mig-A/E were described previously.35 pCDNA6-HA-AML1-ETO was constructed by digesting pLNSX-HA-AML1-ETO38 with XbaI and cloning HA-AML1-ETO DNA fragment into pCDNA6 (Clontech, Palo Alto, CA) digested with XbaI. pCMV5-AML1-ETO, pCMV5-AML1-ETO-L148D, pCMV5-CBFβ, and pCMV5 were received from Dr S. Hiebert (Vanderbilt University, Vanderbilt, TN).39 pCDNA6-HA-AML1-ETO-R174Q was constructed by polymerase chain reaction (PCR) mutagenesis with primers 5′-GAT GGG CCC CAA GAA CCT CGA-3′ (AML1-R174Q) and 5′-CTG GTG CCA TTA GTT AAC GTT G-3′ (ETO-HpaI), digesting the product with ApaI/HpaI, and cloning this into pCMV5-AML1-ETO ApaI/HpaI. This plasmid was further digested with HindIII/HpaI and cloned by a 3-way ligation of HindIII/HindIII and HpaI/HindIII fragments of pCDNA6-HA-AML1-ETO. The point mutation was confirmed by sequencing. The luciferase reporter plasmids for the p21WAF1 promoter in pGL2-basic (p21pGL2) and deletion mutant (Δ1.9p21pGL2) was provided by Dr X.-F. Wang (Duke University, Durham, NC).40 The pRL-null reporter is from Promega (Madison, WI).
Infection of K562 cells, cell cycle, and Western blot analysis
These procedures were carried out as described previously.35
Northern blot analysis
Cells were harvested into RNA-Bee and RNA extracted according to the manufacturer's instructions (Tel-Test, Friendswood, TX). Northern blot analysis was performed as described previously.19 The blot was hybridized with random primed labeled p21WAF1 cDNA excised with NotI from pBS-p21WAF1 received from Dr P. Sun (The Scripps Research Institute, La Jolla, CA).
Southern blot analysis
gDNA was isolated from splenic-derived leukemic cells and Southern analysis performed as previously described.35
Reporter assays and chromosome immunoprecipitation
K562 cells were cultured to 4 to 6 × 105 cells/mL and transfected by electroporation with 2.5 × 106 cells in 400 μL plain RPMI at 260 V and 960 μF on a Gene Pulser II (Bio-Rad, Hercules, CA), with a total of 20 μg DNA. Then, 10 μg of the reporter plasmid p21-pGL2 was mixed with 5.6 μg herring sperm DNA, 2 μg pCMV5-CBFβ, and 400 ng pRL-null (Promega), to which either 2 μg pCMV5, pCDNA6-HA-AML1-ETO, pCDNA6-HA-AML1-ETO-R174Q, pCMV5-AML1-ETO, or pCMV5-AML1-ETO-L148D was added. The cells were cultured overnight in 4 mL RPMI-10%FBS-2 mM glutamine-1% penicillin/streptomycin between 16 and 23 hours. Following the culture they were harvested and washed with PBS and lysed into 200 μL passive lysis buffer (Promega). The dual-luciferase measurements were performed as described by the manufacturer (Promega). Chromosome immunoprecipitations (ChIPs) were performed as previously described41 with 293T cells transfected with 10 μg of either p21pGL2 or Δ1.9p21pGL2 and 2 μg of either pcDNA6, pCDNA6-HA-AML1, or pCDNA6-HA-AML1ETO. Primers for amplifying the p21pGL2 fragments were 21F1 5′-ctgcaaatgagggttatttggc-3′ and 21R1 5′-ctccctacaccctacactcacctg-3′, and for amplifying Δ1.9p21pGL2 they were GL2F1 5′-tgtatcttatggtactgtaactg-3′ and 21R2 5′-cggaacctcgcgtgctgcagg-3′.
Viral infection, bone marrow transplantation, and analysis of leukemic mice
These procedures were carried out as described previously.35
Results
AML1-ETO regulates the expression of the cell cycle inhibitor p21WAF1
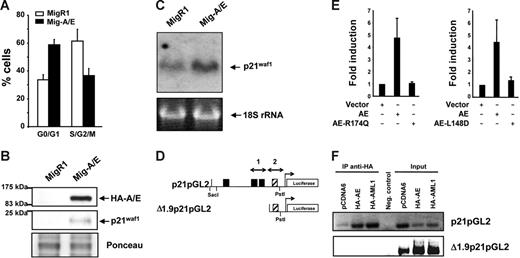
We have previously shown that the expression of AML1-ETO in myeloid cell lines promotes growth arrest,28,35 observing various cell cycle regulators affected at their protein expression level, including p21WAF1. Herein, we further investigate this p21WAF1 induction by AML1-ETO expression by transducing K562 cells with the AML1-ETO gene using the Mig-A/E retrovirus reconfirming growth arrest induction (Figure 1A). 35 The level of p21WAF1 protein was again increased in AML1-ETO–expressing cells (Figure 1B). Furthermore, we examined if the increase of p21WAF1 in the presence of AML1-ETO is regulated at the RNA level by preparing RNA from MigR1- and Mig-A/E–infected cells and performed Northern blot hybridization with radiolabeled p21WAF1 cDNA. As shown in Figure 1C, AML1-ETO promoted an increase of p21WAF1 mRNA. Thus, these observations suggest that AML1-ETO may directly enhance p21WAF1 mRNA expression. Such an increase of p21WAF1 RNA has also been observed in primary murine primitive hematopoietic cells expressing AML1-ETO under conditions without growth arrest,42 supporting the possibility that the increase of p21WAF1 mRNA is due to the expression of AML1-ETO and not cell cycle arrest.
AML1-ETO regulates the cell cycle inhibitor p21WAF1. (A) K562 cells infected with MigR1 and Mig-A/E were sorted by flow cytometry 2 days after infection for EGFP+ cells and cultured overnight at 2 × 105 cells/mL and harvested for cell cycle analysis by propidium iodide staining by flow cytometry. Percentages of G0/G1 and cells in S/G2/M phase of cells are presented from 3 individual infection experiments. (B) Following infection and flow cytometry sorting were analyzed by Western transfer with 20 μg protein the blots were probed with anti-HA or anti-p21waf1 to analyze their expression. Ponceau staining of the blot depicts loading. (C) K562 cells were infected, sorted, and cultured as described and RNA was prepared. RNA (10 μg) was analyzed by Northern blot with the full-length p21WAF1 cDNA. Equal loading is depicted by the 18S ribosomal RNA. (D) Schematic diagram of the 2.3-kb promoter region of the p21WAF1 gene cloned into the pGL2 reporter vector; ▪, AML1 DNA-binding sites; ▨, putative AML1-binding site (TGTGGG). (E) K562 cells were transfected with 10 μg p21-pGL2, with either 2 μg empty vector or those expressing HA-AML1-ETO, HA-AML1-ETO-R174Q, AML1-ETO, or AML1-ETO-L148D brought to a total of 20μg with 5.6 μg herring sperm DNA, 2 μg of an expression plasmid for CBFβ and 400 ng pRL-null as internal control for transfection, and cultured for 16 to 23 hours. Experiments depict the average and SD of 5 individual experiments with 2 different batches of DNA. (F) ChIP assay for association of AML1 and AML1-ETO to the distal and proximal regions of the p21WAF1 promoter. Cells were transfected with 10 μg p21pGL2 or Δ1.9p21pGL2 with either 2 μg pCDNA6, pCDNA6-HA-AML1, or pCDNA6-HA-AML1-ETO and following ChIP the isolated DNA fragments were subjected to PCR with specific primers that amplify regions 1 and 2 depicted in panel D, and analyzed by DNA gel electrophoresis.
AML1-ETO regulates the cell cycle inhibitor p21WAF1. (A) K562 cells infected with MigR1 and Mig-A/E were sorted by flow cytometry 2 days after infection for EGFP+ cells and cultured overnight at 2 × 105 cells/mL and harvested for cell cycle analysis by propidium iodide staining by flow cytometry. Percentages of G0/G1 and cells in S/G2/M phase of cells are presented from 3 individual infection experiments. (B) Following infection and flow cytometry sorting were analyzed by Western transfer with 20 μg protein the blots were probed with anti-HA or anti-p21waf1 to analyze their expression. Ponceau staining of the blot depicts loading. (C) K562 cells were infected, sorted, and cultured as described and RNA was prepared. RNA (10 μg) was analyzed by Northern blot with the full-length p21WAF1 cDNA. Equal loading is depicted by the 18S ribosomal RNA. (D) Schematic diagram of the 2.3-kb promoter region of the p21WAF1 gene cloned into the pGL2 reporter vector; ▪, AML1 DNA-binding sites; ▨, putative AML1-binding site (TGTGGG). (E) K562 cells were transfected with 10 μg p21-pGL2, with either 2 μg empty vector or those expressing HA-AML1-ETO, HA-AML1-ETO-R174Q, AML1-ETO, or AML1-ETO-L148D brought to a total of 20μg with 5.6 μg herring sperm DNA, 2 μg of an expression plasmid for CBFβ and 400 ng pRL-null as internal control for transfection, and cultured for 16 to 23 hours. Experiments depict the average and SD of 5 individual experiments with 2 different batches of DNA. (F) ChIP assay for association of AML1 and AML1-ETO to the distal and proximal regions of the p21WAF1 promoter. Cells were transfected with 10 μg p21pGL2 or Δ1.9p21pGL2 with either 2 μg pCDNA6, pCDNA6-HA-AML1, or pCDNA6-HA-AML1-ETO and following ChIP the isolated DNA fragments were subjected to PCR with specific primers that amplify regions 1 and 2 depicted in panel D, and analyzed by DNA gel electrophoresis.
AML1-ETO regulates p21WAF1 promoter activity
The p21WAF1 promoter is regulated by AML1 in a cell context-dependent manner.43 It contains 3 consensus AML1-DNA–binding sites (Figure 1D). We thus performed transient transfection-luciferase reporter assays using p21pGL2 that contains a 2.3-kb fragment of the p21 promoter region.40 AML1 heterodimer partner CBFβ was coexpressed with AML1-ETO to enhance AML1-ETO activity. In the presence of AML1-ETO expression, a 4.5-fold increase of p21WAF1 promoter activity was observed (Figure 1E), indicating the positive effect of AML1-ETO on the regulation of p21WAF1 promoter activity. We included as controls 2 DNA-binding domain mutants of AML1-ETO, namely, AML1-ETO-R174Q and AML1-ETO-L148D. The R174Q mutant is a reported point mutation found in a FPD/AML patient that resulted in loss of the ability to bind DNA.44,45 The L148D mutant is a synthetic mutant that lost its ability to bind to DNA and interact with CBFβ.39 In contrast to wild-type AML1-ETO, the 2 mutants did not show any enhancement of p21WAF1 promoter activity (Figure 1E). Together, these results show that AML1-ETO activates the p21WAF1 promoter and that the DNA-binding domain of AML1-ETO is critical for the activation. However, closer examination of a deletion mutant of p21pGL2 to define the region of activation precisely shows that the deletion of the 3 upstream AML1-binding sites was not sufficient to eliminate transactivation of the promoter (data not shown) that contains a putative (TGTGGG) AML1 binding site within the Δ1.9pGL2 promoter region (Figure 1D). Chromatin immunoprecipitation was performed to investigate if this site is functional with the promoter constructs p21pGL2 and Δ1.9p21pGL2 in cells expressing either HA-AML1 or HA-AML1-ETO. As shown in Figure 1F, AML1-ETO and AML1 are able to bind the 2 AML1-binding sites within p21pGL2, but are not able to bind to the Δ1.9p21pGL2 region containing the putative AML1 site, suggesting that AML1-ETO is able to activate/inactivate an unknown factor or factors that regulate this region of the synthetic p21WAF1 promoter in these cells.
AML1-ETO promotes leukemia in p21Waf1-deficient mice
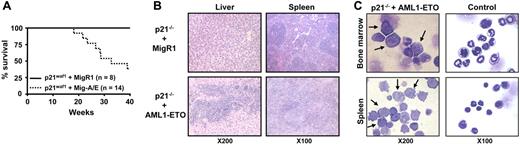
It is well documented that AML1-ETO does not promote leukemia development in mouse models without a secondary mutagenic event. As described (see “AML1-ETO regulates the expression of the cell cycle inhibitor p21WAF1”), we observed the induction of the cell cycle regulator p21WAF1 by AML1-ETO, consistent with its growth arrest function in myeloid cells. These observations suggest that a mechanism of AML1-ETO–induced leukemogenesis could be the bypass of the p21Waf1 pathway. p21Waf1-deficient mice display an increase in stem cells compared to their littermate controls, implicating it in the regulation of stem cell maintenance.46 However, no leukemogenesis was observed in p21Waf1-deficient mice. We thus tested if AML1-ETO would cooperate with the loss of p21WAF1 in promoting leukemia. For this we used fetal liver hematopoietic cells isolated from p21Waf1-deficient mice and performed retroviral transduction and hematopoietic cell transplantation with MigR1 control and Mig-A/E. Three weeks after transplantation, these mice were monitored for signs of leukemia development, including their physical appearance, parameters of their blood cells, and the percentage of EGFP+ cells in peripheral blood. Mice receiving transplants of MigR1-infected cells did not show any obvious abnormalities during the entire experimental period. In contrast, mice receiving transplants of Mig-A/E–infected cells showed the increase of EGFP+ cells in their blood. Near 17 weeks after transplantation, some of the mice began to show symptoms of anemia and labored breathing (Figure 2A). They had increased immature cells in their peripheral blood and eventually became moribund displaying infiltration of myeloid blast cells into the liver and spleen (Figure 2B). Increased immature myeloid cells were observed in the bone marrow and spleen of these diseased mice as compared to their respective controls (Figure 2C). A differential count showed an overall increase of more than 20% in immature blasts cells in their bone marrow compared to control mice (Table 1). We analyzed the expression of AML1-ETO in 2 independent mice in bone marrow and spleen samples and detected the expression of AML1-ETO (Figure 3A). We further analyzed the expression of myeloperoxidase (MPO), a characteristic marker of immature leukemic blast cells, which are large cells with large nuclei and very little cytoplasm,47 and observed that leukemic blasts present in both bone marrow and spleen were positive for cytoplasmic MPO, compared to neutrophilic MPO+ cells in the control bone marrow and MPO− splenocyte samples (Figure 3B). We further confirmed retroviral integrations of the Mig-A/E retrovirus in the leukemic cells by Southern blot analysis using the specific human ETO probe on BamHI digested gDNA and found these to be oligoclonal, suggestive of the outgrowth of more than one specific leukemic clone associated with the integrations (Figure 3C). Flow cytometry analysis of bone marrow and spleen cells showed that the majority of AML1-ETO expressing (EGFP+) bone marrow and spleen cells in leukemic mice are Gr-1 and CD11b double-positive cells (Figure 4). Furthermore, a moderate increase in c-kit+ cells was also detected in the EGFP+ population in both spleen and bone marrow. Lastly, we also confirmed that the leukemia is transplantable into secondary recipients giving rise to the same phenotypic aspects 2 months after transplantation (data not shown). Thus, these results indicate that the expression of AML1-ETO in p21Waf1-deficient cells promotes the development of AML.
AML1-ETO promotes leukemia in p21Waf1-deficient mice. (A) Kaplan-Meier survival plot of mice receiving transplants of MigR1- or Mig-A/E–infected p21Waf1 cells. Mice were monitored for leukemia development 3 weeks after transplantation. (B) Hematoxylin and eosin staining of liver and spleen of a control mouse and a leukemic mouse, showing the disruption of the tissue architecture by infiltrating leukemic cells. (C) Accumulation of blast cells derived from AML1-ETO–expressing p21Waf1-deficient cells in hematopoietic compartments. Cells isolated from spleen and bone marrow from a recipient mouse of transplanted p21WAF1-deficient cells expressing AML1-ETO and a control were cytospun and stained with Wright-Giemsa for the analysis of immature blasts indicated by the arrows. Microscopy was performed with a Leica DMLB microscope (Leica Microsystems, Wetzlar, Germany) using an HC PLAN 10×/22 eyepiece and 10×/0.25 or 20×/0.4 objective lenses, or a 100×/1.30 oil-immersion objective lens (Richard Allan Scientific, Kalamazoo, MI). Pictures were captured using Spot software from Diagnostic Instruments (Sterling Heights, MI).
AML1-ETO promotes leukemia in p21Waf1-deficient mice. (A) Kaplan-Meier survival plot of mice receiving transplants of MigR1- or Mig-A/E–infected p21Waf1 cells. Mice were monitored for leukemia development 3 weeks after transplantation. (B) Hematoxylin and eosin staining of liver and spleen of a control mouse and a leukemic mouse, showing the disruption of the tissue architecture by infiltrating leukemic cells. (C) Accumulation of blast cells derived from AML1-ETO–expressing p21Waf1-deficient cells in hematopoietic compartments. Cells isolated from spleen and bone marrow from a recipient mouse of transplanted p21WAF1-deficient cells expressing AML1-ETO and a control were cytospun and stained with Wright-Giemsa for the analysis of immature blasts indicated by the arrows. Microscopy was performed with a Leica DMLB microscope (Leica Microsystems, Wetzlar, Germany) using an HC PLAN 10×/22 eyepiece and 10×/0.25 or 20×/0.4 objective lenses, or a 100×/1.30 oil-immersion objective lens (Richard Allan Scientific, Kalamazoo, MI). Pictures were captured using Spot software from Diagnostic Instruments (Sterling Heights, MI).
Bone marrow cell differential counts
| Sample . | Myeloblasts/promyelocytes . | Metamyelocytes . | Neutrophils . | Lymphocytes . | Erythroids . | Monocytes . | Eosinophils . |
|---|---|---|---|---|---|---|---|
| MIGR1 | 7.0 ± 1.6 | 14.7 ± 2.5 | 40.0 ± 2.4 | 15.3 ± 2.9 | 19.3 ± 4.5 | 2.7 ± 1.2 | 1.0 ± 0.8 |
| AE | 28.3 ± 3.2 | 16.7 ± 3.2 | 27.3 ± 1.5 | 6.7 ± 2.9 | 5.7 ± 1.5 | 5.3 ± 2.0 | 10.0 ± 2.0 |
| Sample . | Myeloblasts/promyelocytes . | Metamyelocytes . | Neutrophils . | Lymphocytes . | Erythroids . | Monocytes . | Eosinophils . |
|---|---|---|---|---|---|---|---|
| MIGR1 | 7.0 ± 1.6 | 14.7 ± 2.5 | 40.0 ± 2.4 | 15.3 ± 2.9 | 19.3 ± 4.5 | 2.7 ± 1.2 | 1.0 ± 0.8 |
| AE | 28.3 ± 3.2 | 16.7 ± 3.2 | 27.3 ± 1.5 | 6.7 ± 2.9 | 5.7 ± 1.5 | 5.3 ± 2.0 | 10.0 ± 2.0 |
Bone marrow cytospins were prepared from p21−/− + MIGR1 (n = 3) and p21−/− + AML1-ETO (n = 4) mice, and the percentage of denoted hematopoietic lineages was determined. Results are shown ± standard deviation.
Expression of AML1-ETO, MPO, and clonality in leukemic cells. (A) Western blot analysis of 2 independent mice spleen cell samples and one bone marrow cell sample showing the expression of AML1-ETO. Kasumi cells serve as a positive control. (B) Bone marrow and spleen cells from recipient mice of p21Waf1-deficient cells expressing AML1-ETO and a control mouse were cytospun and stained for MPO and counterstained with Wright-Giemsa for the analysis of immature blasts. The arrows indicate MPO+ immature blasts in the leukemic mice and in the control sample MPO+ neutrophils in the bone marrow. Image acquisition was performed as described for Figure 2, using a 100×/1.30 oil-immersion objective lens. (C) gDNA was isolated from 3 leukemic p21Waf1−/− mice expressing AML1-ETO. The DNA was digested with BamHI and a Southern blot prepared. The blot was hybridized with the ETO probe 5′ of the BamHI site in ETO, washed, and exposed to film. The black dots indicate the integration signals.
Expression of AML1-ETO, MPO, and clonality in leukemic cells. (A) Western blot analysis of 2 independent mice spleen cell samples and one bone marrow cell sample showing the expression of AML1-ETO. Kasumi cells serve as a positive control. (B) Bone marrow and spleen cells from recipient mice of p21Waf1-deficient cells expressing AML1-ETO and a control mouse were cytospun and stained for MPO and counterstained with Wright-Giemsa for the analysis of immature blasts. The arrows indicate MPO+ immature blasts in the leukemic mice and in the control sample MPO+ neutrophils in the bone marrow. Image acquisition was performed as described for Figure 2, using a 100×/1.30 oil-immersion objective lens. (C) gDNA was isolated from 3 leukemic p21Waf1−/− mice expressing AML1-ETO. The DNA was digested with BamHI and a Southern blot prepared. The blot was hybridized with the ETO probe 5′ of the BamHI site in ETO, washed, and exposed to film. The black dots indicate the integration signals.
Flow cytometric analysis of AML1-ETO–expressing p21WAF1-deficient cells. Bone marrow and spleen cells were collected from leukemic mice transplanted with p21Waf1−/−-expressing AML1-ETO cells and analyzed for the expression of CD3/B220, Gr-1/CD11b, and c-kit/Sca-1. Depicted are the EGFP+ and EGFP− cells within a particular mouse.
Flow cytometric analysis of AML1-ETO–expressing p21WAF1-deficient cells. Bone marrow and spleen cells were collected from leukemic mice transplanted with p21Waf1−/−-expressing AML1-ETO cells and analyzed for the expression of CD3/B220, Gr-1/CD11b, and c-kit/Sca-1. Depicted are the EGFP+ and EGFP− cells within a particular mouse.
Discussion
Various experimental data indicate that AML1-ETO displays a dichotomy as a leukemogenic factor that is directly involved in leukemogenesis in the presence of additional mutations. Most recently, we have reported an increase in p21WAF1 protein in AML1-ETO–expressing cells.35 In this report, we demonstrate that AML1-ETO activates the p21WAF1 promoter dependent on the AML1-ETO DNA-binding runt homology domain. More importantly, we have shown that a lack of p21Waf1 expression cooperates with AML1-ETO to induce AML1-ETO–involved leukemogenesis. These results indicate that mutations that disrupt p21WAF1 cellular functions are able to cooperate with AML1-ETO in leukemogenesis. However, not 100% of mice receiving transplants of AML1-ETO–expressing p21Waf1-deficient cells developed leukemia and the time required for leukemia development was relatively long, indicating that disruption of the p21 pathway, next to being able to enhance the population of transduced cells,48 contributes or facilitates AML1-ETO–involved leukemia development possibly requiring additional mutations.
AML1-ETO has almost the entire ETO protein fused to the AML1 DNA-binding domain, which interacts with histone deacetylase complexes and can function as a repressor in gene expression.12 However, AML1-ETO also activates gene expression demonstrated via gene expression profiling and promoter-reporter transfection assays.38,49–52 The molecular mechanism of AML1-ETO–involved activation of gene expression is not yet clear. Recently, p21WAF1 overexpression has been observed in t(8;21) leukemia patients compared to other patients with AML, suggestive of a role of this protein in AML1-ETO–induced leukemogenesis.53 Here, we report that in addition to increased p21WAF1 protein and RNA expression, the activity of the p21WAF1 promoter is enhanced by AML1-ETO. Furthermore, R174Q and L148D mutations of AML1-ETO that disrupt either DNA binding or both DNA binding and CBFβ interaction, respectively, do not have any effect on the p21WAF1 promoter, indicating that AML1-ETO may bind directly to the p21WAF1 promoter to activate gene expression. However, deletion of 3 AML1 consensus-binding sites in the p21WAF1 promoter did not fully eliminate the positive effect of AML1-ETO on the p21WAF1 promoter (data not shown). The AML1 runt homology domain has been reported to bind DNA sequences that do not perfectly match the 6 base pairs of the consensus-binding sequence TGT/CGGT.54,55 Besides CBFβ, the runt homology DNA-binding domain can also interact with other transcription regulators.12 Therefore, it is also possible that other sites besides the 3 well-matched AML1-binding sites on the p21WAF1 promoter can bind AML1-ETO. However, ChIP suggests that AML1-ETO is not able to bind to a putative AML1-binding site within the 0.4 kb of the p21WAF1 promoter that is still transactivated by AML1-ETO in the absence of the 3 AML1 consensus sites. Thus, these results suggest that AML1-ETO activates/inactivates other factors that transactivate/inhibit the p21WAF1 promoter independent of the 3 AML1-binding sites. What factors precisely warrant future study.
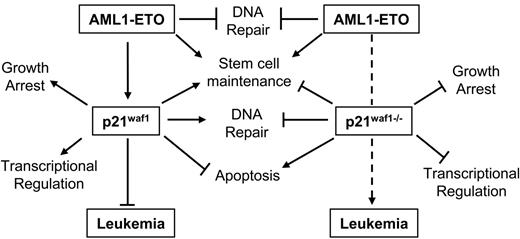
Besides the inhibitory effect of p21WAF1 on the cell cycle through blocking cyclin/CDK activation, p21WAF1 is also known to regulate the G1/S cell cycle transition by stabilizing the cyclin D/CDK4 association,36 to protect against apoptosis in various leukemia cell lines,56–58 to regulate DNA repair in vivo through its interaction with PCNA,59,60 negatively regulating DNA methylation during DNA repair by blocking the methyltransferase DNMT3A binding to PCNA,61 and to regulate the activity of various transcription regulators62–65 and coactivators.64,66–68 Furthermore, p21Waf1-deficient mice show an increase in stem cell number and proliferation compared to littermate controls.46 A role of p21WAF1 in leukemogenesis is also suggested through its up-regulation by leukemogenic mutant FLT3-expressing cells via Stat5 activation.69 The p21WAF1 gene is rarely found inactivated in solid or liquid tumors.70 One point mutant was identified in a ductal breast carcinoma, which disrupted its ability to regulate cell cycle-dependent kinases.71 In AML no disruption of the p21WAF1 gene has been reported specifically.70,72,73 Furthermore, epigenetic regulation of p21WAF1 is rare.72,74,75 However, bypass of p21WAF1 growth-arresting potential is exemplified by cyclin A1 and cyclin E deregulation in myeloid leukemia.76–78 It is also evident that regulation of the p21WAF1 protein itself through proteolytic cleavage by proteinase-3 positively affects myeloid cell growth.79 Therefore, the important message from this study involving AML1-ETO leukemogenesis, is that disruption of the pathways through which p21WAF1 regulates cellular processes could counteract the up-regulation of p21WAF1 by AML1-ETO and contribute to leukemogenesis. Furthermore, our model seems not only dependent on the loss of p21Waf1 and the expression of AML1-ETO but also on the acquisition of additional mutagenic events, as the onset of leukemia is delayed by more than 100 days. These events are putatively involved in survival signals, DNA repair, or epigenetic regulation by DNA methylation, or its role in regulating crucial transcription factors such as E2F and Stat3, since these p21WAF1 pathways are lost in this model. Thus these observations suggest that the deregulation of the p21WAF1 pathway through the different factors associated with it is an important route of leukemogenesis. Contrarily, the increase of p21WAF1 in t(8;21) patients at the RNA level suggests that the survival role of p21WAF1 pathway may be positively manipulated in these patients; however, further studies are required in patient samples to determine if the p21WAF1 protein is increased and how it is localized to determine how posttranslational events influence the availability of normal functional p21WAF1 protein. Furthermore, the Stat3 or E2F negative regulatory functions of p21WAF1 may be bypassed in t(8;21) patients, or alternatively their role in regulating p300 activities is modulated, promoting aberrant growth. Specifically Stat3 is often hyperactive in AML.80 Although it seems that a dichotomy exists between increased expression of p21WAF1 in t(8;21) and our model of loss of p21Waf1 cooperating with AML1-ETO in leukemia development, a model emerges based on our observations as shown in Figure 5. Herein we hypothesize that the induction of p21WAF1 by AML1-ETO in stem cells promotes stem cell maintenance, whereas p21WAF1 function in survival is bypassed as cells undergo apoptosis in the presence of AML1-ETO; however, in primary bone marrow cells expressing AML1-ETO and inducing p21WAF1 an expansion of stem cells42 is observed. Following the removal of p21WAF1, AML1-ETO will still influence stem cell maintenance, while its growth suppressive ability through p21WAF1 is lost. In addition, the loss of the protective role of p21WAF1 in DNA repair and the deregulation of other DNA repair genes by AML1-ETO52 creates an advantage for the acquisition of additional mutagenic events supportive of leukemogenesis. Furthermore, the loss of controlled transcriptional programs regulated by p21WAF1 through factors such as the Stats, E2F, and p300/CBP should enhance further leukemogenic potential of the cells. This mouse model thus delineates that AML1-ETO–associated leukemia is a complex mechanism that relies on many factors and pathways that can bypass pathways regulated by the fusion protein itself, which impede AML development.
Model of AML1-ETO leukemogenesis through the p21WAF1 pathway. Here we propose a model whereby the bypass of the p21WAF1-regulated pathway(s) cooperates with AML1-ETO in promoting leukemia development. Specifically, in the presence of p21WAF1 AML1-ETO ability to promote leukemia is limited by p21WAF1 role in growth regulation, genome stability, regulating transcription networks, and in primary cells' apoptosis evasion. However, both p21WAF1 and AML1-ETO regulate stem cell maintenance and expansion. The loss of p21WAF1 role in growth regulation, DNA repair, and regulation of transcription establishes an advantageous situation for leukemia development.
Model of AML1-ETO leukemogenesis through the p21WAF1 pathway. Here we propose a model whereby the bypass of the p21WAF1-regulated pathway(s) cooperates with AML1-ETO in promoting leukemia development. Specifically, in the presence of p21WAF1 AML1-ETO ability to promote leukemia is limited by p21WAF1 role in growth regulation, genome stability, regulating transcription networks, and in primary cells' apoptosis evasion. However, both p21WAF1 and AML1-ETO regulate stem cell maintenance and expansion. The loss of p21WAF1 role in growth regulation, DNA repair, and regulation of transcription establishes an advantageous situation for leukemia development.
Authorship
Contribution: L.F.P. designed and performed the experiments, analyzed data, and wrote the paper; M.Y. designed and performed the experiments and analyzed data; and D.E.Z. designed the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests:
L.F.P. and M.Y. contributed equally to this work.
Correspondence: Dong-Er Zhang, MEM-L51, Scripps Research Institute, 10550 N Torrey Pines Rd, La Jolla, CA 92037; e-mail: dzhang@scripps.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by National Institutes of Health grant CA96735 (D.E.Z.). The Stein Endowment Fund has partially supported the Molecular and Experimental Medicine departmental molecular biology service laboratory for DNA sequencing and oligonucleotide synthesis.
We wish to thank Drs Scott Hiebert (Vanderbilt University School of Medicine, Nashville, TN), Jeffery Kudok (Harvard Medical School, Boston, MA), Nancy Speck (Dartmouth Medical School, Hanover, NH), and Xiao-Fan Wang (Duke University Medical Center, Durham, NC) for valuable discussion and DNA constructs, as well as Joseph Biggs for editing the manuscript. In addition, we thank Drs Tobias Berg and Michael Lübbert (University of Freiburg Medical Center, Freiburg, Germany) for sharing and discussing valuable prepublished data with us.
This is manuscript no. 18122-MEM from the Scripps Research Institute.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal