Abstract
Human papilloma virus (HPV)–like particles (VLPs) have been used as a vaccine to prevent HPV infection. Recent studies demonstrate that VLPs bind to dendritic cells and induce the expression of antiviral cytokines such as interferon-α (IFN-α), interleukin-10 (IL-10) and IFN-γ. In the present study, we evaluated the effect of VLPs on HIV-1 replication in peripheral blood mononuclear cells (PBMCs), CD4+ T cells, and macrophages. Here, we show that VLPs suppress the replication of both X4 and R5 HIV-1 without affecting the expression of CD4, CXCR4, and CCR5. Soluble factor(s) released by PBMCs and macrophages on VLPs treatment inhibited HIV-1 replication. To determine the inhibitory factors, DNA microarray analysis was performed using VLP-treated PBMCs and macrophages. VLPs induced the genes associated with IFN induction, immune responses, and antiviral responses, among with the recently described cytokine IL-27. Subsequently, IL-27 was found to be a potent inhibitor of HIV-1 replication in PBMCs, CD4+ T cells, and macrophages. Taken together, our studies identify a novel role of IL-27 in restricting HIV-1 replication and suggest that further examination of the inhibitory property of IL-27 may pave the way for a novel therapy for HIV-1 infection.
Introduction
It has been demonstrated that both innate and adaptive immune response regulate HIV-1 replication,1–10 and certain cell types secrete soluble HIV-1–suppressing proteins in response to various stimuli.11–15 For example, secretion of anti-HIV β-chemokines and CD8 antiviral factor (CAF) from human T-lymphotropic virus (HTLV)–type 1 and herpes virus saimiri–infected cell line have been described.11–15 Although a combination of antibodies and suppressive factors was able to reverse the inhibitory activity of CAF, complete reversal of HIV-1 suppression was not observed, suggesting that additional unknown suppressive factors were involved.16
Certain microbial organisms elicit immune responses that reduce clinical HIV-1 infection presumably through the induction of soluble suppressive factors. Some persons who are simultaneously infected with HIV-1 and either dengue, Orienta tsutsugamushi, hepatitis G/GB virus C, or measles morbilli virus have reduced viral loads compared with patients who are infected with HIV-1 alone or with HIV-1 and other pathogens.17–25 In the case of hepatitis G virus (HGV), the mechanism of action is postulated to be via binding of the serum HGV E2 protein to CD81, leading to increased regulated on activation normal T expressed and secreted protein (RANTES) secretion and reduced CCR5 expression.24 In vitro studies have shown that HTLV-type 2 infection might up-regulate the production of macrophage inflammatory protein-1α (MIP-1α).26
Recent studies demonstrate that human papillopma virus-like particles (HPV-VLPs) bind to dendritic cells and are able to induce a range of responses in immune cells, notably the expression of anti–HIV-1 cytokines such as interferon-α (IFN-α), interleukin-10 (IL-10), interferon-γ (IFN-γ), and monocyte chemotactic protein 2 (MCP-2).27,28 The HPV-VLP vaccine has been demonstrated to be protective against HPV infection.29–31 HPV-type 16 (HPV16) represents the primary causative agent of cervical cancer.32 The HPV16 VLPs, composed of the L1 major capsid protein, form nonenveloped icosahedral particles that lack viral DNA but both morphologically and immunologically resemble native virions.33 Studies suggest that a potent immune response induced by VLPs is through the Toll-like receptor (TLR)/MyD88 pathway in dendritic cells.27
In the current work, we evaluated the impact of VLPs on HIV-1 replication. Our results demonstrated that VLPs strongly inhibit replication of X4 and R5 HIV-1 in peripheral blood mononuclear cells (PBMCs) and monocyte-derived macrophages (MDMs). It was discovered that the anti–HIV-1 activity of VLPs involved the release of suppressive factors. Gene expression profiles of PBMCs and MDMs demonstrated that VLP treatment up-regulated numerous potential antiviral genes along with the induction of recently described cytokine interleukin-27 (IL-27). Subsequent experiments demonstrated that recombinant IL-27 was able to inhibit X4 and R5 HIV replication. Taken together, these studies identify IL-27 as a novel antiviral cytokine and imply that further investigation of the inhibitory properties of IL-27 may have implications for HIV-1 therapeutics.
Materials and methods
Approval for these studies was granted by the institutional review board of NIH.
Cells and reagents
A cloned proviral X4 HIV-1 DNA, pNL4.3,34 was obtained from Dr M. A. Martin through the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health; Dr M. A. Martin kindly provided a cloned R5 HIV-1 proviral DNA pAd8.35 Recombinant infectious HIVNL4.3 and HIVAd8 were prepared as previously described.36 The recombinant VLPs were expressed in the baculovirus-infected Sf9 insect cells (Novavax, Rockville, MD). Production of clinical lots of recombinant HPV16 L1 VLP vaccines (endotoxin free) was performed in accordance with GMP guidelines at the Vaccine Production Facility of Novavax, as reported elsewhere.37 Formulated L1 VLPs from a single lot were dispensed aseptically into sterile vials (3.0-mL size, type 1 borosilicate glass, silanized, depyrogenated; Wheaton Glass, Millville, NJ) as a single-unit dose, and VLPs were stored at −20°C.38 PBMCs were isolated from leukapheresed blood from healthy donors using lymphocyte separation medium (ICN Biomedical, Aurora, OH).36
CD4+ T cells and CD3−/CD14+ monocytes were purified from PBMCs using CD4 MicroBeads (Miltenyi Biotec, Auburn, CA) and CD14 MicroBeads (Miltenyi Biotec) according to the manufacturer's instructions. The purity of both cell types was at least 90%, based on flow cytometric analysis. CD8+ T-cell–depleted PBMCs (CD8− PBMCs) were prepared from the isolated PBMCs using a combination of CD8 MicroBeads and a depletion column (both from Miltenyi Biotech). The depletion process decreased the fraction of CD8+ T lymphocytes to less than 1% in PBMCs as determined by flow cytometry. MDMs were prepared as follows: isolated CD3−/CD14+ monocytes were plated in a tissue culture flask at 5 × 106 cells/mL in the presence of 10% (vol/vol) human AB serum (Cambrex, Baltimore, MD) in Dulbecco Modified Eagle Medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 2 mM glutamine and 50 μg/mL gentamicin (Invitrogen), 10 mM N-2-hydroxyethypiperazine-N′-2-ethanesulfonic acid (HEPES; Quality Biochemical, Gaithersburg, MD), 10% (vol/vol) fetal bovine serum (FBS; Quality Biochemical, Logan, UT), and then differentiated into macrophages for 7 days. Flow cytometric analysis confirmed that more than 80% of the adherent cells were macrophages. Cell viability was determined using trypan blue exclusion method.
HIV replication assay
The levels of HIV-1 replication under various culture conditions were determined as follows. PBMCs, CD4+ T cells, or CD8− PBMCs were stimulated with 5 μg/mL phytohemagglutinin (PHA; Sigma-Aldrich, St Louis, MO) in RPMI-1640 supplemented with 10 mM HEPES, 32 U/mL native IL-2 (Roche Molecular Biology, Indianapolis, IN), 10% FBS, and 50 μg/mL gentamicin. The PHA-stimulated PBMCs or CD4+ T cells (10 × 106 cells) were infected with 10 000 TCID50 HIV-1NL4.3 or HIV-1Ad8 for 2 hours at 37°C and then cultured at 1 × 106 cells/mL for 7 days at 37°C.36 Half of the culture supernatants was exchanged with fresh RPMI after 4 days of incubation. MDMs (5 × 106 cells) were infected with 5000 TCID50 HIV-1Ad8 for 2 hours and then cultured at 1 × 106 cells/mL in DMEM for 10 days at 37°C. Half of the culture supernatants was changed with fresh DMEM on the 4th and 7th days. Each culture was performed in quadruplicate. HIV-1 replication was determined by measuring p24 antigen amount in the culture supernatants using a p24 antigen capture assay (Beckman-Coulter, Miami, FL).
Generation of conditioned medium
Conditioned media derived from untreated cells (Control-Sup) and VLP-treated cells (VLP-Sup) were prepared as follows. The PHA-stimulated PBMCs or MDMs were cultured in the presence or absence of 1 μg/mL VLPs at 1 × 106 cells/mL. At appropriate time points (36 hours for PBMCs, 24 hours for MDMs), crude supernatants were clarified by low-speed centrifugation (500g for 5 minutes), filtered through a Millex-GV 0.22-μm PVDF membrane (Millipore, Billerica, MA) to remove cellular debris, and stored at −70°C until used.
Neutralization of cytokines and immunodepletion of IL-27
To neutralize IFNs (IFN-α and IFN-β), IL-10, MCP-2, or CCR5 ligands in VLP-Sup from PBMCs and MDMs, 10 μg/mL neutralizing antibody (all antibodies were purchased from R&D Systems, Minneapolis, MN) were added in the culture. To neutralize CCR5 ligands in VLP-Sup, an antibody mixture containing neutralizing antibody to MIP-1α, MIP-1β, and RANTES (10 μg of each antibody/mL) was used. As a control for the assay, the same amount of normal IgG (R&D Systems) was added in the culture. As a positive control for the neutralization, 10 μg/mL neutralizing antibody or the antibody cocktail was added in the culture of HIV-infected cells in the presence of 100 ng/mL recombinant cytokines or a mixture of 100 ng/mL of each chemokine. Immunodepletion of IL-27 was performed as follows39 : 0.4 mL VLP-Sup from PBMCs and MDMs was incubated with 5 μg goat polyclonal anti–IL-27 IgG (R&D Systems) or normal goat IgG (Santa Cruz Biotechnology, Santa Cruz, CA) at 4°C for 2 hours with rocking, followed by immunoprecipitation (IP) using Protein G PLUS agarose (Santa Cruz Biotechnology) at 4°C for 2 hours. To obtain the immunoabsorbed VLP-Sup, the suspension was centrifuged at 300g at 4°C for 1 minute to pellet the IP complexes bound to Protein G PLUS agarose.
Quantitation of cytokine concentration in conditioned medium
Concentrations of IFN-α, IFN-β, IL-10, and MCP-2 in conditioned media were determined using commercial sandwich enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems) following the vendor's protocol. The concentration of IL-27 was measured using a sandwich ELISA developed in this laboratory. Briefly, each well of 96-well plates (Nunc-immunoplate; Nalgene Nunc International, Rochester, NY) was coated with 100 μL of 5 μg/mL of a capture protein WSX-Fc (R&D Systems) in phosphate-buffed saline (PBS) pH 7.4 at 4°C overnight. Each well of the plates was then blocked by the addition of 300 μL PBS containing 1% (wt/vol) bovine serum albumin (BSA; Sigma-Aldrich), 5% (wt/vol) sucrose, and 0.05% (wt/vol) NaN3 and incubated at room temperature for 3 hours. The plates were washed 5 times with wash buffer (0.05% [vol/vol] Tween 20 [Roche Molecular Biology] in PBS, pH 7.4). Samples or various concentrations of recombinant human IL-27 (R&D Systems) were incubated at room temperature for at least 2 hours in final volumes of 100 μL. After washing, the plates were incubated with 100 μL of 200 ng/mL rabbit anti–human IL-27 IgG (Imagenex, San Diego, CA) for 2 hours at room temperature. To detect the bound rabbit anti–human IL-27 antibody, horseradish peroxidase conjugate of an antirabbit antibody (R&D Systems) was used. The bound horseradish peroxidase was measured using 3,3′,5,5′-tetramethylbenzidine (Sigma-Aldrich) as substrate, and the reaction was stopped with stop solution (R&D Systems) after 30 minutes. Optical density was determined at 450 nm. The detection limit was 500 pg/mL.
VLP binding assay
Green fluorescent protein (GFP)–tagged HPV-VLPs were kindly provided by Dr J. T. Schiller (National Cancer Institute, Bethesda, MD); they consist of HPV16 L1 VLPs containing L2 bovine papillomavirus–GFP fusion proteins. These were produced as previously reported.40 Cell-surface markers were identified with the following antibodies: FITC-conjugated anti-CD3 (BD Biosciences, San Jose, CA), PE-conjugated anti-CD8 (BD Biosciences) or anti-CD14 (BD Biosciences), and PC5-conjugated anti-CD4 (Beckman-Coulter). PHA-stimulated PBMCs were incubated with GFP-tagged VLPs (10 μg/mL) or media for 1 hour at 37°C in PBS, supplemented with 2% (vol/vol) FBS and 1 mM CaCl2 (assay buffer; Sigma-Aldrich). After incubation, cells were washed with the assay buffer and then analyzed by flow cytometry (at least 5000 cells were counted).
Flow cytometry
PHA-stimulated PBMCs were incubated with 1 μg/mL VLPs at 37°C for 24 hours. As a positive control for down-regulation of chemokine receptors, the PBMCs were incubated with 100 ng/mL stromal cell-derived factor 1 (SDF-1; R&D Systems) or a mixture of 100 ng/mL RANTES (R&D Systems) and 100 ng/mL MIP-1α (R&D Systems) in RPMI at 4°C overnight and then cultured at 37°C for 10 minutes. After the incubation, the treated cells were washed with cold PBS and then fixed with 1% (wt/vol) paraformaldehyde (Sigma-Aldrich) in PBS. The fixed cells were stained for 15 minutes at room temperature in PBS containing 2% (wt/vol) BSA with the following antibodies: APC-conjugated anti-CD3 (BD Biosciences), FITC-conjugated anti-CD4 (BD Biosciences), PC5-conjugated anti-CD14 (Beckman-Coulter), PE-conjugated anti-CCR5 (BD Pharmingen, San Diego, CA), PE-conjugated anti-CXCR4 (BD Pharmingen), or PE-conjugated anti-CD8 (BD Biosciences). The cell preparations were analyzed with a B-D fluorescence-activated cell sorting (FACS) Calibur flow cytometer (BD Biosciences). Forward and orthogonal light scatter gates were drawn to include either lymphocytes or monocytes, and a fluorescence gate was drawn to include either CD3+ lymphocytes or CD3−/CD14+ monocytes. These gated events were further analyzed for the expression of CD4 and CCR5 or CD4 and CXCR4. Positives and negatives were determined through the use of matching isotype controls.
Microarray analysis
A microarray gene expression analysis was conducted using the Affymetrix GeneChip system (Affymetrix, Santa Clara, CA). The Affymetrix human genome U133 plus a 2.0 array containing more than 47 000 transcripts was used. For the analysis, 5 × 106 cells of PHA-stimulated PBMCs and MDMs were cultured at 1 × 106/mL in the presence of 1 μg/mL VLPs for 36 and 24 hours, respectively. RNA was isolated from cells using the RNeasy Isolation kit (Qiagen, Valencia, CA) and quantitated following the manufacturer's recommended protocols (Affymetrix). Biotin-labeled cRNA was generated as previously described.36 Gene expression levels were determined following laser scanning of the GeneChip at 570 nm. Gene expression values were determined using GCOS1.3 (Affymetrix). Differentially expressed genes were identified as P values less than .05 (1-way ANOVA) and an absolute fold change difference was greater than 2. Gene subsets were determined by using a bioinformatics analysis tool, DAVID.41
RT-PCR
PHA-stimulated PBMCs or MDMs (1 × 106cells/mL) were cultured in the absence or presence of 1 μg/mL VLPs for 24 or 36 hours at 37°C. Cells were washed using cold PBS, and RNA was isolated from the cells using the RNeasy Isolation kit (Qiagen) and on-column DNA digestion using RNase free DNase, following the vendor's protocol. The cDNA was synthesized using the Superscript First Strand Synthesis system for reverse transcriptase–polymerase chain reaction (RT-PCR; Invitrogen) with oligo (dT). PCR was performed using human IL-27 PCR primer (p28 PCR Primer Pair; R&D Systems) and β-actin (Clontech, Mountain View, CA) following the manufacturer's protocol as described before.36 The reaction products were separated on a 1% (wt/vol) agarose gel (Biowhittaker Molecular Applications, Walkersville, MD) in the presence of 0.5 μg/mL ethidium bromide (Invitrogen) and photographed.
Quantitation of transcribed HIV RNA
PHA-stimulated CD4+ T cells were infected with X4-virus and then cultured at 1 × 106 cells/mL for 7 days in the absence or presence of 100 ng/mL IL-27. A total cellular RNA was isolated using Trizol reagent (Invitrogen), according to the manufacturer's instructions. Real-time quantitative RT-PCR was performed as previously described.42,43 Cell equivalents were estimated via real-time PCR using CCR5 sequence, located on chromosome 3 in human cells,43,44 instead of the porphobilinogen deaminase gene used previously.43
Statistical analysis
Differences in HIV inhibition and cytokine production between the untreated and VLP-treated cells were calculated by t test using the StarView program (Abacus Concepts, Berkeley, CA). P values less than .05 were considered statistically significant.
Results
HPV-VLPs inhibit HIV-1 replication in PBMCs
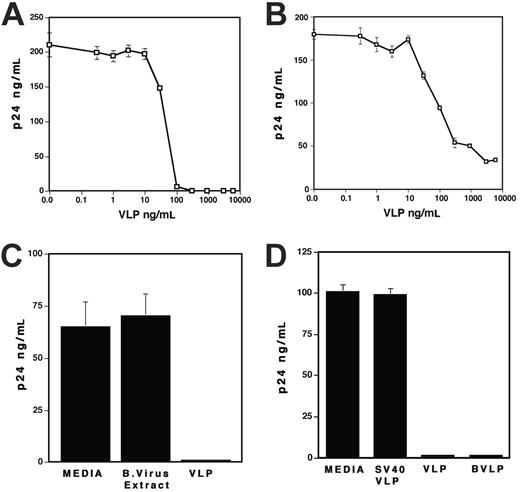
To characterize the effects of HPV-VLPs on replication of X4 and R5 viruses, PHA-stimulated PBMCs were infected with HIV-1NL4.3 (X4 virus) or HIV-1Ad8 (R5 virus) and then cultured in the presence of VLPs. Figure 1A shows that VLPs significantly inhibited X4 virus replication in a dose-dependent manner. In the presence of 1 μg/mL VLPs, replication of X4 virus was inhibited nearly 100%. More than 90% inhibition was also observed in replication of R5 virus (Figure 1B). On the contrary, baculovirus (1 μg/mL) lysate had no effect on replication of X4 virus (Figure 1C) or R5 virus (data not shown) in PBMCs, which eliminated the possibility that the inhibitory effect of VLPs is due to contaminating baculovirus cell lysate in VLPs. Of interest, the bovine papilloma virus VLPs (BVLPs) also suppressed X4 virus replication in PBMCs; however, SV40 VLPs had no influence on replication of the virus (Figure 1D). VLPs at a concentration of 1 μg/mL had no effect on cell viability using trypan blue staining (data not shown). These results suggest that VLPs impede HIV-1 replication in a dose-dependent manner, regardless of the tropism of HIV-1 strains.
VLPs inhibit HIV-1 replication. (A-B) PHA-stimulated PBMCs were infected with (A) X4 virus (HIV-1NL4.3) or (B) R5 virus (HIV-1Ad8) as described in “Materials and methods.” The infected cells were cultured for 7 days in the presence of various concentrations of VLPs. (C) PHA-stimulated PBMCs were infected with X4 virus for 2 hours and then cultured for 7 days in the absence or the presence of 1 μg/mL baculovirus extract or VLPs. (D) PHA-stimulated PBMCs were infected with X4 virus for 2 hours and then cultured for 7 days in the absence or the presence of 1 μg/mL VLPs, SV40-VLPs, or BVLPs. In all experiments, half of the culture supernatants was changed on the 4th day with fresh culture media alone. HIV-1 replication was measured by p24 antigen capture assay. Data show means ± SDs and are representative of at least 3 independent experiments.
VLPs inhibit HIV-1 replication. (A-B) PHA-stimulated PBMCs were infected with (A) X4 virus (HIV-1NL4.3) or (B) R5 virus (HIV-1Ad8) as described in “Materials and methods.” The infected cells were cultured for 7 days in the presence of various concentrations of VLPs. (C) PHA-stimulated PBMCs were infected with X4 virus for 2 hours and then cultured for 7 days in the absence or the presence of 1 μg/mL baculovirus extract or VLPs. (D) PHA-stimulated PBMCs were infected with X4 virus for 2 hours and then cultured for 7 days in the absence or the presence of 1 μg/mL VLPs, SV40-VLPs, or BVLPs. In all experiments, half of the culture supernatants was changed on the 4th day with fresh culture media alone. HIV-1 replication was measured by p24 antigen capture assay. Data show means ± SDs and are representative of at least 3 independent experiments.
Pretreatment of PBMCs with VLPs blocks HIV-1 infection without impacting HIV-1 receptor expression
To further characterize the impact of VLPs on HIV-1 replication, PHA-stimulated PBMCs were pretreated with 1 μg/mL VLPs at 37°C for 2 or 24 hours before infection. After washing the treated cells, they were then infected with X4 virus and cultured for 7 days in the absence of VLPs. When p24 levels were determined on the 7th day, HIV-1 replication was strongly inhibited (99%), despite removal of the VLPs prior to infection (Figure 2A). This result demonstrated that the pretreatment was adequate to block HIV-1 infection of the cells, raising the possibility that interaction with VLPs was leading to an alteration in the expression pattern of HIV-1 receptors and coreceptors. Thus, we examined whether the pretreatment down-regulated the expression of CD4, CXCR4 (for X4 virus), and/or CCR5 (for R5 virus) on the cell surface. PHA-stimulated PBMCs were incubated with or without VLPs at 1 μg/mL for 24 hours, and then flow cytometric analysis was carried out. As a positive control for down-regulation of chemokine receptors, PBMCs were treated with SDF-1 alone (for CXCR4) or a mixture of RANTES and MIP-1α (for CCR5). VLPs had no significant effect on the expression of CD4, CXCR4, and CCR5 on CD3+ and CD3−/CD14+ cells in PBMCs (Figure 2B). These results suggest that VLPs may not directly affect HIV-1 infection.
Pretreatment of PBMCs with VLPs inhibit HIV-1 replication without changing in the expression of HIV receptors. (A) PHA-stimulated PBMCs were pretreated with media alone or 1 μg/mL VLPs for 2 or 24 hours and then washed with fresh media to remove unbound VLPs. The pretreated cells were infected with X4 virus for 2 hours and cultured for 7 days without addition of VLPs. Half of the culture supernatants was changed on the 4th day with fresh culture media alone. HIV-1 replication was assessed by p24 antigen capture assay. Data show means ± SDs and are representative of at least 2 independent experiments. (B) PHA-stimulated PBMCs were cultured in the absence (red) or presence (black) of VLPs (1 μg/mL) for 24 hours. The surface expression of CD4, CXCR4, and CCR5 was then assessed by flow cytometry as described in “Materials and methods.” For a positive control of down-regulated receptors, the cells were treated with 100 ng/mL of a mixture of RANTES and MCP-1 for CCR5 or SDF-1 for CXCR4, and then the expression level was analyzed. Left panels show the staining of CD3+ T lymphocytes; right panels show CD14+ monocytes. Upper, middle, and bottom panels show CD4, CXCR4, and CCR5 staining, respectively. The staining pattern of isotype control antibodies are shown in blue, and the positive control of down-regulation are shown in green. The x-axis and y-axis show florescence intensity and cell count, respectively. The data are representative of 3 independent experiments with similar outcomes.
Pretreatment of PBMCs with VLPs inhibit HIV-1 replication without changing in the expression of HIV receptors. (A) PHA-stimulated PBMCs were pretreated with media alone or 1 μg/mL VLPs for 2 or 24 hours and then washed with fresh media to remove unbound VLPs. The pretreated cells were infected with X4 virus for 2 hours and cultured for 7 days without addition of VLPs. Half of the culture supernatants was changed on the 4th day with fresh culture media alone. HIV-1 replication was assessed by p24 antigen capture assay. Data show means ± SDs and are representative of at least 2 independent experiments. (B) PHA-stimulated PBMCs were cultured in the absence (red) or presence (black) of VLPs (1 μg/mL) for 24 hours. The surface expression of CD4, CXCR4, and CCR5 was then assessed by flow cytometry as described in “Materials and methods.” For a positive control of down-regulated receptors, the cells were treated with 100 ng/mL of a mixture of RANTES and MCP-1 for CCR5 or SDF-1 for CXCR4, and then the expression level was analyzed. Left panels show the staining of CD3+ T lymphocytes; right panels show CD14+ monocytes. Upper, middle, and bottom panels show CD4, CXCR4, and CCR5 staining, respectively. The staining pattern of isotype control antibodies are shown in blue, and the positive control of down-regulation are shown in green. The x-axis and y-axis show florescence intensity and cell count, respectively. The data are representative of 3 independent experiments with similar outcomes.
VLPs suppress X4-virus replication in macrophages but not in CD4+ T cells
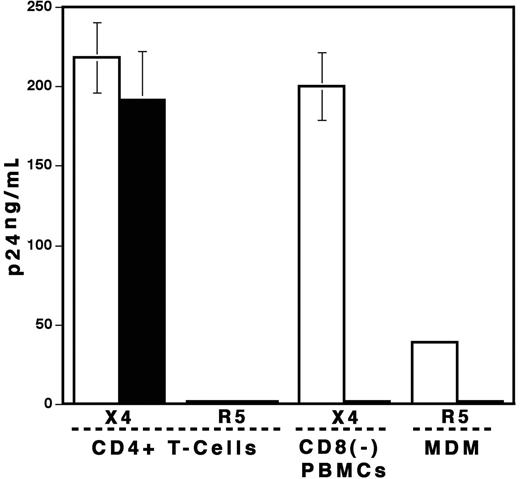
The finding that the treatment of PBMCs with VLPs inhibited HIV-1 replication prompted us to investigate the effect of VLPs on subsets of PBMCs. For this purpose, CD4+ T cells and CD8+-depleted PBMCs (CD8− PBMCs) were stimulated with PHA and then infected with X4 virus at 37°C for 2 hours. The infected cells were cultured for 7 days in the absence or presence of 1 μg/mL VLPs. Surprisingly, VLPs had no impact on X4 virus replication in CD4+ T cells (P > .05); however, they inhibited viral replication in CD8− PBMCs (Figure 3). These results indicate that VLPs do not directly inhibit X4 virus replication in CD4+ T cells, and CD8+ T cells are not required for the HIV-1 suppressive effect of VLP treatment. To evaluate the impact of VLPs on replication of R5 virus, the stimulated CD4+ cells or MDMs were infected with R5 virus and then cultured in the absence or presence of VLPs. Although R5 replication in CD4+ T cells was relatively low because of low level of the expression of CCR5, VLPs did not inhibit the virus replication in this cell type. In contrast, R5 replication in MDMs was dramatically (98.4%) inhibited by VLP treatment (Figure 3).
VLPs inhibit HIV replication in CD8− PBMCs and MDMs but not in CD4+ T cells. CD4+ T cells, CD8− PBMCs, and MDMs were prepared as described in “Materials and methods.” CD4+ T cells and CD8− PBMCs were stimulated with PHA and then infected with X4 or R5 viruses. The infected cells were cultured in the absence (□) or the presence of 1 μg/mL VLPs (▪) for 7 days. MDMs were infected with R5 virus and then cultured for 10 days. Half of the culture supernatants was changed on the 4th day and the 7th day with fresh culture media alone. HIV-1 replication was measured by p24 antigen capture assay. Data show means ± SDs and represent 3 independent experiments.
VLPs inhibit HIV replication in CD8− PBMCs and MDMs but not in CD4+ T cells. CD4+ T cells, CD8− PBMCs, and MDMs were prepared as described in “Materials and methods.” CD4+ T cells and CD8− PBMCs were stimulated with PHA and then infected with X4 or R5 viruses. The infected cells were cultured in the absence (□) or the presence of 1 μg/mL VLPs (▪) for 7 days. MDMs were infected with R5 virus and then cultured for 10 days. Half of the culture supernatants was changed on the 4th day and the 7th day with fresh culture media alone. HIV-1 replication was measured by p24 antigen capture assay. Data show means ± SDs and represent 3 independent experiments.
To determine whether VLP binding has any cell tropism, VLP binding assay was performed using the GFP-tagged VLPs. VLPs strongly bound to CD3−/CD14+ monocyte/macrophages; however, weak binding was seen with CD4+, CD8+, or natural killer (NK) cells (data not shown). Altogether, these data indicate that VLP per se has a direct effect on monocytes/macrophages in PBMCs.
Soluble factors secreted by HPV-VLPs treated PBMCs inhibit HIV-1 replication in T lymphocytes
Because VLPs exerted the inhibitory effect on PBMCs but not on CD4+ T cells (Figure 3), it was speculated that VLPs might induce anti-HIV soluble factor(s) from monocytes/macrophages in PBMCs. To address this possibility, PHA-stimulated PBMCs were cultured in the presence of 1 μg/mL VLPs for 36 hours, and then the conditioned media (VLP-Sup) were acquired. As a control, culture supernatants from untreated cells (Control-Sup) were also collected. The X4-infected CD4+ T cells were cultured with various concentrations (vol/vol) of VLP-Sup or Control-Sup. The VLP-Sup significantly inhibited HIV-1 replication, although not to the same extent as active VLP itself (Figure 4A). Interestingly, the inhibitory effect seen with VLP-Sup from PBMCs varied from donor to donor (data not shown); at 12.5% of the VLP-Sup, virus replication was inhibited by 84% ± 4.4% (n = 6). VLP-Sup from MDMs also inhibited HIV replication by a dose-dependent manner (Figure 4B); a final 12.5% of the Sup inhibited viral replication by 65% ± 2.7% (n = 7). Although VLP-Sup from PBMCs was more potent than that from MDMs (P < .01), these results suggest that VLPs are able to induce soluble suppressive factor(s) from both PBMCs and MDMs
Conditioned media derived from VLP-treated PBMCs or MDMs inhibit HIV replication in CD4+ T cells. Control-Sup (□) and VLP-Sup (▪) from PBMCs (A) and MDMs (B) were collected as described in “Materials and methods.” PHA-stimulated CD4+ T cells were infected with X4 virus and then cultured in the presence of different percentages of the Sup for 7 days. Half of culture supernatants were changed on the 4th day with fresh culture media alone. HIV-1 replication was measured by p24 antigen capture assay. Data show means ± SDs. Representative results from 3 independent results are shown.
Conditioned media derived from VLP-treated PBMCs or MDMs inhibit HIV replication in CD4+ T cells. Control-Sup (□) and VLP-Sup (▪) from PBMCs (A) and MDMs (B) were collected as described in “Materials and methods.” PHA-stimulated CD4+ T cells were infected with X4 virus and then cultured in the presence of different percentages of the Sup for 7 days. Half of culture supernatants were changed on the 4th day with fresh culture media alone. HIV-1 replication was measured by p24 antigen capture assay. Data show means ± SDs. Representative results from 3 independent results are shown.
VLPs induce transcriptional changes in PBMCs and macrophages
To investigate the underlying host factors responsible for VLP-mediated HIV-1 inhibition, we analyzed the gene expression profiles in PBMCs and MDMs on VLP treatment using a DNA microarray. The results of this analysis revealed that VLPs significantly induced IFN, IFN-stimulated genes, and a number of other cytokine and/or chemokine genes: IL10, IL15, IL27, IFN inducible protein-10 (IP10/CXCL10), IFN-inducible T-cell chemoattractant (ITAC/CXCL11), tumor necrosis factor–related apoptosis-inducing ligand (TNSF10/TRAIL), and MCP2 (Table 1). VLP treatment also significantly activated genes encoding members of a gene family associated with cellular antiviral proteins. VLPs up-regulated several members of the tripartite motif (TRIM) family in both PBMCs and MDMs: TRIM5 (∼ 5-fold), TRIM14 (3-fold), TRIM19 (∼ 5-fold), TRIM22 (∼ 3-fold), TRIM34 (∼ 3-fold), TRIM38 (∼ 2-fold), and TRIM56 (∼ 2-fold). The treatment activated a member of the apolipoprotein B mRNA editing complex 3 (APOBEC3) family, APOBEC3A, in PBMCs and MDMs by 52- and 27-fold, respectively.
Effect of VLP treatment on the expression of cytokine genes
| Gene ID . | Gene name . | Fold increase* . | |
|---|---|---|---|
| PBMC . | MDM . | ||
| 24678 | IL27 | 14.0 | 20.0 |
| 6355 | MCP2 | 31.0 | 30.0 |
| 10673 | TNSF13B | 2.6 | 2.5 |
| 3586 | IL10 | NS | 2.3 |
| 8743 | TNSF10 | 6.4 | 111.0 |
| 3600 | IL15 | NS | 4.7 |
| 6373 | CXCL11 | 85.0 | 54.0 |
| 3627 | CXCL10 | 10.0 | 58.0 |
| 6347 | MCP1 | 15.0 | NS |
| 3469 | IFNA | 143.0 | NS |
| 3456 | IFNB | 135.0 | NS |
| Gene ID . | Gene name . | Fold increase* . | |
|---|---|---|---|
| PBMC . | MDM . | ||
| 24678 | IL27 | 14.0 | 20.0 |
| 6355 | MCP2 | 31.0 | 30.0 |
| 10673 | TNSF13B | 2.6 | 2.5 |
| 3586 | IL10 | NS | 2.3 |
| 8743 | TNSF10 | 6.4 | 111.0 |
| 3600 | IL15 | NS | 4.7 |
| 6373 | CXCL11 | 85.0 | 54.0 |
| 3627 | CXCL10 | 10.0 | 58.0 |
| 6347 | MCP1 | 15.0 | NS |
| 3469 | IFNA | 143.0 | NS |
| 3456 | IFNB | 135.0 | NS |
PHA-stimulated PBMCs and MDMs were cultured in the absence or presence of 1 μg/mL VLPs for 36 and 24 hours, respectively. A total RNA was extracted from the cells, and then DNA microarray analysis was performed as described in “Microarray analysis.” Data show genes known to be associated with cytokines which were selected using the DAVID bioinformatics tool.41
NS indicates not significant; TNSF10, TRAIL (tumor necrosis factor–related apoptosis-inducing ligand).
The data show means of fold increases in VLP-treated cells compared with those in untreated cells from 3 independent experiments.
As shown in Table 1, VLPs significantly enhanced transcription of cytokines: IFNA, IFNB, MCP2, and IL10. It was reported that IFN-α and IFN-β inhibit both R4 and R5 viruses,45 whereas MCP-2 and IL-10 inhibit replication of R5 but not X4 virus.46,47 To elucidate whether VLP treatment produces the antiviral proteins in culture supernatants, quantitative analysis was performed. VLP treatment significantly increased the production of IFN-α and IL-10, but not IFN-β and MCP-2 from PBMCs (Table 2). Of note, SV40 VLPs, which lack the inhibitory effect on HIV replication, did not induce a detectable level of either IFN-α or IFN-β from PBMCs (data not shown). To define the relative role of IFNs (IFN-α and IFN-β), MCP-2, or IL-10 in VLP-Sup, a neutralization assay was performed. The X4-infected CD4+ T cells or R5-infected PBMCs were cultured in the presence of VLP-Sup from PBMCs with neutralizing antibodies or isotype antibody. None of the treatments had an impact on HIV inhibition by the VLP-Sup. In addition, to directly address whether CCR5 ligands (MIP-1α, MIP-1β, and RANTES) were released from PBMCs and then contributed to the antiviral activity in VLP-Sup, the ligands were neutralized using a mixture of neutralizing antibody to each chemokine (10 μg of each antibody/mL). VLP-Sup with control and the neutralizing antibody inhibited R5 replication in PBMCs to 25% ± 2.8% and 26% ± 2.9%, respectively (Figure 5A), whereas the same concentration of the mixture significantly decreased anti-HIV effect by a mixture of 100 ng/mL CCR5 chemokines (data not shown). This result suggests that other factor(s) in VLP-Sup rather than those cytokines/chemokines may play a role of HIV inhibition.
Quantitation of anti-HIV cytokines in conditioned media
| Cytokines . | PBMCs . | MDMs . | ||
|---|---|---|---|---|
| Control-Sup . | VLP-Sup . | Control-Sup . | VLP-Sup . | |
| IFN-α | ND | 0.112 ± 0.02 | 0.042 ± 0.004 | 0.070 ± 0.03* |
| IFN-β | ND | ND | ND | ND |
| IL-10 | 0.014 ± 0.003 | 0.022 ± 0.001* | 0.013 ± 0.002 | 0.032 ± 0.002* |
| MCP-2 | ND | ND | ND | 27 ± 1.5 |
| IL-27 | 2.2 ± 0.17 | 12 ± 1.4† | 3.0 ± 0.29 | 25 ± 3.0† |
| Cytokines . | PBMCs . | MDMs . | ||
|---|---|---|---|---|
| Control-Sup . | VLP-Sup . | Control-Sup . | VLP-Sup . | |
| IFN-α | ND | 0.112 ± 0.02 | 0.042 ± 0.004 | 0.070 ± 0.03* |
| IFN-β | ND | ND | ND | ND |
| IL-10 | 0.014 ± 0.003 | 0.022 ± 0.001* | 0.013 ± 0.002 | 0.032 ± 0.002* |
| MCP-2 | ND | ND | ND | 27 ± 1.5 |
| IL-27 | 2.2 ± 0.17 | 12 ± 1.4† | 3.0 ± 0.29 | 25 ± 3.0† |
PHA-stimulated PBMCs and MDMs were cultured in the absence or presence of 1 μg/mL VLP for 36 and 24 hours, respectively. Conditioned media from untreated cells (Control-Sup) and VLP-treated cells (VLP-Sup) were obtained as described in “Generation of conditioned medium.” Concentrations of each cytokine (ng/mL) were quantitated by ELISA. Data show means ± SEs from at least 3 different conditioned media.
ND indicates not detected.
P < .01.
P < .05.
IL-27 suppresses HIV replication. (A) PHA-stimulated CD4+ T cells infected with X4 virus and PHA-stimulated PBMCs infected with R5 virus were cultured for 7 days in the absence or presence of 12.5% of VLP-Sup from PBMCs. To neutralize IFNs (IFN-α and IFN-β), MCP-2, IL-10, or CCR5 ligands, 10 μg/mL of each neutralizing antibody was added in the culture. To neutralize CCR5 ligands, an antibody mixture containing 10 μg/mL of each antibody to MIP-1α, MIP-1β, and RANTES was used. IL-27 in VLP-Sup from PBMCs was immunodepleted as described in “Materials and methods.” As a replication control, the infected cells were cultured in the absence of the VLP-Sup. On the 4th day after infection, 50% of culture supernatants were changed with fresh culture media alone. HIV-1 replication was measured by p24 antigen capture assay. The neutralization and the immunodepletion assay were performed at least 3 times, and results show the means ± SEs. (B) Relative expression of IL-27 mRNAs in PBMCs and MDMs were assessed by RT-PCR. PHA-stimulated PBMCs and MDMs were cultured in the absence or presence of 1 μg/mL VLPs for 36 and 24 hours, respectively. A total cellular RNA was isolated, and RT-PCR was performed as described in “Materials and methods.” As a control, RT-PCR was performed using RNA from SV40VLP-treated PHA-simulated PBMCs. (C-E) X4-infected CD4+ T cells (C) or PBMCs (D) were cultured in the presence of different concentrations of recombinant human IL-27. On the 4th day after infection, 50% of culture supernatants were changed with fresh culture media containing IL-27. HIV-1 replication was measured on 7th day by p24 antigen capture assay. MDMs were infected with R5 virus (E) and then cultured for 10 days in the presence of IL-27. Half of the culture supernatants were changed on the 4th and 7th day with fresh media with IL-27. HIV-1 replication was measured by p24 antigen capture assay. HIV-1 replication was measured by p24 antigen capture assay. Data show means ± SDs and are representative of 5 experiments.
IL-27 suppresses HIV replication. (A) PHA-stimulated CD4+ T cells infected with X4 virus and PHA-stimulated PBMCs infected with R5 virus were cultured for 7 days in the absence or presence of 12.5% of VLP-Sup from PBMCs. To neutralize IFNs (IFN-α and IFN-β), MCP-2, IL-10, or CCR5 ligands, 10 μg/mL of each neutralizing antibody was added in the culture. To neutralize CCR5 ligands, an antibody mixture containing 10 μg/mL of each antibody to MIP-1α, MIP-1β, and RANTES was used. IL-27 in VLP-Sup from PBMCs was immunodepleted as described in “Materials and methods.” As a replication control, the infected cells were cultured in the absence of the VLP-Sup. On the 4th day after infection, 50% of culture supernatants were changed with fresh culture media alone. HIV-1 replication was measured by p24 antigen capture assay. The neutralization and the immunodepletion assay were performed at least 3 times, and results show the means ± SEs. (B) Relative expression of IL-27 mRNAs in PBMCs and MDMs were assessed by RT-PCR. PHA-stimulated PBMCs and MDMs were cultured in the absence or presence of 1 μg/mL VLPs for 36 and 24 hours, respectively. A total cellular RNA was isolated, and RT-PCR was performed as described in “Materials and methods.” As a control, RT-PCR was performed using RNA from SV40VLP-treated PHA-simulated PBMCs. (C-E) X4-infected CD4+ T cells (C) or PBMCs (D) were cultured in the presence of different concentrations of recombinant human IL-27. On the 4th day after infection, 50% of culture supernatants were changed with fresh culture media containing IL-27. HIV-1 replication was measured on 7th day by p24 antigen capture assay. MDMs were infected with R5 virus (E) and then cultured for 10 days in the presence of IL-27. Half of the culture supernatants were changed on the 4th and 7th day with fresh media with IL-27. HIV-1 replication was measured by p24 antigen capture assay. HIV-1 replication was measured by p24 antigen capture assay. Data show means ± SDs and are representative of 5 experiments.
Identification of IL-27 as a novel antiviral cytokine
The DNA microarray analysis indicated that VLPs significantly increased IL27 gene activation in PBMCs and MDMs. IL-27, a recently identified heterodimeric member of the IL-6/IL-12 family of type I cytokines, is produced in response to pathogenic and host-derived inflammatory signals. Studies indicate that IL-27 directly acts on T cells, thereby regulating Th1-type response.48,49 To confirm the up-regulation of gene activation and production of IL-27, RT-PCR and quantitative assays were performed. HPV VLPs, but not SV40 VLPs, significantly increased the activation of IL27 gene in both PBMCs and MDMs (Figure 5B) and increased production of IL-27 from PBMCs and MDMs by 6- and 8-fold, respectively (Table 2). VLP-Sup from MDMs contained higher amounts of IL-27 than that from PBMCs.
To determine the relative role of IL-27 in virus replication, IL-27 was immunodepleted from VLP-Sup using anti-IL27 antibody. As a control, VLP-Sup was treated with normal antibody. The X4-infected CD4+ T cells or R5-infected PBMCs were cultured in the presence of the treated VLP-Sup. The VLP-Sup treated with a control antibody inhibited replication of X4 and R5 virus to 26% ± 1.0% and 17% ± 2.9%, respectively. The immunodepletion of IL-27 significantly reduced the anti-X4 and anti-R5 effect to 61% ± 3.5% (P < .01) and 40% ± 4.1% (P < .01), respectively (Figure 5A). A similar result was seen in the immunoabsorbed VLP-Sup from MDMs (data not shown). These results suggested that IL-27 might be one of the suppressive factors in VLP-Sup from PBMCs and MDMs
To verify the inhibitory activity of IL-27 on virus replication, X4-infected CD4+ T cells were cultured for 7 days in the presence of recombinant IL-27. IL-27 inhibited X4 viral replication in a dose-dependent manner (Figure 5C). The maximal inhibitory effect (70%∼80%) was observed at 100 ng/mL. Trypan blue exclusion assay indicated that 100 ng/mL IL-27 treatment did not induce significant cytotoxicity after 7 days of incubation; the viability of untreated and IL-27–treated cells was 89% ± 2.1% and 88% ± 2.7%, respectively (P > .05). IL-27 also inhibited X4 replication in PBMCs (Figure 5D) with a similar fashion. R5 virus in MDMs was also inhibited by IL-27 (Figure 5E). Of note, 10 ng/mL IL-27 inhibited R5 replication nearly 100% without significant effects on cell viability (data not shown). Taken together, IL-27 is able to inhibit X4 and R5 HIV replication without induction of cell death.
To define whether the IL-27–mediated HIV inhibition is associated with the suppression of transcription of HIV RNA, real-time quantitative RT-PCR was performed. In the presence of 100 ng/mL IL-27, HIV transcription was inhibited by 60% to approximately 80% in accordance with the inhibition of viral replication (Table 3). To further assess the mechanism underlying the HIV inhibition, a gene expression profile was analyzed. PHA-stimulated CD4+ T cells were cultured with or without 100 ng/mL IL-27 for 24 hours, and then DNA microarray assay was carried out. The results from the analysis demonstrated that IL-27 treatment significantly activated genes encoding antiviral proteins, for example, IFN-induced protein, IFN regulatory factor 1 (IRF1), IRF8, myxovirus resistance protein, and the 2′-5′-oligoadenylate synthetase.
Effect of IL-27 on HIV replication and HIV transcription
| IL-27 treatment . | HIV replication . | Transcription . |
|---|---|---|
| Experiment 1 | ||
| Absence | (1300 ± 135 (100) | 15.6 ± 0.2 (100) |
| Presence | 478 ± 29 (36) | 5.7 ± 0.2 (39) |
| Experiment 2 | ||
| Absence | 234 ± 30 (100) | 4.96 ± 0.16 (100) |
| Presence | 49 ± 3.0 (21) | 1.03 ± 0.034 (21) |
| IL-27 treatment . | HIV replication . | Transcription . |
|---|---|---|
| Experiment 1 | ||
| Absence | (1300 ± 135 (100) | 15.6 ± 0.2 (100) |
| Presence | 478 ± 29 (36) | 5.7 ± 0.2 (39) |
| Experiment 2 | ||
| Absence | 234 ± 30 (100) | 4.96 ± 0.16 (100) |
| Presence | 49 ± 3.0 (21) | 1.03 ± 0.034 (21) |
X4-infected CD4+ T cells were cultured for 7 days in the absence or presence of 100 ng/mL recombinant IL-27. HIV replication was monitored using p24 antigen capture assay. Data show means ± SD (ng/mL). A total cellular RNA was extracted from the cultured cells as described in “Quantitation of transcribed HIV RNA,” and then the copy number of HIV transcripts was determined by a real-time quantitative RT-PCR. Copy numbers determined from real-time quantitative PCR experiments were adjusted for cell equivalents using CCR5. Data show means ± SD (× 108 copies/106 cells). Values in parentheses show percentage of activity compared with untreated cells.
Discussion
It has been reported that HPV-VLPs bind to TLRs on dendritic cells. TLR signaling links the innate and adaptive arms of immune response against invading pathogens50,51 and induces anti–HIV-1 cytokines such as IFN-α, IL-10, IFN-γ, and MCP-2.27,28 In the present study, we demonstrated that VLPs can bind to monocytes/macrophages and induced a novel anti–HIV-1 cytokine, IL-27. To our knowledge, this is the first report that IL-27 is able to inhibit replication of both X4 and R5 HIV in CD4+ T cells and MDMs, although IL-27–related cytokines, IL-6 and IL-12, are known to induce HIV replication.52,53
HPV-VLPs, but not SV40-VLPs, induced a strong anti-HIV effect. This difference in the anti-HIV effect between HPV-VLPs and SV40-VLPs may be associated with the difference in binding affinity to the receptors or receptor usage. In agreement with another report,27 HPV-VLPs increased transcription of genes encoding IFN-induced protein, myxovirus (influenza virus) resistance proteins, and the 2′-5′-oligoadenylate synthetase in PBMCs and MDMs. Of note, our data indicated that VLPs activated genes encoding TRIM family proteins, including TRIM19. TRIM19, promyelocytic leukemia protein, has been documented to interfere with the early steps of HIV-1 replication.54 VLPs also enhanced the gene encoding APOBEC3A which is currently described as a potent antiretroviral protein.55,56 This is the first report that VLPs are able to activate TRIM family genes. These results suggest that HPV-VLP treatment may inhibit replication of not only HIV but also other retroviruses. Thus, elucidating the mechanism of VLP-mediated viral inhibition may provide insights into the mechanism underlying the protection against HPV infection as well as other virus infection in VLP-vaccinated persons.
Although bacterial products directly induce IL-27 production, a variety of host-derived factors, including CD40 ligand, IFN-β, and IFN-γ, can promote IL-27 expression.57–60 Induction of IL-27 by lipopolysaccharide indicates that TLR4 could trigger IL-27 induction.51,58–61 IL-27 is a heterodimer of an IL-27 p28 with a group 3 soluble cytokine receptor Epstein-Barr virus-induced gene 3 (EBI3).50 The IL-27 receptor (IL-27R) is composed of the group 2 cytokine receptors (gp130 and WSX-1).50,61 Although coexpression of both IL-27R subunits can be detected in a variety of immune cell types (spleen, thymus, lungs, intestine, liver, PBMCs, and lymph nodes), production of IL-27 has only been reported in endothelial cells, dendritic cells, and monocytes.61 Binding of IL-27 to the receptor directly regulates T-cell function by shifting the T-cell response toward the Th1-type response62 with phosphorylation of Janus kinases 1 (Jak1), signal transducer and activator of transcription 1, 2, 3, and 5 (STAT1, STAT2, STAT3, and STAT5).58,63,64 Our preliminary study indicated that IL-27 suppressed HIV transcription and significantly enhanced gene activation of the IFN-inducible antiviral proteins. Therefore, IL-27–mediated HIV inhibition may involve a similar mechanism with IFN-mediated HIV inhibition through the Jak/STAT pathway. Further study may be needed to address the mechanism by which IL-27 inhibits HIV replication.
Although 12.5% of VLP-Sup from PBMCs inhibited HIV replication by 84%, Control-Sup from PBMCs at the same concentration had no impact on HIV replication. Unlike native IL-27 (a heterodimer protein),50 the recombinant IL-27 is a fusion protein. On the basis of theses structural differences, one may not be able to precisely compare the anti-HIV effect between both proteins on a milligram-to-milligram basis. It is, however, noteworthy that 1.5 ng/mL recombinant IL-27, which is an extrapolation of the amount of IL-27 in 12.5% VLP-Sup, inhibited HIV by 40%, whereas 0.28 ng/mL recombinant IL-27, an extrapolated amount of IL-27 in Control-Sup, suppressed HIV replication less than 1%. It is thus possible that other factors in VLP-Sup may play a role in the observed viral inhibition.
The PBMC-Sup was more potent in antivirus activity than MDM-Sup, despite VLP-Sup from PBMCs containing a lower amount of IL-27 than that from MDMs. The immunodepletion of IL-27 partially decreased the anti-X4 and R5 effect in VLP-Sup; thus, it is postulated that other factor(s) in the PBMC-Sup may add to the effect of IL-27, or coexisting factor(s) in the MDM-Sup may reduce the IL-27 effect. The neutralization of IFNs, MCP-2, IL-10, or CCR5 ligands in VLP-Sup did not significantly reduce the antiviral effect in VLP-Sup; therefore, the role of these factors on HIV-1 inhibition may be modest if anything at all. Additional studies may address the impact of coexisting factors in VLP-Sup on HIV replication.
In this study, we have demonstrated that HPV-VLP, a vaccine for cervical cancer, inhibits replication of both X4 and R5 viruses in vitro, and VLP treatment induces IL-27 production. Here, we have identified that IL-27 is a novel antiviral cytokine. A number of soluble anti-HIV factors have been identified from tissue culture supernatants,11–15 and it has been proposed that HIV replication might be immunologically controlled by soluble factors.65 Understanding the mechanism of IL-27–mediated HIV inhibition may delineate an invaluable insight into immunologic control of HIV replication and possible strategies for therapeutic invention.
Authorship
Contribution: T.I. designed the study, performed the research, analyzed the data, and wrote the paper; J.M.F. performed the research, analyzed the data, and wrote the paper; R.J.G. and A.J.G.-P. performed the research and analyzed the data; R.A.L., J.Y., and J.W.A. analyzed the data; L.A.P. provided reagents and contributed to the intellectual work; and H.C.L. contributed to the intellectual work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tomozumi Imamichi, Bldg 550, Rm 126, Sultan Dr, SAIC-Frederick, Inc, NCI-Fredrick, PO Box B, Frederick, MD 21702-1124; e-mail: timamichi@mail.nih.gov.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank H. Imamichi, C. Watkins, C. Yeager, T. Brann, and B. Fullmer for technical help. We also thank Z. Howard, T. Yoshimura, and J. Lifson for suggestions and helpful discussion and R. Viscidi for providing the SV40 VLPs.
This work was supported by federal funds from the National Cancer Institute, National Institutes of Health (contract N01-CO-12400, Article H.36 of the Prime Contract).




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal