Abstract
CD4+ cells of most individuals infected with HIV-1 harbor a C-terminally truncated and constitutively activated form of signal transducer and activator of transcription-5 (STAT5Δ). We report that the chronically HIV-infected U1 cell line expresses STAT5Δ but not full-length STAT5. Granulocyte-macrophage colony-stimulating factor (GM-CSF) stimulation of U1 cells promoted early activation of STAT5Δ and of extracellular signal regulated kinases (ERKs), followed by later activation of activator protein 1 (AP-1) and HIV expression. Inhibition of ERK/AP-1 by PD98,059 abolished, whereas either tyrphostin AG490 or a STAT5 small interfering RNA (siRNA) enhanced, virion production in GM-CSF–stimulated U1 cells. Chromatin immunoprecipitation demonstrated the induction of STAT5Δ binding to STAT consensus sequences in the HIV-1 promoter together with a decreased recruitment of RNA polymerase II after 1 hour of GM-CSF stimulation of U1 cells. Down-regulation of STAT5Δ by siRNA resulted in the up-regulation of both HIV-1 gag-pol RNA and p24 Gag antigen expression in CD8-depleted leukocytes of several HIV-positive individuals cultivated ex vivo in the presence of interleukin-2 but not of interleukin-7. Thus, the constitutively activated STAT5Δ present in the leukocytes of most HIV-positive individuals acts as a negative regulator of HIV expression.
Introduction
Infection with HIV-1 is associated with a progressive depletion of CD4+ T lymphocytes, a generalized immunologic dysfunction, and chronic activation involving several proinflammatory and immunoregulatory cytokines.1,2 In this context, it has been previously reported that peripheral blood mononuclear cells (PBMC) of most HIV-1-infected individuals are characterized by the constitutive activation of the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway, predominantly accounted for by STAT5,3 a host factor triggered by several type I cytokines either belonging to the γ-common (γc) (interleukin-2 [IL-2], IL-7, IL-9, IL-15, IL-21) or βc- (IL-3, IL-5 and granulocyte-macrophage colony stimulating factor [GM-CSF]) families.4,5 Of note is the fact that the predominant form of activated STAT5 expressed in unstimulated PBMC of HIV-positive individuals was found to be truncated at the C terminus (STAT5Δ).3,6 Similar C-terminally truncated forms of STAT5 have been previously described as endowed with DNA-binding activity but lacking transcriptional activation, thereby often functioning as dominant negative factors.7–9
HIV replication is under the control of both viral and host factors affecting either preintegration, transcriptional or posttranscriptional events in its life cycle.1,2 In particular, regulation of HIV expression from integrated provirus is strongly influenced by host transcription factors binding to the 5′ virus long terminal repeats (LTR), including constitutively activated specific protein-1, cytokine-inducible nuclear factor kB (NF-kB), nuclear factor of activated T cells-1, and activator protein-1 (AP-1).2,10,11 Functional binding sites for other cellular transcription factors such as cAMP response element binding12 and interferon (IFN) regulatory factor-113 have also been described in the U3 region of the virus LTR. In addition, AP-1 binds to a consensus sequence present in an intragenic enhancer downstream from the transcription start site.14–17
In this scenario, the role of STAT proteins in controlling HIV transcription has been demonstrated primarily as a consequence of the antiviral effect of IFNs activating STAT1 and STAT2.5,18 However, the observation that several STAT5-activating cytokines exert upregulatory effects on HIV replication in either T cells or mononuclear phagocytes19 suggested a potential role of STAT5 in controlling viral expression. In this regard, 3 independent regions in the HIV LTR have been recently identified as close matches to the STAT5 consensus-binding sequence along with functional evidence of a direct positive effect of full-length STAT5 (STAT5FL) activation on viral transcription.20
We investigated the functional consequences of STAT5Δ activation and inactivation on HIV replication in vitro and provide substantial evidence for its negative role in HIV expression in both a chronically infected cell line (U1) and in CD4+ PBMCs of HIV-positive individuals cultivated ex vivo.
Materials and methods
Approval was obtained from the Institutional Review boards of the San Raffaele Scientific Institute of Milano and of the Hospital of Lodi, Italy. Informed consent was obtained in accordance with the Declaration of Helsinki.
Cell lines and reagents
The ACH-2 and U1 cell lines were maintained in RPMI 1640 medium (Bio-Whittaker, Verviers, Belgium) containing 10% fetal bovine serum (FBS) (Bio-Wittaker) and supplemented with penicillin-streptomycin and glutamine (PSG; complete medium). The 293T cell line was propagated in D-MEM supplemented with 10% fetal bovine serum (FBS)3 and PSG. Phorbol 12-myristate, 13-acetate (PMA) was used at a concentration of 10-8M (Sigma Chemical Corp., St. Louis, MO). Recombinant GM-CSF was used at 20 ng/mL (R&D Systems, Minneapolis, MN). AG490 (Calbiochem, San Diego, CA) and PD98,059 (Sigma Chemical Corp.) were used at concentrations of 200 and 20 μM, respectively. Recombinant IL-2 (Chiron, Emeryville, CA) and IL-7 (R&D Systems) were used at 400 and 20 ng/mL, respectively, based on previous references.3,21,22
PBMCs from HIV-positive individuals
PBMCs were isolated from peripheral venous blood collected from HIV-positive individuals on heparin, EDTA, or acid citrate dextrose by centrifugation on Ficoll-Paque Plus gradients (Amersham Bioscience, Uppsala, Sweden). PBMC were then depleted of CD8+ cells using anti-CD8 monoclonal antibody (mAb) conjugated magnetic microbeads (Miltenyi Biotec Inc., Sunnyvale, CA) according to the manufacturer's instructions. Cells were maintained in complete medium supplemented with 400 international units/ml of recombinant IL-2 (Chiron).
Plasmids
HA-tagged STAT5A cDNA (donated by B. Mathey-Prevot, Harvard University, Boston, MA) was subcloned into the expression vector pXM to generate the pXM-HA-STAT5A plasmid. Their sequences and construction are detailed in Supplemental Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article.23,24
LTR-GFP assay
The eGFP-PΔN vector was obtained by inserting the enhanced green fluorescent protein (eGFP) polymerase chain reaction (PCR)-amplified ORF into the ClaI site of the previously described HIV-1 based PΔN vector.25 In this configuration, eGFP is driven by the unmodified 5′ LTR of the vector. 293T cells were seeded at a concentration of 105 cells/well in 24-well plastic plates (Corning Inc., New York, NY) 24 hours before transfection. Cells were cotransfected with eGFP-PΔN (100 ng) and pXM, pXM-HA-STAT5A-P, or pXM-HA-STAT5A-Δ710-P (900 ng) by Fugene6 according to the manufacturer's instructions (Roche Diagnostics Corp., Indianapolis, IN). For detection of LTR-driven GFP activity 24 hours posttransfection, cells were acquired by FACScan BD (Becton Dickinson, Franklin Lakes, NJ) and analyzed with FlowJo software (Tree Star, Ashland, OR4 ).
Whole-cell extracts and immunoblotting
Whole cell extracts (WCE) preparation and Western blot analyses were performed as previously described.8 Anti-STAT5 mouse mAb (BD Biosciences, Transduction Laboratories, Heidelberg, Germany), anti-phospho-STAT5 mAb (Y694-Y699; Upstate, Charlottesville, VA) and goat anti-mouse horseradish peroxidase Ab (Promega Corp., Madison, WI) were diluted 1:500, 1:1,000 and 1:15,000, respectively. Rabbit polyclonal anti-ERK2 (C-14) Ab (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), goat anti-rabbit horseradish peroxidase Ab (Dako Cytomation, Glostrup, Denmark), anti-phospho-ERK (E-4) Ab (Santa Cruz), and rabbit anti-mouse Ab (Dako Cytomation) were diluted 1:1000, whereas an anti-actin Ab (Sigma) was diluted 1:500. The immunoblots were developed by the enhanced chemiluminescence detection system (Amersham, Little Chalfont, UK).
LTR nucleotide sequence analysis
The U3 region of the subtype B HIV-1 LTR nucleotide sequences was analyzed for the identification of potential STAT-binding sites by the TFSEARCH software (www.cbrc.jp/research/db/TFSEARCH.html) and COMPEL databases (www.bionet.nsc.ru/TRRD).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed as described previously26 with some modifications. The methodology is described in detail in Document S1.
Reverse transcriptase–polymerase chain reaction analysis for HIV-1 mRNA
To verify the levels of transcription induction after PMA or GM-CSF stimulation of U1 cells, isolated mRNA (RNeasy kit; QIAGEN GmbH, Hilden, Germany) was reverse-transcribed and quantified by TaqMan real time reverse transcriptase-polymerase chain reaction (RT-PCR) using a specific probe that maps in the HIV-1 gag gene (U1A). The levels of viral mRNA were normalized against a housekeeping gene, 18S rRNA (Applied Biosystems), and were represented as fold induction over control unstimulated cells. The sequences of primers and probes used for real-time PCR are available in Document S1.
Transfection of STAT5 small interfering RNA
U1 cells were transfected by the Amaxa electroporator Nucleofector I, using the Nucleofector Kit V (program V-01) according to the manufacturer's procedure (AMAXA Biosystems, Cologne, Germany). To evaluate the transfection efficiency, cells were transfected with the BLOCK-iT Fluorescent Oligo (Invitrogen Corp., Paisley, PA); efficiency of transfection was estimated to be approximately 90% in most experiments. Small interfering RNA (siRNA) STAT5 was performed with STAT5A Stealth RNAi kit (HSS110,284-85-86; Invitrogen Corp.) and with STAT5B Stealth RNAi validated Stealth RNAi DuoPak (12,937-20, Invitrogen Corp.). U1 cells (2×106 cells resuspended in 1 mL) were transfected by electroporation with scramble Low GC Duplex or Med GC Duplex Stealth RNAi Negative Control (12,935-200/300; Invitrogen Corp.) or with STAT5AB RNAi (2 μM each; Invitrogen Corp.). Twenty-four hours posttransfection, cells were seeded in 96-well plates at the concentration of 2×105 cells/mL and stimulated with 20 ng/mL of GM-CSF (R&D Systems, Minneapolis, MN).
CD8-depleted PBMC from HIV-positive individuals were transfected by the Amaxa electroporator Nucleofector I and Human T cell Nucleofector Kit (program U-14) according to the manufacturer's procedure. Transfection efficiency (approximately 80%) was evaluated with the BLOCK-iT Fluorescent Oligo kit (Invitrogen Corp.). Four million cells were transfected by electroporation with either a scramble Low GC Duplex or Med GC Duplex Stealth RNAi Negative Control (12,935-200/300; Invitrogen Corp.) or with anti-STAT5AB RNAi (4 μM each; Invitrogen Corp.). Twenty-four hours posttransfection, cells (106 cells/mL) were seeded in 24-well plastic plates (Corning Inc.) in complete medium enriched with IL-2 (400 IU/mL).
Intracellular staining and cytofluorimetric analysis for expression of HIV-1 gag-pol RNA
Four days after transfection of the different siRNA, approximately 106 cells were harvested from the cell culture, centrifuged (1600 rpm for 5 minutes), resuspended in phosphate-buffered saline (PBS), permeabilized, and stained for HIV-1 RNA (gag-pol) with the Virotect Plus kit according to the manufacturer (Invirion Diagnostics, Oak Brook, IL).27 Samples were acquired by cytoflurorimetry and lymphocytes were gated by forward scatter/side scatter. The results were analyzed by FlowJo (Free Star).
Intracellular staining and cytofluorimetric analysis for expression of HIV-1 p24 Gag antigen (Ag)
Five days after siRNA transfection, 5×105 cells were harvested from the culture, centrifuged 5 minutes at 1500 rpm, resuspended in 0.5 mL, and fixed with 2% paraformaldehyde for 15 minutes. Cells were washed with PBS containing 2% FCS and 0.1% Na-azide, and permeabilized with saponin buffer (PBS containing 2% FCS and 0.1% Na-azide, 0.5% saponin). Cells were then incubated with 1 μg of a phycoerythrin-conjugated anti-p24 Gag mAb KC57-RD1 (Coulter Clone, Fullerton, CA) or with 1 μg of phycoerythrin-conjugated mouse IgG2a, kappa isotype control (BD Biosciences) for 20 minutes at room temperature and acquired by a FACScan apparatus (Becton Dickinson, Franklin Lakes, NJ). During analysis by FlowJo software, a lymphocyte gate in the forward scatter/side scatter diagram was adopted.
Reverse transcriptase activity assay
HIV production was measured in the cell culture supernatants by a liquid-phase Mg++-dependent RT activity assay, as described previously.3
Results
Expression of a similar C-terminally truncated STAT5 isoform in PBMC of HIV-positive individuals and in chronically infected U1 cells
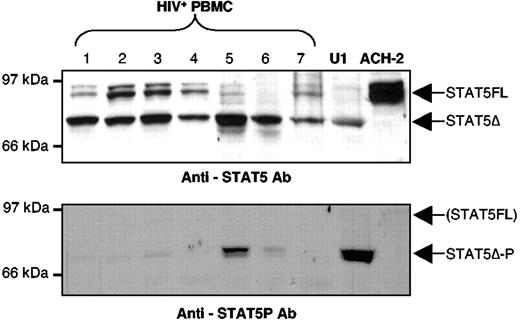
Several HIV-positive individuals (Table 1), most of whom are under highly active antiretroviral therapy and distinct from those originally studied,3,6 showed the presence of a STAT5Δ isoform characterized by a molecular weight lower than that of STAT5FL. Some patients (patients no. 1, 2, 3, 4, and 7) showed variable levels of expression of STAT5FL, whereas others (patients no. 5 and 6) were characterized by the presence of virtually only STAT5Δ (Figure 1, upper panel). Of note is the fact that variable levels of constitutive STAT5Δ activation were detected by an Ab recognizing only the phosphorylated (activated) form of STAT5 with a prominence in PBMC of patients no. 5 and 6 (Figure 1, lower panel) despite the concomitant expression of STAT5FL and STAT5Δ. These findings confirm and extend our original observation that was based on electrophoretic mobility shift assays (EMSA) experiments.3,6
Clinical features of HIV-positive individuals
| Patient no. . | Year of birth . | Seroconversion (year) . | CDC stage* . | CD4+ T cells/μL . | CD8+ T cells/μL . | CD4/CD8 ratio . | Viremia (RNA copies/mL) . |
|---|---|---|---|---|---|---|---|
| 1 | 1969 | 1995 | C3 | 131 | 1223 | 0.10 | 386,000 |
| 2 | 1962 | 1983 | C3 | 210 | 1479 | 0.14 | 42,600 |
| 3 | 1955 | 1992 | C3 | 215 | 1507 | 0.14 | ≤50 |
| 4 | 1969 | 1995 | C2 | 327 | 2244 | 0.15 | 12,000 |
| 5 | 1963 | 1994 | C3 | 238 | 1129 | 0.21 | 156 |
| 6 | 1960 | 1994 | C2 | 378 | 1533 | 0.25 | ≤50 |
| 7 | 1969 | 1991 | C2 | 433 | 921 | 0.47 | ≤50 |
| 8 | 1947 | 1991 | C3 | 116 | ND | ND | 195,000 |
| 9* | 1961 | 1997 | A2 | 360 | 1041 | 0.35 | 18,157 |
| 10 | 1957 | 1988 | A2 | 267 | 978 | 0.27 | 40,304 |
| 11* | 1973 | 2003 | A1 | 474 | 1062 | 0.45 | 135,315 |
| 12 | 1968 | 1995 | C1 | 1,167 | 2237 | 0.52 | 67,555 |
| Patient no. . | Year of birth . | Seroconversion (year) . | CDC stage* . | CD4+ T cells/μL . | CD8+ T cells/μL . | CD4/CD8 ratio . | Viremia (RNA copies/mL) . |
|---|---|---|---|---|---|---|---|
| 1 | 1969 | 1995 | C3 | 131 | 1223 | 0.10 | 386,000 |
| 2 | 1962 | 1983 | C3 | 210 | 1479 | 0.14 | 42,600 |
| 3 | 1955 | 1992 | C3 | 215 | 1507 | 0.14 | ≤50 |
| 4 | 1969 | 1995 | C2 | 327 | 2244 | 0.15 | 12,000 |
| 5 | 1963 | 1994 | C3 | 238 | 1129 | 0.21 | 156 |
| 6 | 1960 | 1994 | C2 | 378 | 1533 | 0.25 | ≤50 |
| 7 | 1969 | 1991 | C2 | 433 | 921 | 0.47 | ≤50 |
| 8 | 1947 | 1991 | C3 | 116 | ND | ND | 195,000 |
| 9* | 1961 | 1997 | A2 | 360 | 1041 | 0.35 | 18,157 |
| 10 | 1957 | 1988 | A2 | 267 | 978 | 0.27 | 40,304 |
| 11* | 1973 | 2003 | A1 | 474 | 1062 | 0.45 | 135,315 |
| 12 | 1968 | 1995 | C1 | 1,167 | 2237 | 0.52 | 67,555 |
All individuals, except nos. 9 and 11 (*), were receiving various highly active antiretroviral therapy protocols, including RT and protease inhibitors, at the time of analysis. None of the patients had received either IFN or IL-2-based therapies.
ND indicates not determined.
CDC stage.79
Expression and activation of STAT5 isoforms in PBMC of HIV-positive individuals and in the chronically infected U1 cell line. Expression of both STAT5FL and STAT5Δ (upper panel) and of constitutively activated STAT5Δ in WCE of PBMC from HIV-positive individuals (lower panel) as detected by Western blotting using specific anti-STAT5 and antiphosphorylated STAT5 Ab, respectively. PBMC from all HIV-positive individuals studied are characterized by the dominant (patient nos. 1, 2, 3, 4, and 7) or exclusive (patient nos. 5 and 6) presence of STAT5Δ (upper panel). Variable levels of constitutive activation of STAT5Δ were detected by using anti-phospho-STAT5 (lower panel). Exclusive expression of either STAT5Δ or STAT5FL was detected in U1 and ACH-2 cells, respectively (upper panel). GM-CSF stimulation (15 minutes) of U1 cells led to the phosphorylation of STAT5Δ (U1, lower panel), whereas no stimuli were identified as capable of activating STAT5FL in ACH-2 cells (see text).
Expression and activation of STAT5 isoforms in PBMC of HIV-positive individuals and in the chronically infected U1 cell line. Expression of both STAT5FL and STAT5Δ (upper panel) and of constitutively activated STAT5Δ in WCE of PBMC from HIV-positive individuals (lower panel) as detected by Western blotting using specific anti-STAT5 and antiphosphorylated STAT5 Ab, respectively. PBMC from all HIV-positive individuals studied are characterized by the dominant (patient nos. 1, 2, 3, 4, and 7) or exclusive (patient nos. 5 and 6) presence of STAT5Δ (upper panel). Variable levels of constitutive activation of STAT5Δ were detected by using anti-phospho-STAT5 (lower panel). Exclusive expression of either STAT5Δ or STAT5FL was detected in U1 and ACH-2 cells, respectively (upper panel). GM-CSF stimulation (15 minutes) of U1 cells led to the phosphorylation of STAT5Δ (U1, lower panel), whereas no stimuli were identified as capable of activating STAT5FL in ACH-2 cells (see text).
To investigate the potential expression of STAT5Δ in HIV-infected cells, we examined the expression of STAT5 in the U1 (promonocytic) and ACH-2 (T lymphocytic) cells chronically infected with HIV-1 and characterized by inducibility of virus expression by several cytokines and PMA.19 ACH-2 cells displayed abundant levels of STAT5FL (Figure 1A, upper panel), whereas U1 cells expressed virtually exclusively a shorter STAT5 isoform indistinguishable in size from the one detected in PBMC of HIV-positive individuals (Figure 1A, upper panel).
GM-CSF induces the activation of STAT5Δ in U1 cells
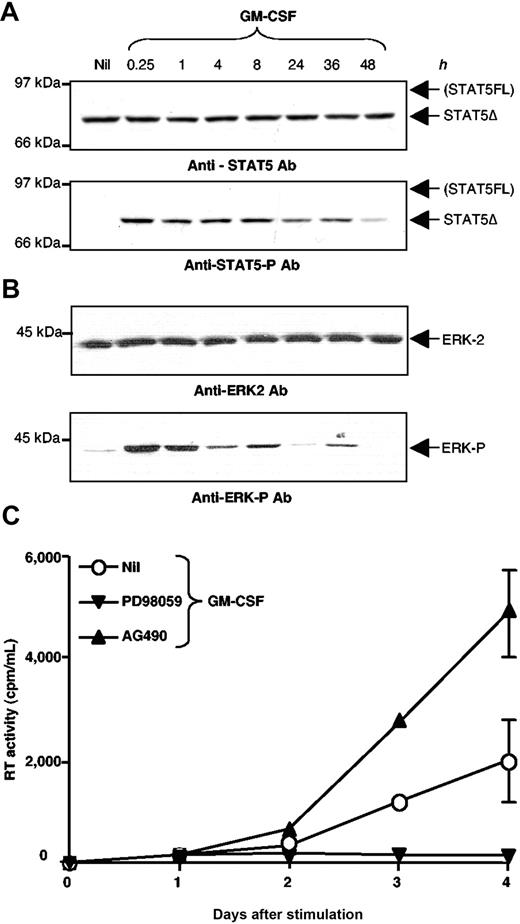
Because chronically infected U1 cells show the expression of a STAT5Δ isoform similar in size to that observed in PBMC of HIV-positive individuals, we investigated the potential relevance of STAT5Δ activation in this model of inducible HIV expression. U1 cells were stimulated with GM-CSF (20 ng/mL), a known inducer of STAT5 phosphorylation,33 from 15 minutes to 48 hours and their WCE were analyzed by Western blot after incubation with an anti-STAT5 Tyr-P Ab recognizing only the phosphorylated form of STAT5. The filters were then stripped and reprobed with an anti-STAT5 Ab specific for an internal region of the molecule. STAT5Δ was readily phosphorylated 15 minutes after GM-CSF stimulation and it remained activated in U1 cells for at least 48 h (Figures 1 and 2A, lower panel). No evidence of expression and activation of STAT5FL was obtained in U1 cells even when analyzed at different time points (Figures 1 and 2A, upper panel).
Unlike U1 cells, no STAT5 activating factors have been reported thus far as capable of inducing HIV expression in ACH-2, a cell line responsive to cytokines acting through NF-kB such as tumor necrosis factor-α/-β or CD30 ligand.28,29 Nonetheless, we attempted to stimulate ACH-2 cells with STAT5-inducing cytokines such as IL-2, IL-3, IL-7, and GM-CSF. These cytokines did not induce HIV expression or activate STAT5FL in ACH-2 cells (data not shown) and, therefore, experiments with this cell line were not pursued further.
GM-CSF induces the early activation of ERK and a later activation of AP-1 and HIV-1 expression in U1 cells
In addition to STAT5, GM-CSF is known to trigger the activation of multiple intracellular signaling pathways, including Ras-Raf-extracellular signal regulated kinases (ERK)-AP-1, phosphatidyl-inositol 3-kinase, stress-activated protein kinases, and p38 mitogen-activated protein kinases32 as well as NF-kB and specific protein-1 in the Ba/F3 murine cell line.33 ERK1/2 are known activators of Jun/Fos (AP-1), a transcription factor previously linked to the activation of HIV transcription,14–17,32 as we have recently observed in the case of IL-6 stimulated U1 cells.34,35 Indeed, GM-CSF induced rapid activation of ERK-2 that remained detectable up to 36 hours after stimulation (Figure 2B), whereas activation of AP-1 became detectable only after several hours (Figure 1, Figure S1), as already observed in IL-6-stimulated U1 cells.35 Basal activation of Jun N-terminal kinase, protein kinase B, and stress-activated and mitogen-activated p38 protein kinases was observed in unstimulated cells but their state of activation was not increased after GM-CSF stimulation (data not shown). Furthermore, no up-regulation of NF-kB activation was detected by stimulation of U1 cells with GM-CSF by EMSA (data not shown).
GM-CSF induces an early and long-lasting activation of STAT5Δ, ERK2, and a later up-regulation of HIV expression in U1 cells. (A) STAT5Δ expression (upper panel) and activation (lower panel) was assessed by incubation with the same Ab indicated in Figure 1, and by Western blotting of WCE of U1 cells stimulated with GM-CSF and collected at the indicated times after stimulation. STAT5Δ activation was detected for at least 48 hours after cytokine stimulation. (B) ERK-2 activation persisting up to 36 h after GM-CSF stimulation of U1 cells was demonstrated by Western blotting, using either anti-ERK-2 (upper panel) or anti-phosphorylated-ERK (lower panel) Ab. (C) Opposite effects of PD98,059 and AG490 on GM-CSF induced HIV-1 expression in U1 cells. U1 cells were stimulated with GM-CSF (20 ng/mL) in the presence or absence of the ERK inhibitor PD98,059 (20 μM) or the JAK2/JAK3 inhibitor tyrphostin AG490 (200 μM). Culture supernatants were collected daily to measure RT activity production and accumulation. The results shown were obtained in a single experiment representative of three independently performed in duplicate cultures (mean ± standard deviation).
GM-CSF induces an early and long-lasting activation of STAT5Δ, ERK2, and a later up-regulation of HIV expression in U1 cells. (A) STAT5Δ expression (upper panel) and activation (lower panel) was assessed by incubation with the same Ab indicated in Figure 1, and by Western blotting of WCE of U1 cells stimulated with GM-CSF and collected at the indicated times after stimulation. STAT5Δ activation was detected for at least 48 hours after cytokine stimulation. (B) ERK-2 activation persisting up to 36 h after GM-CSF stimulation of U1 cells was demonstrated by Western blotting, using either anti-ERK-2 (upper panel) or anti-phosphorylated-ERK (lower panel) Ab. (C) Opposite effects of PD98,059 and AG490 on GM-CSF induced HIV-1 expression in U1 cells. U1 cells were stimulated with GM-CSF (20 ng/mL) in the presence or absence of the ERK inhibitor PD98,059 (20 μM) or the JAK2/JAK3 inhibitor tyrphostin AG490 (200 μM). Culture supernatants were collected daily to measure RT activity production and accumulation. The results shown were obtained in a single experiment representative of three independently performed in duplicate cultures (mean ± standard deviation).
GM-CSF has been previously shown to induce virus replication in both primary monocyte-derived macrophages30 and U1 cells.31 Indeed, GM-CSF stimulation of U1 cells induced the release of HIV-1 particles (measured as release of RT activity in culture supernatants) after 36 to 48 hours of stimulation (Figure 2C). The HIV-inductive effect of GM-CSF was, however, significantly lower and characterized by delayed kinetics in comparison to that observed in PMA-stimulated cells (Figure 2, Figure S2).
Dichotomous effect of pharmacologic inhibitors on GM-CSF–induced HIV-1 expression in U1 cells
To explore the potentially distinct contribution of ERK-AP1 and STAT5Δ to HIV expression in GM-CSF-stimulated U1 cells, experiments were conducted in the presence of noncytotoxic/cytostatic (data not shown) concentrations of relatively specific pharmacologic inhibitors, PD98,059 and AG490. PD98,059 is a MEK and ERK1/2 inhibitor that blocks AP-1 activation,32,33,35 whereas AG490 has been reported to inhibit activation of JAK2 and JAK3, thereby preventing the phosphorylation of STAT5.36–38 PD98,059 completely suppressed, whereas AG490 enhanced, HIV-1 expression in GM-CSF-stimulated U1 cells in terms of both kinetics and peak viral production (Figure 2C).
Silencing STAT5Δ expression enhances HIV production in GM-CSF stimulated U1 cells
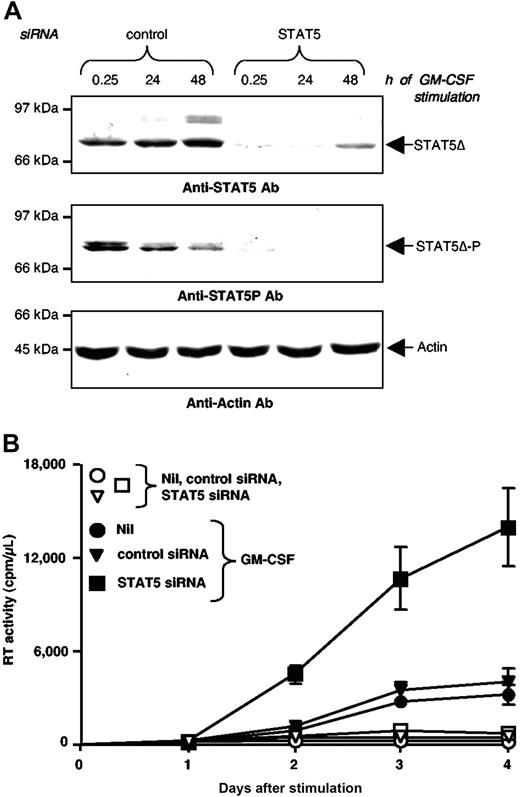
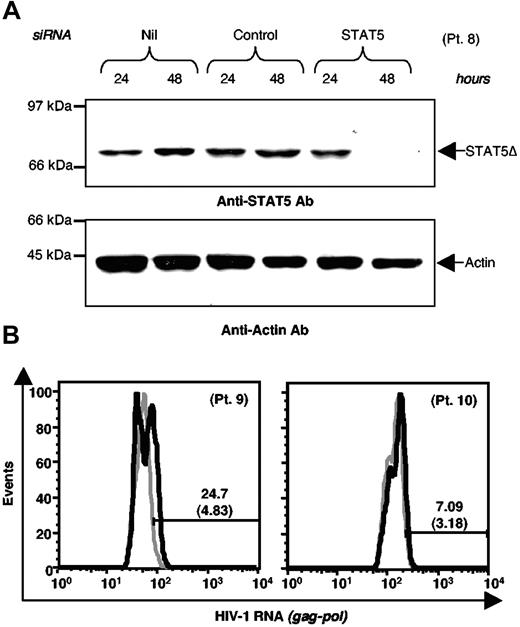
To further demonstrate the functional role of STAT5Δ on GM-CSF stimulation of U1 cells, we used siRNA to suppress STAT5 expression. U1 cells were electroporated with either a specific STAT5 siRNA or a scrambled control siRNA. Twenty-four hours later, cells were stimulated with GM-CSF and the expression of both STAT5Δ and HIV were monitored. STAT5 siRNA, but not control siRNA, inhibited the expression of STAT5Δ (Figure 3A, middle panel) resulting in a reduction in the levels of activated STAT5Δ after GM-CSF stimulation (Figure 3A, upper panel). Either transfection or control siRNA did not alter the basal and GM-CSF stimulated levels of RT activity relative to untransfected cells; in contrast, siRNA-mediated inhibition of STAT5 expression induced a significant enhancement of HIV expression in GM-CSF-stimulated U1 cells (Figure 3B).
Silencing of STAT5Δ expression enhances GM-CSF stimulated HIV production in U1 cells. (A) Western blot analysis of U1 cells electroporated with either control or STAT5 siRNA. Twenty-four hours after siRNA electroporation, U1 cells were stimulated with GM-CSF and their WCE were prepared at the indicated time points. Membranes were hybridized with anti-STAT5 mAb (upper panel), anti-phospho-STAT5 mAb (middle panel), and anti-α-actin Abs (lower panel). (B) U1 cells were electroporated with STAT5 or control siRNA and 24 hours later they were either left unstimulated (Nil) or were stimulated with GM-CSF (20 ng/mL). HIV-1 production was measured by RT activity in culture supernatants. The results shown were obtained in a single experiment representative of five independently performed in triplicate cultures (mean ± standard deviation).
Silencing of STAT5Δ expression enhances GM-CSF stimulated HIV production in U1 cells. (A) Western blot analysis of U1 cells electroporated with either control or STAT5 siRNA. Twenty-four hours after siRNA electroporation, U1 cells were stimulated with GM-CSF and their WCE were prepared at the indicated time points. Membranes were hybridized with anti-STAT5 mAb (upper panel), anti-phospho-STAT5 mAb (middle panel), and anti-α-actin Abs (lower panel). (B) U1 cells were electroporated with STAT5 or control siRNA and 24 hours later they were either left unstimulated (Nil) or were stimulated with GM-CSF (20 ng/mL). HIV-1 production was measured by RT activity in culture supernatants. The results shown were obtained in a single experiment representative of five independently performed in triplicate cultures (mean ± standard deviation).
Phosphorylated STAT5Δ binds in vivo to STAT5 consensus sequences in the HIV-LTR
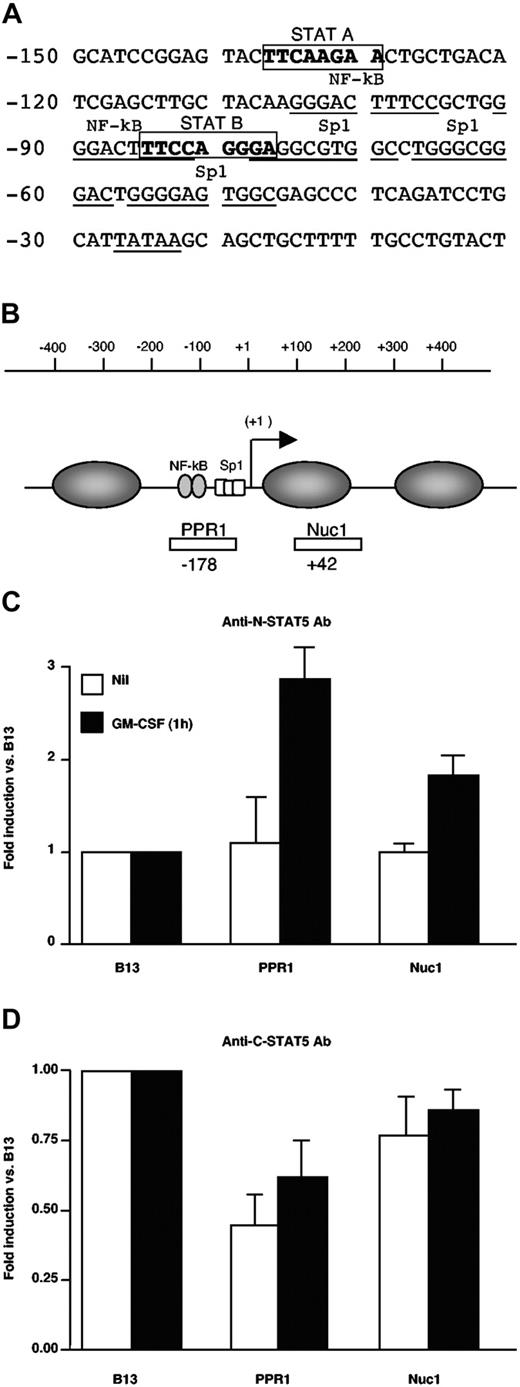
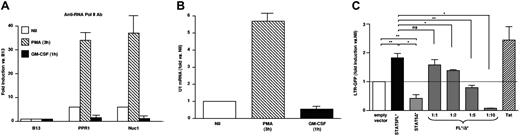
To investigate the mechanism by which STAT5Δ could inhibit HIV expression, we have searched for putative STAT-binding sites in the HIV-LTR using TFSEARCH software (see “Materials and Methods”). Two potential STAT-consensus elements (A and B) were identified at positions -137 to -130 and -85 to -77 in the U3 LTR of the subtype B NL4-3 provirus (Figure 4A) as reported by independent investigators.39 To determine whether STAT5Δ was actually present in living cells and whether it could actually bind to the HIV LTR, we performed ChIP assays.26 U1 cells were stimulated or left unstimulated for 1 hour with GM-CSF, and then sonicated chromatin was immunoprecipitated with either anti-N-terminal or C-terminal5 STAT5 polyclonal Ab. The DNA content of the immunoprecipitated material was then quantified by real-time PCR. In particular, three regions were investigated, two of which mapped to the U3 LTR region of the HIV-1 proviral DNA (Figure 4B). The third region, named B13, mappped to chromosome 19 and was selected as internal negative control because it is a gene-poor region not containing binding sites for known transcription factors.26 The levels of binding of different Ab to the regions of interest were therefore normalized over those detected using B13. U1 cell stimulation with GM-CSF induced binding of STAT5Δ to the PPR1 region of the viral promoter as detected by the anti-STAT5 N-terminus Ab with a 3-fold increase over B13, whereas lower levels of STAT5Δ were also detected within the Nuc1A region (Figure 4C). In contrast, the anti-STAT5 C-terminus Ab recognized any bound protein (Figure 4D), supporting the previous evidence that STAT5Δ is the only STAT5 isoform present in detectable concentrations in U1 cells (Figures 1 and 2).
STAT5Δ binds to the HIV-1 LTR in living U1 cells. (A) Identification of 2 putative STAT binding sites (STAT A and STAT B) in the U3 region of the HIV-1 LTR according to TFSEARCH software (see “Materials and methods”). (B) Positions of primers and TaqMan probes selected to amplify 2 regions in the LTR promoter represented with regard to the transcription start site (+1) and encompassing the putative STAT5 binding sites. The numbers below the investigated segments indicate the location of the 5′ primer used for amplification. For each analyzed region, the amounts of chromatin immunoprecipitated after 1 hour of GM-CSF stimulation vs. unstimulated cells by polyclonal Ab against STAT5-N (C) or STAT5-C (D) are shown. The percentage of input chromatin for the 2 LTR regions indicated in panel B was normalized to that of the unrelated genomic region B13. The results are expressed as fold enrichment over B13-related levels. These results were obtained in a single experiment representative of two independently performed. Real-time PCR (TaqMan) was performed in triplicate in each experiment (mean ± standard deviation).
STAT5Δ binds to the HIV-1 LTR in living U1 cells. (A) Identification of 2 putative STAT binding sites (STAT A and STAT B) in the U3 region of the HIV-1 LTR according to TFSEARCH software (see “Materials and methods”). (B) Positions of primers and TaqMan probes selected to amplify 2 regions in the LTR promoter represented with regard to the transcription start site (+1) and encompassing the putative STAT5 binding sites. The numbers below the investigated segments indicate the location of the 5′ primer used for amplification. For each analyzed region, the amounts of chromatin immunoprecipitated after 1 hour of GM-CSF stimulation vs. unstimulated cells by polyclonal Ab against STAT5-N (C) or STAT5-C (D) are shown. The percentage of input chromatin for the 2 LTR regions indicated in panel B was normalized to that of the unrelated genomic region B13. The results are expressed as fold enrichment over B13-related levels. These results were obtained in a single experiment representative of two independently performed. Real-time PCR (TaqMan) was performed in triplicate in each experiment (mean ± standard deviation).
U1 cell stimulation with PMA, but not with GM-CSF, induced an early increase in RNA polymerase II recruitment to the viral promoter. Worthy of note, the levels of RNA polymerase II recruited to the HIV promoter in U1 cells stimulated for 1 hour with GM-CSF were lower than those of unstimulated cells as revealed by ChIP analysis (Figure 5A). Consistently, PMA induced an increase in viral mRNA levels (6- to 10-fold over control untreated sample), whereas cells stimulated with GM-CSF did not show any change in HIV-1 mRNA levels when compared with unstimulated cells (Figure 5B).
Opposite effects on early HIV transcription in U1 cells stimulated with PMA or GM-CSF. (A). Chromatin preparations from control (unstimulated), PMA, and GM-CSF stimulated U1 cells were immunoprecipitated with a polyclonal Ab against total RNA-polymerase II. The two LTR regions and the B13 region were analyzed by real-time PCR as described in Figure 6. (B) Real-time PCR analysis of HIV RNA levels at early time points after either PMA or GM-CSF stimulation of U1 cells. PMA, but not GM-CSF, stimulation increases HIV RNA accumulation versus the basal levels observed in control, unstimulated cells. (C). Opposite effects of constitutively phosphorylated STAT5 versus STAT5Δ on HIV-1 LTR transactivation. 293T cells were cotransfected with 100 ng of an LTR-GFP reporter construct and 900 ng of either an empty vector or constitutive phosphorylated (*) STAT5FL or STAT5Δ-expressing vectors or with STAT5FL*/STAT5Δ* vectors at the indicated molar ratios. The results are expressed as fold induction of the percentage of GFP+ cells normalized to the levels observed after transfection with an empty vector (mean ± standard error of mean of four independent experiments for empty vector versus STAT5FL* versus STAT5Δ* and of three independent experiments for the competition experiments, respectively). *P <.05; **P<.005; ns: not significant (t test for paired samples).
Opposite effects on early HIV transcription in U1 cells stimulated with PMA or GM-CSF. (A). Chromatin preparations from control (unstimulated), PMA, and GM-CSF stimulated U1 cells were immunoprecipitated with a polyclonal Ab against total RNA-polymerase II. The two LTR regions and the B13 region were analyzed by real-time PCR as described in Figure 6. (B) Real-time PCR analysis of HIV RNA levels at early time points after either PMA or GM-CSF stimulation of U1 cells. PMA, but not GM-CSF, stimulation increases HIV RNA accumulation versus the basal levels observed in control, unstimulated cells. (C). Opposite effects of constitutively phosphorylated STAT5 versus STAT5Δ on HIV-1 LTR transactivation. 293T cells were cotransfected with 100 ng of an LTR-GFP reporter construct and 900 ng of either an empty vector or constitutive phosphorylated (*) STAT5FL or STAT5Δ-expressing vectors or with STAT5FL*/STAT5Δ* vectors at the indicated molar ratios. The results are expressed as fold induction of the percentage of GFP+ cells normalized to the levels observed after transfection with an empty vector (mean ± standard error of mean of four independent experiments for empty vector versus STAT5FL* versus STAT5Δ* and of three independent experiments for the competition experiments, respectively). *P <.05; **P<.005; ns: not significant (t test for paired samples).
Opposite effects of constitutively phosphorylated STAT5FL and STAT5Δ on HIV-LTR transactivation
To further characterize the differential effect of STAT5 vs. STAT5Δ binding to the HIV-1 LTR on HIV-1 transcription, 293T cells were cotransfected with the reporter vector eGFP-PΔN (carrying the eGFP gene under the control of the LTR of the HIV-based vector) together with either a control empty plasmid or with an expression vector containing either constitutively phosphorylated (*) STAT5FL23,24 or STAT5Δ. STAT5FL* induced the expression of eGFP 24 hours after transfection, albeit at levels lower than those induced by transfection of HIV-1 Tat (Figure 5C). In contrast, STAT5Δ* decreased the level of basal eGFP expression observed in cells transfected with an empty vector. Furthermore, a progressive decrease of eGFP+ cells was observed after cotransfection with increasing ratios of STAT5Δ* over STAT5FL* (Figure 5C).
Altogether, these results support the hypothesis that STAT5Δ is a negative regulator with transdominant potential over STAT5FL-dependent HIV transcription.
STAT5Δ silencing enhances HIV-1 RNA and p24 Gag Ag expression in CD8-depleted PBMC of HIV-positive individuals
We next investigated whether a negative effect of activated STAT5Δ on HIV-1 expression could also be observed in cells isolated from HIV-positive individuals. For this purpose, PBMC isolated from 4 HIV-positive individuals were available for these experiments. To favor HIV expression from CD4+ cells, CD8+ cells (including T lymphocytes and a substantial fraction of natural killer cells, both known to secrete several soluble inhibitors of HIV infection and replication40–43 ) were removed by immunomagnetic beads. CD8-depleted PBMC were then transfected with either a scramble control or a specific STAT5 siRNA. STAT5 siRNA led to a significant inhibition of STAT5Δ expression relative to cells transfected with a control siRNA 48 hours after transfection. In contrast, we did not detect either interference with the synthesis of the housekeeping cellular protein such as α-Actin (Figure 6A) or evidence of cytotoxic/cytostatic effects in these primary cells (data not shown). Both transfected and control CD8-depleted PBMC were maintained in culture for 4 days in medium containing IL-2 before cytofluorimetric analysis of HIV-1 gag-pol RNA expression. STAT5 siRNA led to enhanced expression of the HIV-1 RNA in cultures of 2 (patients no. 9 and 10) of 4 individuals tested, although with variable intensity (from 4.83 to 24.7 and from 3.18 to 7.09, respectively; Figure 6B).
STAT5 silencing enhances HIV RNA expression in IL-2 stimulated CD8-depleted PBMC of HIV-positive individuals. (A) Western blot analysis of WCE of PBMC from individual number 8 (patient no. 8) after electroporation with either control or STAT5 siRNA. Membranes were sequentially hybridized with anti-STAT5 mAb (upper panel) and a control anti-α-actin Ab (lower panel). (B). CD8-depleted PBMC from patient nos. 9 and 10 were electroporated with either control or STAT5 siRNA; 24 hours later, the cells were resuspended in a medium enriched with IL-2 and intracellular HIV RNA expression was analyzed by flow cytometry after 4 days of cultivation; gray thick histograms: scramble control siRNA; black thick histograms: STAT5 siRNA.
STAT5 silencing enhances HIV RNA expression in IL-2 stimulated CD8-depleted PBMC of HIV-positive individuals. (A) Western blot analysis of WCE of PBMC from individual number 8 (patient no. 8) after electroporation with either control or STAT5 siRNA. Membranes were sequentially hybridized with anti-STAT5 mAb (upper panel) and a control anti-α-actin Ab (lower panel). (B). CD8-depleted PBMC from patient nos. 9 and 10 were electroporated with either control or STAT5 siRNA; 24 hours later, the cells were resuspended in a medium enriched with IL-2 and intracellular HIV RNA expression was analyzed by flow cytometry after 4 days of cultivation; gray thick histograms: scramble control siRNA; black thick histograms: STAT5 siRNA.
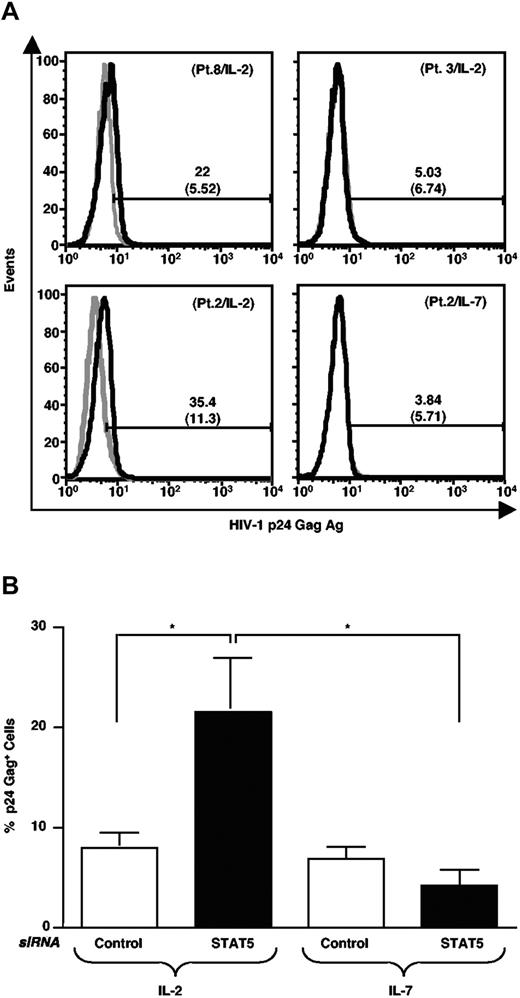
CD8-depleted PBMC of 6 additional HIV-positive individuals were maintained in culture for 5 days in medium containing IL-2 and then analyzed for intracellular (IC) HIV-1 p24 Gag antigen (Ag) expression by cytofluorimetry. STAT5 siRNA led to enhanced expression of p24 Gag Ag in CD8-depleted PBMC of 4 of 6 individuals (Figure 7A, top panels, and data not shown). Enhancement of HIV-1 p24 Gag Ag expression was observed in cells of both individuals expressing exclusively STAT5Δ (patients no. 6 and 8) and of 2 of 4 individuals with a mixture of STAT5Δ and STAT5FL (patients no. 2 and 4); no up-regulation of viral Ag expression was observed in PBMC of patients no. 1 and 3 carrying a mixture of STAT5 isoforms.
STAT5 silencing enhances HIV-1 p24 Gag Ag expression in IL-2 stimulated, but not in IL-7-stimulated, CD8-depleted PBMC of HIV-positive individuals. (A) CD8-depleted PBMC from patient nos. 8 and 3 were electroporated with either control or STAT5 siRNA; 24 hours later, the cells were resuspended in a medium enriched with IL-2 and p24 Gag Ag expression was analyzed by intracellular flow cytometry after 5 days of cultivation; gray histograms: scramble control siRNA; black histograms: STAT5 siRNA (upper panels). Cells from patient no. 8, but not those from patient no. 3, showed an up-regulation of p24 Gag Ag expression after STAT5 siRNA. CD8-depleted PBMC of patient no. 2 were stimulated with IL-2 or IL-7 after transfection with either STAT5 or control siRNA. Induction of p24 Gag Ag expression was observed in cells stimulated with IL-2, but not with IL-7 after 5 days of cultivation (lower panels). (B) Selective up-regulation of p24 Gag Ag expression by STAT5 siRNA in IL-2, but not IL-7, stimulated CD8-depleted PBMC cultures established from patient nos. 1, 2, 3, 4, 6, and 8 for IL-2 and from patient nos. 1, 2, 3, 4, and 6 for IL-7. *P<.05 (t test for paired values).
STAT5 silencing enhances HIV-1 p24 Gag Ag expression in IL-2 stimulated, but not in IL-7-stimulated, CD8-depleted PBMC of HIV-positive individuals. (A) CD8-depleted PBMC from patient nos. 8 and 3 were electroporated with either control or STAT5 siRNA; 24 hours later, the cells were resuspended in a medium enriched with IL-2 and p24 Gag Ag expression was analyzed by intracellular flow cytometry after 5 days of cultivation; gray histograms: scramble control siRNA; black histograms: STAT5 siRNA (upper panels). Cells from patient no. 8, but not those from patient no. 3, showed an up-regulation of p24 Gag Ag expression after STAT5 siRNA. CD8-depleted PBMC of patient no. 2 were stimulated with IL-2 or IL-7 after transfection with either STAT5 or control siRNA. Induction of p24 Gag Ag expression was observed in cells stimulated with IL-2, but not with IL-7 after 5 days of cultivation (lower panels). (B) Selective up-regulation of p24 Gag Ag expression by STAT5 siRNA in IL-2, but not IL-7, stimulated CD8-depleted PBMC cultures established from patient nos. 1, 2, 3, 4, 6, and 8 for IL-2 and from patient nos. 1, 2, 3, 4, and 6 for IL-7. *P<.05 (t test for paired values).
IL-2, but not IL-7, enhances HIV-1 p24 Gag expression in STAT5Δ silenced CD8-depleted PBMC of HIV-positive individuals
Because IL-7, a γc cytokine like IL-2, has been reported to increase HIV-1 replication in CD4+ T lymphocytes infected either in vitro or in vivo,21,22 we next investigated the potential effect of STAT5 silencing in CD8-depleted PBMC of 5 HIV-positive individuals transfected with either control or STAT5 siRNA. The cells were maintained in culture for 5 days in medium containing either IL-2 or IL-7 at concentrations previously established as optimal for these culture conditions.3,21,22 Cultures from patient no. 8 and CD8-depleted PBMC cultures of patient no. 2 transfected with a STAT5 siRNA showed an increased expression of p24 Gag Ag in the presence of IL-2. In contrast, parallel autologous cultures transfected with a STAT5 siRNA and stimulated with IL-7 did not show an up-regulation of HIV-1 p24 Gag Ag expression versus cultures transfected with a control siRNA (Figure 7A, bottom panels). The selective effect of IL-2 vs. IL-7 in the primary cell cultures established from PBMC of several HIV-positive individuals were statistically significant (Figure 7B) suggesting that IL-7 may not up-regulate HIV expression through a STAT5-dependent pathway, at least in these experimental conditions.
Discussion
In the present study, we have demonstrated that the C-terminally truncated form of STAT5 (STAT5Δ) constitutively activated in PBMC of most HIV-positive individuals, and phosphorylated on GM-CSF stimulation in chronically infected U1 cells, acts as a suppressor of HIV expression. In addition to STAT5Δ, GM-CSF stimulation of U1 cells activated an ERK/AP-1-dependent transcriptional pathway responsible for the weak induction of HIV expression induced by the cytokine. Indeed, a pharmacologic inhibitor of the ERK/AP1 pathway (PD98,059) shut off GM-CSF-mediated virus expression, whereas, in sharp contrast, prevention of STAT5Δ activation by either AG490 or STAT5 siRNA enhanced viral replication in GM-CSF-stimulated U1 cells. ChIP experiments demonstrated that STAT5Δ is actually present in living U1 cells where, after stimulation by GM-CSF, it binds to two previously identified39 putative STAT5 binding sites in the U3 region of the HIV-1 LTR. STAT5Δ binding, however, leads to repression rather than activation of HIV transcription as demonstrated by direct measure of the levels of RNA polymerase II recruitment to the viral promoter and by competition assays between STAT5Δ and STAT5FL for LTR-driven activation in 293T cells. STAT5 siRNA up-regulated the ex vivo expression of both HIV RNA and p24 Gag Ag in CD8-depleted PBMC of several infected individuals maintained in culture in the presence of IL-2, but not of IL-7. Thus, activated STAT5Δ acts as a suppressor of HIV expression in chronically infected U1 cells stimulated with GM-CSF, in 293T cells transfected with a constitutively phosphorylated STAT5 and, finally, in CD4+ cells isolated from HIV-positive individuals stimulated with IL-2.
Apart from HIV infection, naturally occurring C-terminally truncated forms of STAT5 have been described in both animal and human cells,44,45 usually associated with the development and differentiation of different tumors.8,46–48 STAT5 truncation occurs after translation of the protein independently of its phosphorylated state.9 The proteolytic activity associated with STAT5 cleavage has been characterized as a 25 kDa nuclear protein in murine cells49,50 and by a serine-dependent enzyme present both in the nucleus and in the cytoplasm of human cells.51 STAT5 processing has been linked to both myelomonocytic differentiation52,53 and cytokine-induced polymorphonuclear cell activation.54 Of interest is the fact that STAT5Δ isoforms show increased DNA binding capacity together with resistance to proteasome-mediated degradation,55 thereby exerting dominant negative effects on STAT5-dependent gene transcription.9 The observation that U1 cells, but not ACH-2 cells (that were infected with the same subtype B strain, ie, HIV-1LAI/IIIB, used to infect U937 cells from which the U1 cell line was derived31,56 ), express exclusively a STAT5Δ isoform likely indicates that viral infection alone is not responsible for STAT5 truncation. In support of this interpretation, no correlation was observed between expression of FL and truncated STAT5 isoforms and the levels of HIV replication in vivo (Table 1). Furthermore, uninfected U937 cells, the parental cell line of U1, expressed different levels of both STAT5FL and STAT5Δ (data not shown), as previously reported,57 suggesting that these isoforms existed in this cell line before its infection with HIV.
HIV replication is under the control of both viral and host factors.1,2 Although no “latency genes” are encoded by the virus, properties inherent to retroviruses, such as integration into the host chromosomes as a provirus, allows an epigenetic regulation of viral transcription, translation, virion assembly, and release.2 In vivo, both active replication and latent infection have been unequivocally demonstrated in CD4+ T lymphocytes, circulating monocytes, tissue macrophages, dendritic cells, and astrocytes of the central nervous system.1,2,19 Conversion from a latent state to active viral expression can be triggered by either heterologous infections or host-related factors. In the case of T cells, activation signals usually promote HIV replication as well as factors inducing monocyte differentiation into macrophages.2,19,58,59 In this regard, several γc-cytokines such as IL-2, IL-7, and IL-15 (which trigger JAK1/JAK3 and STAT5A/STAT5B) induced HIV replication in primary CD4+ T cells or unfractionated PBMC.60,61 GM-CSF, which belongs to the βc-cytokines and promotes STAT5 phosporylation through activation of JAK2,62 has been reported to either induce30,63–65 or suppress66–68 HIV replication in primary human macrophages. The reasons for these discrepant results are unclear but they may relate to the stage of monocytic differentiation at the time of in vitro infection. Whether a differential expression of STAT5 isoforms,69 including STAT5Δ, may account for these different experimental outcomes has not been explored.
The molecular link between STAT5-activating factors and the induction of HIV replication has been recently elucidated by an independent study indicating that STAT5FL binds directly to one or more putative DNA binding sites present in the U3 region of the HIV LTR.20 We have confirmed these findings by both a bioinformatics analysis and, experimentally, by transient transfection experiments of HIV-LTR-reporter genes in the 293T cell line. Therefore, activation of STAT5FL by any γc- or βc-cytokines could potentially lead to triggering or enhancing viral transcription.20 However, the observation that most HIV-positive individuals harbor constitutively activated STAT5Δ as the predominantly expressed isoform of STAT5 challenges the concept that STAT5-inducing cytokines may ultimately increase viral replication in vivo. Our current evidence obtained in living U1 cells, that STAT5Δ promotes a decrease in RNA polymerase II complex formation early (1 hour) after GM-CSF stimulation, before the activation of the ERK-AP-1 HIV activatory pathway, is direct evidence supporting this hypothesis. Furthermore, competition experiments suggest that STAT5Δ may be considered a transdominant isoform of STAT5.
Our observations may help explain the apparent paradox that “constitutively activated” PBMC of infected individuals are extremely inefficient in sustaining productive HIV replication unless potently stimulated with mitogens70 or cytokine combinations.60,61 In this regard, a recent ex vivo stimulation study of resting memory peripheral CD4+ T cells with IL-2 alone did not note induction of HIV replication, but did observe HIV replication induction with IL-7 or with PHA plus IL-2 stimulation.21 These independent observations are consistent with a negative role of STAT5Δ on viral expression in IL-2-stimulated cells of infected individuals. The potent effect of IL-7 on HIV replication in latently infected CD4+ T cells, as well as its in vitro effect of quiescent T cells,22 was likely mediated through a STAT5-independent pathway. In support of this hypothesis, no up-regulation of HIV p24 Gag expression was observed when CD8+ cell-depleted PBMC cultures were transfected with a STAT5 siRNA and stimulated with IL-7, unlike what is observed frequently in the presence of IL-2.
The role of either γc- or βc-cytokines on HIV replication might therefore be different in the case of in vitro infection of PBMC or PBMC subsets obtained from healthy uninfected individuals, not expressing STAT5Δ, and when PBMC are isolated from infected individuals, as in our study. IL-2, a γc-cytokine activating STAT5,4 is currently in a phase III efficacy trial as a potentially effective therapy in HIV disease based on its capacity to expand CD4+ T lymphocytes without increasing viral load.71–74 When we compared the impact of intermittent IL-2 administration on STAT5 activation in individuals who responded either well or poorly in terms of CD4+ T cell count increase, we observed that STAT5Δ activation was more prominent in the good versus the poor responder.6 Like IL-2, GM-CSF is also currently being studied for its hematopoietic reconstitution capacity and as a potential adjuvant to antiretroviral therapy.75,76 GM-CSF has been shown to enhance peripheral CD4+ T cell counts and reduce plasma viremia,77 although this latter claim has been challenged by a recent study.78 Our current observation that STAT5Δ likely acts as an inhibitor rather than an activator of HIV expression sheds new light on the rationale for the clinical testing of STAT5-inductive cytokines in HIV-positive individuals.6
Presented in abstract form at the 2006 International Meeting of the Institute of Human Virology, Baltimore, MD, November 17-21, 2006.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants of the VI° National Program against AIDS of the Istituto Superiore di Sanità, Rome, Italy (C.B., G.P.).
We thank Elisa Vicenzi, Claudia Cicala, and Jim Arthos for critical reading of the manuscript and Silvia Ghezzi for her help in collecting the clinical data of HIV-positive individuals, who we also thank for their generous participation in this study.
A.C. is a PhD candidate of the Vita-Salute University of Milano and of the Open University of London, UK.
Authorship
Contribution: A.C. conceived and performed most of the experiments leading to the results shown; he also significantly contributed to the writing of the manuscript; M.L. conceived and executed the chromatin immunoprecipitation experiments and helped with the interpretation of the results and their overall meaning in the context of the manuscript; R.L. performed a number of crucial experiments either alone or with A.C. P.M.J.L. and E.L. expressed and preliminarily tested the constitutively phosphorylated STAT5 vectors; G.D.C. performed several new experiments leading to the additional results present in the revised manuscript; M.T. provided the blood samples from and the clinical information on additional HIV-positive individuals; he also participated in the discussion of the experimental results; A.L. provided the blood samples from and the clinical information on HIV-positive individuals; he also participated in the discussion of the experimental results. B.K.P. helped perform the experiments dealing with HIV RNA expression from both the cell line and primary lymphocytes, and contributed to the interpretation of the results and of the overall manuscript. M.G. conceived and supervised the chromatin immunoprecipitation experiments and helped with the interpretation of the results and their overall meaning in the context of the manuscript; C.B. helped to conceive the experimental designs and supervise all the crude results; she also significantly contributed to the writing of the manuscript; G.P. helped to conceive the experimental designs and supervised all the crude results and significantly contributed to the writing of the manuscript.
Conflict-of-interest disclosure: Rossella Lupo and Chiara Bovolenta are employees of Molmed S.p.A. MolMed has no commercial products that are discussed in the present article.
Correspondence: Prof. Guido Poli, P2/P3 Laboratories, DIBIT, Via Olgettina n. 58, 21,032, Milano, Italy; e-mail: poli.guido@hsr.it; Dr. Chiara Bovolenta, Molmed, via Olgettina n. 58, 20,132, Milano, Italy; e-mail: chiara.bovolenta@molmed.com.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal