Abstract
Arsenic trioxide (As2O3) is highly efficacious in acute promyelocytic leukemia (APL). Aquaglyceroporin 9 (AQP9) is a transmembrane protein that may be involved in arsenic uptake. In 10 of 11 myeloid and lymphoid leukemia lines, quantitative polymerase chain reaction (Q-PCR) and Western blotting showed that AQP9 expression correlated positively with As2O3-induced cytotoxicity. As a proof-of-principle, transfection of EGFP-tagged AQP9 to the hepatoma line Hep3B, not expressing AQP9 and As2O3 insensitive, led to membrane AQP9 expression and increased As2O3-induced cytotoxicity. Similarly, the chronic myeloid leukemia line K562 expressed low levels of AQP9 and was As2O3 insensitive. The K562EGFP-AQP9 transfectant accumulated significantly higher levels of intracellular arsenic than control K562EGFP when incubated with As2O3, resulting in significantly increased As2O3-induced cytotoxicity. Pretreatment of the myeloid leukemia line HL-60 with all-trans retinoic acid (ATRA) up-regulated AQP9, leading to a significantly increased arsenic uptake and As2O3-induced cytotoxicity on incubation with As2O3, which might explain the synergism between ATRA and As2O3. Therefore, AQP9 controlled arsenic transport and might determine As2O3 sensitivity. Q-PCR showed that primary APL cells expressed AQP9 significantly (2-3 logs) higher than other acute myeloid leukemias (AMLs), which might explain their exquisite As2O3 sensitivity. However, APL and AML with maturation expressed comparable AQP9 levels, suggesting that AQP9 expression was related to granulocytic maturation.
Introduction
Arsenic trioxide (As2O3) is a standard treatment for acute promyelocytic leukemia (APL)1-3 and is showing promise in multiple myeloma and other hematologic malignancies.4,5 The intracellular actions of As2O3 include interaction with proteins possessing high cysteine and histidine contents,6 generation of superoxides and reactive oxygen species,7,8 disruption of the mitochondrial transmembrane potential with release of cytochrome c and caspases activation, and blockade of cell cycle at the G1/S and G2/M phases.9
As2O3 is detoxified by the glutathione redox system, comprising thiol-rich glutathione (GSH) carrier proteins, glutathione peroxidase, and glutathione S-transferase (GST-π).9-12 GSH forms a transient As(GSH)3 complex with As3+, a process catalyzed by GST-π.11 As(GSH)3 binds the multi-drug–resistant protein 1 (MRP1), inducing a conformational change.13,14 This in turn leads to an ATP-driven transmembrane transport of As(GSH)3 to the extracellular space.13,14 Free reduced GSH carrier protein is then replenished by glutathione peroxidase.
The arsenic GSH redox system might be one of the mechanisms that account for the varying sensitivities of different cell types to As2O3. The sensitivities to As2O3 had been shown to be inversely proportional to the cellular GSH content.15 Furthermore, cell lines treated with ascorbic acid and buthionine sulfoxide (BSO), which decreased intracellular GSH levels, had increased sensitivity to As2O3.15 The role of MRP1 in arsenic sensitivity is more controversial. Overexpression of MRP1 had been shown to confer resistance to As2O3 in lung cancer cells and the HeLa cell line.16 However, an HL-60 variant, HL-60/AR, that overexpressed MRP1 did not show increased arsenic sensitivity.17
The transmembrane protein aquaglyceroporin 9 (AQP9) was first identified in adipose tissue and leukocytes.18,19 It is also expressed in the liver, lung, and spleen.20 AQP9 facilitates the passage of water and glycerol, as well as a large variety of small noncharged solutes (caramides, polyols, purine, pyrimidine, and protonated monocarboxylates).20 Recently, AQP9 was shown to restore arsenic sensitivity to a Saccharomyces cerevisiae mutant that lacked FPS1, the homologue of AQP9.21 Furthermore, Xenopus laevis oocytes microinjected with AQP9 cDNA exhibited increased uptake of As3+ ions.21 Although these results showed that AQP9 facilitated arsenic transport in yeast and Xenopus, further investigations are required to determine the function of AQP9 in arsenic transport in mammalian cells.
In this study, we tested the hypothesis that AQP9 mediated arsenic uptake in leukemic cells and might be a determining factor in arsenic sensitivity.
Patients, materials, and methods
Cell lines
One APL (NB4), 2 myeloid leukemia (HL-60, ML-1), 1 chronic myeloid leukemia (K562), 1 B-cell acute lymphoblastic leukemia (ALL; NALM-20), 1 non-B non-T ALL (NALM-16), and 5 T-cell leukemia (HPB-ALL, JM, JURKAT, KE-37, MOLT-4) cell lines were studied. They were cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS) in 5% CO2 at 37°C. Cells were treated with As2O3 (Sigma-Aldrich, St Louis, MO) and with all-trans retinoic acid (ATRA; Sigma-Aldrich) in selected experiments.
Arsenic cytotoxicity
Cells were washed twice with phosphate-buffered saline (PBS) and resuspended in fresh RPMI-1640 medium supplemented with 10% FBS and various concentration of As2O3 (0, 0.5, 1.0, 2.0, and 4 μM) at 2.5 × 105 cells/mL. Quadruplicates of 100 μL of each arsenic/cell suspension were incubated for 48 hours. MTT reagent (100 μL) was then added and incubated for 4 hours, followed by the addition of 100 μL of solubilizing buffer and incubation overnight. The absorbance at 560 nm was then measured.
Western blotting
Leukemic cells were washed once in PBS supplemented with Complete protease inhibitor (Roche, Manheim, Germany). Washed pellets (4 × 107 to 5 × 107 cells) were resuspended in 5 mL lysis buffer (7.5 mM sodium phosphate, pH 7.0; 0.25 M sucrose; 5 mM EDTA with Complete protease inhibitor; and 1 mM phenylmethylsulphonylfluoride), sonicated on ice for 60 J, and centrifuged at 1000g for 10 minutes at 4°C. The crude membrane protein fraction was then pelleted by ultracentrifugation (200 000g for 90 minutes at 4°C), resuspended in 150 μL buffer (5.0% SDS; 20 mM Tris, pH 8.0; and 5 mM EDTA with Complete protease inhibitor and 1 mM PMSF), sonicated, and incubated at 37°C for 30 minutes. Protein concentrations of each sample were determined (DC protein assay kit II; Bio-Rad, Hercules, CA) and adjusted to 2 μg/μL in lysis buffer and 5 × sample buffer (15% SDS; 50 mM Tris, pH 6.8; 30% glycerol; 0.05% bromophenol blue; and 0.5 M dithiothreitol). Protein samples were boiled at 95°C for 5 minutes and kept at 37°C, and 30 μg was resolved in 12% acrylamide gel with 4 M urea, electrophoretically transferred to PVDF membrane (Amersham Biosciences, Piscataway, NJ), blocked with 5% low-fat milk for 1 hour, incubated with rabbit anti-AQP9 antibody (3.5 μg/mL) or rabbit anti–enhanced green fluorescent protein (EGFP) antibody (1:10 000; Chemicon, Temecula, CA), and probed with an antirabbit secondary antibody (1:1000; Dako Cytomation, Glostrup, Denmark). Immunoblotting was evaluated after chemiluminescence development (Western Lightning Chemiluminescence Reagent; PE Biosystems, Foster City, CA), followed by PointSau staining (Sigma-Aldrich) for normalization of protein loading.
Quantification of AQP9 gene expression
AQP9 expression was quantified by quantitative polymerase chain reaction (Q-PCR). Briefly, the AQP9 primers (forward, 5′ AGT TGT TGG GAG CCT TTG TG 3′; reverse, 5′ GTT CGC CAG AGA TAG ATA CGG AG 3′) and the TaqMan probe (5′ CAA CAG CAC ACA TTT TTG CAA CAT ACC 3′) detecting a 155-bp amplicon were designed by Primer Express software (PE Biosystems). The primers (forward, 5′ GAA GGT GAA GGT CGG AGT CA 3′; reverse, 5′ CTT CTA CCA CTA CCC TAA AG 3′) and TaqMan probe (5′ GTT CGA AGG GCA AGA GTC GG 3′) detecting a 155-bp amplicon for the internal control gene GAPDH were similarly designed. For Q-PCR, 100 ng of RNA was added to 45 μL of Taqman EZ reverse transcription–PCR (RT-PCR) reaction mixture (1 × Taqman EZ buffer, 3 mM MnOAc, 300 μM of dNTP mixture, 200 nM of primers and probe, 0.1U rTth DNA polymerase, 0.01U uracil N-glycosylase). PCR conditions were 50°C for 2 minutes, 60°C for 30 minutes, 95°C for 5 minutes, and 40 cycles of 94°C for 20 seconds and 60°C (AQP9)/62°C (GAPDH) for 1 minute. Q-PCR data were collected continuously and analyzed by an automated sequence analyzer (ABI PRISM 7700; PE Biosystems). Expression of the multi-drug–resistance 1 (MDR1) gene was quantified as previously reported.22 The expression levels of AQP9 among the 11 cell lines were analyzed using the comparative CT (ΔCT) method (User Bulletin no. 2; PE Biosystems),23 with GAPDH as the internal reference gene to control for RNA quantity and integrity and NB4 cells as a calibrator to control for interexperimental variations. All samples were quantified in triplicates. Quality assurance of Q-PCR was as previously described.23
Plasmids pEGFP-AQP9 and pSV-βgal
To construct an EGFP-tagged AQP9 expression plasmid, the complete coding sequence of AQP9 (888 bp) was cloned by PCR using NB4 cell line cDNA as a template with primers (forward, 5′ CCGCCG AGA TCT CCA TGC AGC CTG AGG GAG CA 3′; reverse, 5′ CAGCAG GGA TCC CTA CAT GAT GAC A 3′) containing BglII and BamHI restriction sites (underlined), respectively. The PCR product was cloned into the pEGFP-C2 vector (BD Biosciences Clontech, Mountain View, CA) to give the plasmid pEGFP-AQP9. The nucleotide sequence of AQP9 in pEGFP-AQP9 was analyzed by DNA sequencing to confirm correct nucleotide sequence and in-frame insertion of AQP9 with EGFP. The plasmid pSV-βgal, containing the β-galactosidase gene, was purchased from Promega (Madison, WI).
Transfection of Hep3B with pEGFP-AQP9 for assaying of As2O3 sensitivity
Hep3B cells in MEM cultured to 80% confluence were transfected with pEGFP-C2/pSV-βgal or pEGFP-AQP9/pSV-βgal in Lipofectamine 2000 (Invitrogen, Carlsbad, CA). In order to ensure that cells transfected with pSV-βgal would also express AQP9, a 1:5 ratio of pSV-βgal to pEGFP-AQP9 was used. Hence, β-galactosidase activity could be used as an indicator of viability of cells expressing both β-gal and EGFP-C2 or EGFP-AQP9. The transfectants, designated as Hep3BEGFP-AQP9/β-gal and HEP3BEGFP-C2/β-gal, were further incubated for 48 hours. The cells were then treated with As2O3 at various concentrations (0, 0.5, 1.0, and 2.0 μM) for 24 hours. For assaying of β-galactosidase activity, transfected Hep3B cells were washed with PBS, lysed with reporter lysis buffer (Promega) for 15 minutes at room temperature, transferred to Eppendorf tubes on ice, vortexed for 20 seconds, and centrifuged at 1000g at 4°C for 1 minute. Cell lysates were mixed with an equal volume of 2 × assay buffer and incubated at 37°C for 1 to 2 hours, after which the reaction was stopped by 1 M sodium bicarbonate. Absorbance at 420 nm was then recorded. Untransfected cells not treated with As2O3 served as 100% viability control. All samples were tested in triplicates.
Transfection of K562 cells with pEGFP-AQP9 and fluorescence-activated cell sorting
K562 cells at mid-log phase were transfected with pEGFP-AQP9 or pEGFP-C2 as described for Hep3B cells. The K562 transfectants, designated as K562EGFP-AQP9 and K562EGFP-C2, were washed once with PBS and then resuspended in RPMI-1640 medium supplemented with 500 μg/mL of G418 (Calbiochem, San Diego, CA) at 1 × 106 cells/mL. Cells resistant to G418 were selected for 3 weeks. To further enrich for transfected cells, sorting for green fluorescent cells (K562EGFP-AQP9 and K562EGFP-C2) was performed by an EPICS ALTRA flow cytometer (Beckman Coulter, Fullerton, CA) until a purity of 80% or higher was achieved.
Arsenic-uptake analysis
K562EGFP-AQP9 and K562EGFP cells at 1 × 106 cells/mL were washed twice with PBS; resuspended in fresh RPMI-1640 medium supplemented with 10% FBS and 1 μM As2O3; and incubated in 5% CO2 at 37°C for 0, 30, 60, 90, and 120 minutes. Cells were then harvested into 1 mL ice-cold PBS and pelleted. After 2 washes in ice-cold PBS, the cell pellets were lysed in 0.9 mL double-distilled water by sonication for 10 minutes. HNO3 (2%) with 10 parts per billion (ppb) of yttrium (Chem Service, West Chester, PA) was then added as an internal standard. Samples were vortexed and centrifuged at 3000g for 10 minutes, and the supernatants were collected and assayed for arsenic concentration by inductively coupled plasma–mass spectrometry (ICP-MS) as previously described.24
AQP9 expression in leukemia samples
Archival bone marrow aspirate samples from patients with APL and other subtypes of acute myeloid leukemia (AML) were quantified for AQP9 expression. Leukemic diagnoses were based on the World Health Organization classification system.25 The blast cells were isolated by centrifugation, snap-frozen, and stored at −80°C. For quantification of AQP9 expression, leukemic samples were thawed, total RNA was extracted, and Q-PCR for AQP9 was performed, with the cell line NB4 used as the calibrator in the ΔCT method, using GAPDH as the internal reference. The procurement of materials was approved by the institutional review board at Queen Mary Hospital (Hong Kong, China). Patients gave informed consent and the study was conducted in accordance with the Declaration of Helsinki.
Statistical analysis
Continuous variables were expressed as mean ± standard error of the mean. The correlation between cell viability and AQP9 expression was tested by Pearson correlation test (SPSS software, version 10.0; Chicago, IL). Comparison between groups was performed by Student t test. P values of less than .05 were considered to be statistically significant.
Results
As2O3 cytotoxicity to leukemic cell lines
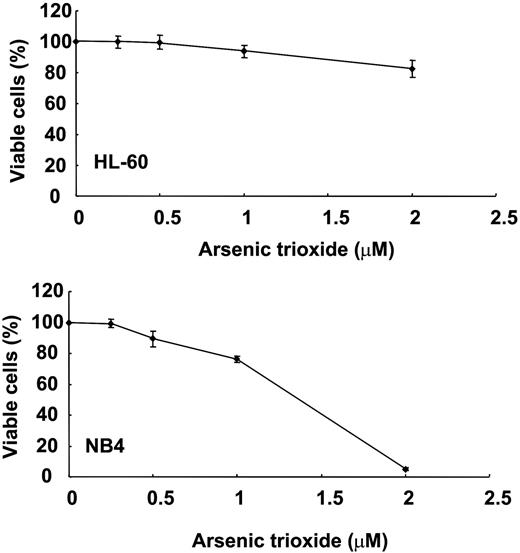
MTT assays were performed on the 11 cell lines tested. The sensitivities of the cell lines differed, although all showed a dose-dependent increase in As2O3 cytotoxicity. Of the 11 cell lines, Jurkat, HL-60, and NALM-20 were most resistant, whereas NB4 was most sensitive (Figure 1; Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
MTT analysis of dose-dependent cytotoxicity of As2O3 in HL-60 and NB4 cells. HL-60 cells were relatively resistant to As2O3, whereas NB4 cells were very sensitive. Error bars indicate the standard error of the mean.
MTT analysis of dose-dependent cytotoxicity of As2O3 in HL-60 and NB4 cells. HL-60 cells were relatively resistant to As2O3, whereas NB4 cells were very sensitive. Error bars indicate the standard error of the mean.
AQP9 expression and sensitivity to As2O3
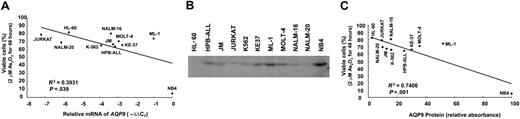
In the 11 leukemia cell lines tested, the expression of AQP9 as determined by Q-PCR was found to be positively correlated with As2O3-induced cytotoxicity as assessed by MTT (P = .039; Figure 2A). The results were confirmed by Western-blot analysis (Figure 2B), showing a direct relationship between AQP9 protein expression and As2O3 sensitivity (P = .001; Figure 2C). However, the cell line ML-1 was an obvious outlier in both experiments. Finally, there was no relationship between AQP9 and MDR1 gene expression (P = .09, Pearson correlation; Table S1).
Correlation of AQP9 expression with As2O3 sensitivity. (A) The quantity of AQP9 mRNA, as determined by Q-PCR, was inversely related to cell survival after treatment in 2 μM As2O3 for 48 hours. ML-1 was an outlier. (B) Western-blot analysis of AQP9 expression. NB4 expressed the highest amount of AQP9 and was used as the standard for comparison in panel C. (C) AQP9 protein expression, as determined by Western-blot analysis using NB4 as the reference, showed an inverse relationship to cell survival. ML-1 was also an outlier.
Correlation of AQP9 expression with As2O3 sensitivity. (A) The quantity of AQP9 mRNA, as determined by Q-PCR, was inversely related to cell survival after treatment in 2 μM As2O3 for 48 hours. ML-1 was an outlier. (B) Western-blot analysis of AQP9 expression. NB4 expressed the highest amount of AQP9 and was used as the standard for comparison in panel C. (C) AQP9 protein expression, as determined by Western-blot analysis using NB4 as the reference, showed an inverse relationship to cell survival. ML-1 was also an outlier.
AQP9 point mutation in ML-1
As ML-1 showed an apparent high level of AQP9 expression but a low As2O3 sensitivity, the entire coding region of AQP9 in ML-1 was amplified by PCR and sequenced. The results showed a homozygous single base substitution, 835G>A, which resulted in an amino-acid change from alanine to serine at codon 279. Whether this might account for the discordant AQP9 expression and As2O3 sensitivity is under further investigations.
AQP9 transfectants of Hep3B and K562
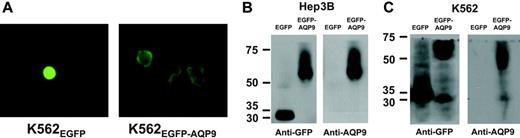
Fluorescence microscopic examination of the Hep3B and K562 AQP9 transfectants showed a difference in distribution of fluorescence. In Hep3BEGFP-C2 and K562EGFP-C2, the green fluorescence was evenly distributed in the cytosol. However, in Hep3BEGFP-AQP9 and K562EGFP-AQP9, the fluorescence was mainly distributed on the cytoplasmic membrane (Figure 3A). The results confirmed selective localization of AQP9 to the plasma membrane. Western blotting was performed to confirm protein overexpression in the transfectants. In K562EGFP-C2 and Hep3BEGFP-C2/β-gal, an EGFP protein band of around 30 kDa was detected with an anti-GFP antibody (Figure 3B-C). In Hep3BEGFP-AQP9/β-gal and K562EGFP-AQP9 lysates, the EGFP-AQP9 fusion protein was detected as a 61-kDa band with antibodies specific to GFP or AQP9, representing a summation of the 30-kDa EGFP and the 31-kDa AQP9 protein (Figure 3B-C). An additional band of approximately 30 kDa was also detected in both the anti-GFP and anti-AQP9 blotting of the K562EGFP-AQP9 lysate, which might be due to the degradation of the EGFP-AQP9 fusion protein to EGFP (30 kDa) and AQP9 peptides (31 kDa).
Transfection of AQP9 into Hep3B and K562 cells. (A) K562EGFP-C2 cells examined under fluorescent microscopy showed even cellular distribution of green fluorescence; however, K562EGFR-AQP9 cells showed selective localization of green fluorescence to the plasma membrane. Images were visualized using an Olympus IX70 microscope equipped with a C plan semi-apochromat 60×/0.7 numerical aperture lens (Olympus, Tokyo, Japan). A Nikon Coolpix 4500 camera (Nikon, Tokyo, Japan) was used to capture the images. (B) Western-blot analysis of Hep3B cells. In anti-GFP immunoblotting, EGFP (30 kDa) and EGFP-AQP9 (61 kDa) were detected in Hep3BEGFP and Hep3BEGFP-AQP9 cells, respectively. With anti-AQP9 immunoblotting, only the protein band of EGFP-AQP9 (61 kDa) was detected in Hep3BEGFP-AQP9 cells, showing that Hep3BEGFP cells did not express detectable levels of AQP9. (C) Western-blot analysis of K562 cells. In anti-GFP immunoblotting, protein bands of EGFP (30 kDa) and EGFP-AQP9 (61 kDa) were detected in K562EGFP and K562EGFP-AQP9 cells. An additional band (30-31 kDa) was observed in the K562EGFP-AQP9 sample. This might be due to degradation of EGFP-AQP9 fusion protein into EGFP and AQP9 fractions. In anti-AQP9 immunoblotting, a predominant band of EGFP-AQP9 (61 kDa) was detected in K562EGFP-AQP9 cells. At the anti-AQP9 antibody concentration and exposure time used to avoid overexposure of the K562EGFP-AQP9 lysate, K562EGFR lysate did not show detectable AQP9, as was shown previously in Figure 2.
Transfection of AQP9 into Hep3B and K562 cells. (A) K562EGFP-C2 cells examined under fluorescent microscopy showed even cellular distribution of green fluorescence; however, K562EGFR-AQP9 cells showed selective localization of green fluorescence to the plasma membrane. Images were visualized using an Olympus IX70 microscope equipped with a C plan semi-apochromat 60×/0.7 numerical aperture lens (Olympus, Tokyo, Japan). A Nikon Coolpix 4500 camera (Nikon, Tokyo, Japan) was used to capture the images. (B) Western-blot analysis of Hep3B cells. In anti-GFP immunoblotting, EGFP (30 kDa) and EGFP-AQP9 (61 kDa) were detected in Hep3BEGFP and Hep3BEGFP-AQP9 cells, respectively. With anti-AQP9 immunoblotting, only the protein band of EGFP-AQP9 (61 kDa) was detected in Hep3BEGFP-AQP9 cells, showing that Hep3BEGFP cells did not express detectable levels of AQP9. (C) Western-blot analysis of K562 cells. In anti-GFP immunoblotting, protein bands of EGFP (30 kDa) and EGFP-AQP9 (61 kDa) were detected in K562EGFP and K562EGFP-AQP9 cells. An additional band (30-31 kDa) was observed in the K562EGFP-AQP9 sample. This might be due to degradation of EGFP-AQP9 fusion protein into EGFP and AQP9 fractions. In anti-AQP9 immunoblotting, a predominant band of EGFP-AQP9 (61 kDa) was detected in K562EGFP-AQP9 cells. At the anti-AQP9 antibody concentration and exposure time used to avoid overexposure of the K562EGFP-AQP9 lysate, K562EGFR lysate did not show detectable AQP9, as was shown previously in Figure 2.
As2O3 sensitivity of Hep3B-AQP9 and K562 transfectants
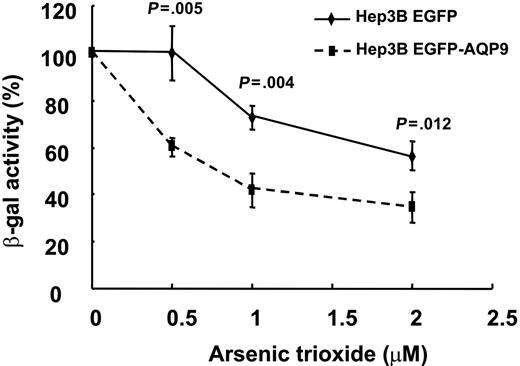
The Hep3B hepatoma line did not express any detectable amount of AQP9 (Figure 3B) and was insensitive to As2O3. In Hep3BEGFP/β-gal and Hep3BEGFP-AQP9/β-gal transfectants, the β-galactosidase activity was used as an indicator of cell viability after treatment with various concentrations of As2O3. As shown in Figure 4, Hep3BEGFP/β-gal cells, similar to the parental Hep3B cells, were resistant to As2O3. On the other hand, As2O3-treated Hep3BEGFP-AQP9/β-gal cells demonstrated a significantly lower β-galactosidase activity compared with Hep3BEGFP/β-gal (Figure 4), showing that overexpression of AQP9 increased As2O3-induced cytotoxicity. Interestingly, the difference of viability of Hep3BEGFP/β-gal and Hep3BEGFP-AQP9/β-gal remained constant with increasing concentrations of As2O3.
β-galactosidase enzyme assay on Hep3BEGFP-AQP9/β-gal cells. Hep3BEGFP-AQP9/β-gal cells exhibited an increase in As2O3 sensitivity compared with Hep3BEGFP/β-gal cells (Student t test for each of the points analyzed). Note that the difference in As2O3 sensitivity remained comparable with increasing concentrations of As2O3.
β-galactosidase enzyme assay on Hep3BEGFP-AQP9/β-gal cells. Hep3BEGFP-AQP9/β-gal cells exhibited an increase in As2O3 sensitivity compared with Hep3BEGFP/β-gal cells (Student t test for each of the points analyzed). Note that the difference in As2O3 sensitivity remained comparable with increasing concentrations of As2O3.
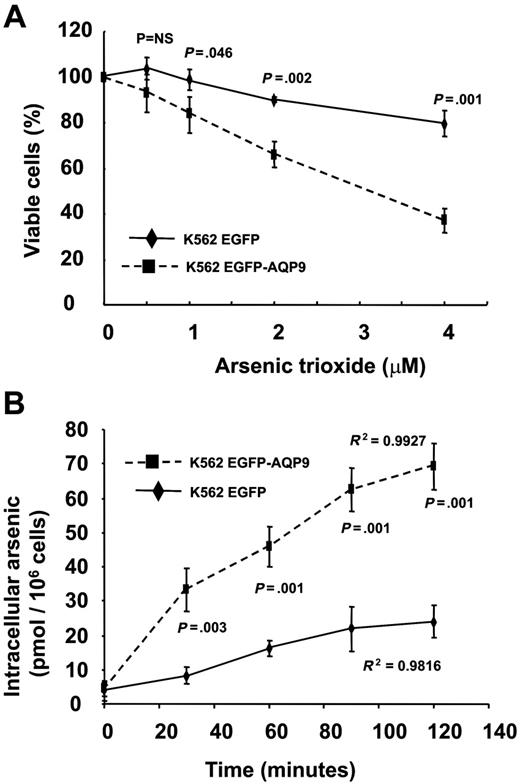
As2O3 sensitivity and arsenic uptake by K562-AQP9 transfectants
The K562 cells expressed very low levels of AQP9 (Figure 3B) and were insensitive to As2O3. The K562EGFP-C2 transfectant was also insensitive to As2O3 (Figure 5A). However, the K562EGFP-AQP9 transfectants showed a significantly increased sensitivity to As2O3 (Figure 5A). Furthermore, in contrast to Hep3B AQP9 transfectants, the difference of viability of K562EGFP-C2 and K562EGFP-AQP9 widened with increasing concentrations of As2O3. The increase in As2O3 sensitivity could be explained by an increased As3+ uptake mediated by the overexpressed AQP9. As shown in Figure 5B, the K562EGFP cells accumulated minimal amounts of arsenic after incubation in 1 μM As2O3 for 2 hours. On the other hand, the K562EGFP-AQP9 showed a time-dependent increase in arsenic accumulation, reaching a significantly higher amount of arsenic (median, 65 pmol/106 cells) after 2 hours.
Transfection of AQP9 in K562 cells. (A) In MTT assay, K562EGFP-AQP9 showed an increase in As2O3 sensitivity compared with K562EGFP-C2 cells (Student t test for each of the points analyzed). Note that the difference of As2O3 sensitivity between K562EGFP-AQP9 and K562EGFP cells widened with an increasing concentration of As2O3. NS indicates not significant. (B) In arsenic-uptake assay, K562EGFP-AQP9 cells accumulated significantly more arsenic than K562EGFP-C2 cells (Student t test for each of the points analyzed). Both lines showed a time-dependent increase in arsenic uptake.
Transfection of AQP9 in K562 cells. (A) In MTT assay, K562EGFP-AQP9 showed an increase in As2O3 sensitivity compared with K562EGFP-C2 cells (Student t test for each of the points analyzed). Note that the difference of As2O3 sensitivity between K562EGFP-AQP9 and K562EGFP cells widened with an increasing concentration of As2O3. NS indicates not significant. (B) In arsenic-uptake assay, K562EGFP-AQP9 cells accumulated significantly more arsenic than K562EGFP-C2 cells (Student t test for each of the points analyzed). Both lines showed a time-dependent increase in arsenic uptake.
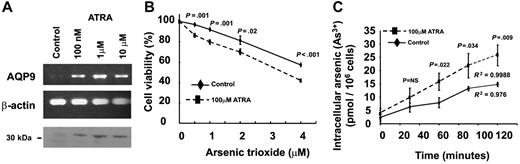
ATRA induced up-regulation of AQP9 and increased arsenic uptake and As2O3 sensitivity
ATRA acts synergistically with As2O3 in vitro and in vivo. The HL-60 cells expressed very low levels of AQP9 (Figure 2A-C) and were insensitive to As2O3. However, treatment with ATRA led to a dose-dependent increase in AQP9 gene transcription and protein expression (Figure 6A). To assess the biologic significance of AQP9 up-regulation, HL-60 cells were pretreated with ATRA (100 nM for 48 hours), washed twice in RPMI to clear the ATRA, and treated with As2O3. ATRA-pretreated HL-60 cells showed an increased sensitivity to As2O3 compared with untreated HL-60 cells (Figure 6B). The increase in As2O3 sensitivity was due to increased arsenic uptake into the cells. Incubation of ATRA-pretreated HL-60 cells in 1 μM As2O3 led to a time-dependent increase in arsenic accumulation, which was significantly higher than in untreated HL-60 cells (Figure 6C).
Induction of AQP9 expression with ATRA treatment of HL-60 cells. (A) Semiquantitative polymerase chain reaction showed that the expression of AQP9 gene in HL-60 was significantly induced after 48-hour incubation of 100 nM, 1 μM, and 10μM ATRA. β-actin gene was used as internal control of the RT-PCR. Western-blot analysis also showed increasing AQP9 protein expression (about 30 kDa) with increasing concentrations of ATRA. The loading amount of protein samples was standardized by PointSau staining. (B) In MTT assay, HL-60 cells pretreated with 100 μM ATRA for 48 hours exhibited an increased sensitivity to the cytotoxicity of As2O3 compared with the untreated control. (C) In arsenic-uptake assay, HL-60 cells pretreated with 100 μM ATRA for 48 hours were incubated with 1 μM As2O3. There was a time-dependent increase in arsenic uptake compared with the untreated control (Student t test for each of the points analyzed).
Induction of AQP9 expression with ATRA treatment of HL-60 cells. (A) Semiquantitative polymerase chain reaction showed that the expression of AQP9 gene in HL-60 was significantly induced after 48-hour incubation of 100 nM, 1 μM, and 10μM ATRA. β-actin gene was used as internal control of the RT-PCR. Western-blot analysis also showed increasing AQP9 protein expression (about 30 kDa) with increasing concentrations of ATRA. The loading amount of protein samples was standardized by PointSau staining. (B) In MTT assay, HL-60 cells pretreated with 100 μM ATRA for 48 hours exhibited an increased sensitivity to the cytotoxicity of As2O3 compared with the untreated control. (C) In arsenic-uptake assay, HL-60 cells pretreated with 100 μM ATRA for 48 hours were incubated with 1 μM As2O3. There was a time-dependent increase in arsenic uptake compared with the untreated control (Student t test for each of the points analyzed).
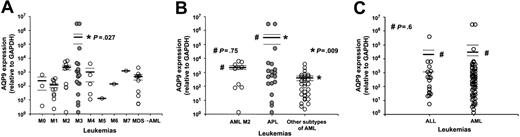
AQP9 expression in leukemic samples
Leukemic samples from 80 patients were studied. These included 19 patients with APL (presentation, 5; relapse, 14), 42 patients with AML (minimally differentiated, 3; without maturation, 11; with maturation, 10; myelomonocytic, 5; monoblastic, 1; erythroid, 1; megakaryoblastic, 1; multilineage dysplasia, 10), and 19 patients with ALL. The mean level of AQP9 was significantly higher in APL compared with all the other subtypes of AML (APL 3.07 × 105 ± 2.08 × 105 versus AML 8.10 × 102 ± 2.26 × 102, P = .027; Figure 7A). Interestingly, in subgroup analysis, when APL was compared with AML with maturation (or AML M2 in the French-American-British classification system26 ; n = 10), the AQP9 expression was similar (APL 3.07 × 105 ± 2.08 × 105 versus AML M2 2.14 × 103 ± 0.72 × 103, P = .75; Figure 7B). Hence, when APL was compared with AML subtypes other than M2 (n = 32), AQP9 expression was even more significantly different (3.07 × 105 ± 2.08 × 105 versus 3.93 × 102 ± 1.37 × 102, P = .009; Figure 7B). However, AQP9 expression in ALL was comparable with AML (ALL 2.05 × 104 ± 1.94 × 104 versus AML 9.62 × 104 ± 6.63 × 104, P = .6; Figure 7C).
AQP9 expression in 80 cases of leukemia. (A) AML M3 (APL) expressed significantly higher AQP9 levels (P = .027) than all the other subtypes of acute myeloid leukemia (M0, minimally differentiated; M1, without maturation; M2, with maturation; M4, myelomonocytic; M5, monoblastic; M6, erythroid; M7, megakaryoblastic; MDS→AML, acute myeloid leukemia evolving from an antecedent myelodysplastic syndrome). The solid line indicates mean; dotted line, standard error of the mean. (B) AQP9 expression was comparable in AML M2 and APL (P = .75) but was significantly different in APL and other subtypes of AML (P = .009). (C) ALL expressed comparable AQP9 levels compared with AML.
AQP9 expression in 80 cases of leukemia. (A) AML M3 (APL) expressed significantly higher AQP9 levels (P = .027) than all the other subtypes of acute myeloid leukemia (M0, minimally differentiated; M1, without maturation; M2, with maturation; M4, myelomonocytic; M5, monoblastic; M6, erythroid; M7, megakaryoblastic; MDS→AML, acute myeloid leukemia evolving from an antecedent myelodysplastic syndrome). The solid line indicates mean; dotted line, standard error of the mean. (B) AQP9 expression was comparable in AML M2 and APL (P = .75) but was significantly different in APL and other subtypes of AML (P = .009). (C) ALL expressed comparable AQP9 levels compared with AML.
Discussion
Our findings showed that AQP9 played a critical role in the transmembrane transport of As2O3. In neutral pH, As3+ exists primarily as As(OH)3, which might be recognized by AQP9 as a small neutral noncharged solute similar to glycerol for facilitated uptake.27 Accordingly, we showed that in leukemia cells of different lineages, in vitro sensitivity to As2O3 was directly proportional to AQP9 expression. The results were confirmed both at the mRNA and protein level. The ML-1 cell line was an exception in that although AQP9 was expressed apparently at high levels, the line was relatively resistant to As2O3. Analysis of the AQP9 coding sequence showed a point mutation resulting in a base substitution. Whether this base substitution is of functional significance is being investigated. An alternative explanation for the poor As2O3 sensitivity would be different intracellular constituents or mechanisms that are distinctive to this cell line. As the MDR1 gene encoding P-glycoprotein also controlled transmembrane drug trafficking, we investigated if the expression of AQP9 and MDR1 genes might be related. Q-PCR showed that their expressions were unrelated. This was consistent with previous observations that although the MDR1 gene might be up-regulated in relapsed APL, the response to As2O3 was unaffected, suggesting that MDR1 did not contribute to As2O3 sensitivity.22 Lastly, for leukemia cell lines of different lineages, the APL cell line NB4 showed the highest expression level of AQP9 and the most exquisite sensitivity to As2O3. Interestingly, APL is also clinically the leukemic subtype that responds best to As2O3 therapy.
To confirm the role of AQP9 in As2O3 entry and sensitivity, a pertinent investigation will be to express AQP9 in an AQP9-negative cell line and to study the response to As2O3 before and after AQP9 expression. Furthermore, transient transfection may be preferable to avoid inadvertent introduction of additional properties acquired unexpectedly during the selection of permanent transfectants. However, a leukemic line with undetectable AQP9 expression was not available. Moreover, the very low transfection-efficiency characteristic of leukemia cells in suspension would make transient transfection and subsequent experiments difficult. We therefore had elected to choose the Hep3B line, which were adherent cells and did not express any detectable AQP9, for this proof-of-principle experiment. In Hep3B cells, the transfected EGFP-AQP9 fusion protein showed the appropriate membrane localization. The selective cell-membrane partition of AQP9 may be due to the hydrophobicity of the AQP9 moiety or the presence of a membrane-localizing motif in AQP9. Besides accurate localization, the function of AQP9 in facilitating As2O3 uptake was also preserved. This was reflected in an increase in sensitivity to As2O3 in the Hep3BEGFP-AQP9 transfectant. The intracellular arsenic was not measured because harvesting the Hep3B cells required trypsinization, which partly permeabilized the cell membrane and resulted in a substantial loss of arsenic.
To further substantiate these observations in leukemia cells, the K562 line was chosen, as it had very low endogenous AQP9 expression and hence was insensitive to As2O3. Due to the low transfection efficiency, stable transfection coupled with flow sorting of fluorescent cells was needed to enrich for AQP9-transfected cells. The stable K562EGFP-AQP9 transfectants showed results similar to those observed in transient transfection of Hep3B, in that K562EGFP-AQP9 also demonstrated an increased As2O3 sensitivity. Moreover, we were able to show that the higher As2O3 sensitivity was due to increased intracellular arsenic accumulation. These observations confirmed that AQP9 facilitated transmembrane transport of arsenic.
Both Hep3BEGFP-AQP9/β-gal and K562EGFP-AQP9 exhibited a higher arsenic sensitivity than Hep3BEGFP/β-gal and K562EGFP-C2. However, the difference in viability of K562EGFP-AQP9 and K562EGFP cells increased progressively with higher concentrations of As2O3, whereas the difference of viability of Hep3BEGFP-AQP9/β-gal and Hep3BEGFP/β-gal remained comparable at different concentrations of As2O3. Therefore, it might be possible that AQP9-mediated arsenic transmembrane transport could be saturable, depending on the amount of AQP9 expressed. Alternatively, in addition to the amount of intracellular arsenic, other intrinsic properties of the cell may also determine its sensitivity to As2O3.
An important prerequisite for putting these observations into therapeutic applications is finding clinically feasible methods for up-regulation of AQP9 in leukemia cells. A synergistic interaction between ATRA and As2O3 has been observed in vitro.28 Furthermore, patients with APL at presentation showed a more efficient leukemic load reduction if treated concomitantly with ATRA and As2O3.29 Patients with APL at presentation or relapse also responded better to combined ATRA and As2O3 treatment.30-34 In the myeloid leukemia line HL-60 we showed that ATRA pretreatment up-regulated AQP9 expression, which was translated into an increase in arsenic uptake and hence sensitivity to As2O3 treatment. As the concentration of ATRA used was as low as 100 nM, which would not induce any significant apoptotic effect on HL-60, the increased cytotoxicity after As2O3 treatment could be attributable to the demonstrable increase in arsenic uptake. These observations provided another mechanistic explanation for the synergism between ATRA and As2O3. We had also conducted similar experiments with the APL cell line NB4. Unfortunately, NB4 was not a good in vitro model. This is because NB4 cells already express very high levels of AQP9 and are extremely sensitive to ATRA and As2O3, even at very low concentrations. Therefore, the exogenous overexpression of AQP9 did not give consistent results of further increases in arsenic uptake or cytotoxicity (data not shown).
To verify the relevance of AQP9 expression in primary leukemia samples, we quantified AQP9 in a series of AML samples. We showed that APL expressed AQP9 significantly (about 2-3 log) more than other subtypes of AML. This observation was consistent with the high sensitivity of APL to As2O3 compared with other AML subtypes. Interestingly, in AML with maturation (AML M2), where the leukemic cells show at least 10% granulocytic maturation, the AQP9 expression was comparable to APL. These results suggested that AQP9 expression might be related to myeloid maturation, with promyelocytes and myelocytes/granulocytes showing the highest expression of AQP9. However, not all APL cases expressed high levels of AQP9. As these cases still responded to As2O3 much more than AML with a similar AQP9 expression, intracellular mechanisms other than membrane AQP9 expression may also determine As2O3 sensitivity. Finally, ALL cells expressed AQP9 levels comparable to AML, implying that the role of As2O3 in ALL treatment might be explored.
Our observations provide useful leads in the research of the therapeutic use of As2O3 in leukemic treatment. The expression level of AQP9 may be one of the molecular determinants predicting the response to As2O3 therapy, although expectedly other cellular mechanisms are also important. Whether AQP9 might also affect other chemotherapeutic agents remains to be determined by experiments measuring drug entry/efflux and AQP9 expression. Secondly, investigations of the transcriptional regulation of the AQP9 gene will be clinically relevant, particularly in AML without granulocytic maturation. Previous studies have shown that steroid hormones and vitamin D3 may induce AQP9 expression.35-38 As the promoter of AQP9 does not contain hormone-responsive elements, the actions of these lipophilic agents may be due to up-regulation of other transcription factors.20 In fact, the AQP9 promoter contains binding sites for C/EBP, NF-κB, and AP-1.20 It is intriguing to note that C/EBP is inducible by retinoids such as ATRA.39,40 Therefore, the definition of agents able to induce AQP9 expression in different cancer cell types may hold promise for increasing the uptake of As2O3 and hence its efficacy in these malignancies. Moreover, the potential contribution of AQP9 dysregulation or mutation to As2O3 resistance needs further investigation. Finally, the organ toxicity of arsenic may also be related to AQP9 expression. In fact, AQP9 is expressed normally at high concentrations in the liver and brain, organs that are most affected by arsenic therapy.6 Agents that may selectively down-regulate AQP9 in these organs may ameliorate arsenic toxicity, which is an important limiting factor in the therapeutic use of As2O3. These propositions will require future validations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: The authors have no financial interest or conflict of interest to disclose.
Contribution: J.L. performed the experiments and wrote and approved the manuscript; A.P. performed the experiments and approved the manuscript; W.-H.Y. performed the experiments and approved the manuscript; Y.-L.K. designed and supervised the study and wrote and approved the manuscript; and E.W.C.T. designed and supervised the study and amended and approved the manuscript.
This work was supported in part by the S. K. Yee Foundation.
The authors thank W. I. Wong for valuable technical assistance and M. F. Yuen for advice on statistical analysis.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal